Despite having structures for eukaryotic large ribosomal subunits, it has been unclear how these ribonucleoprotein complexes are constructed in living cells. Here, Gamalinda et al. used proteomics to assay changes in the levels of ribosomal protein and assembly factors in preribosomes upon depletion of ribosomal proteins that function in 60S assembly. The results show that the eukaryotic 60S ribosomal subunits are assembled in a hierarchical fashion. This study further reveals striking differences and similarities between bacterial and eukaryotic large ribosomal subunits, providing insights into the evolution of RNA–protein particles.
Keywords: ribosome assembly, large ribosomal subunit, ribosomal proteins, preribosomes
Abstract
Despite having high-resolution structures for eukaryotic large ribosomal subunits, it remained unclear how these ribonucleoprotein complexes are constructed in living cells. Nevertheless, knowing where ribosomal proteins interact with ribosomal RNA (rRNA) provides a strategic platform to investigate the connection between spatial and temporal aspects of 60S subunit biogenesis. We previously found that the function of individual yeast large subunit ribosomal proteins (RPLs) in precursor rRNA (pre-rRNA) processing correlates with their location in the structure of mature 60S subunits. This observation suggested that there is an order by which 60S subunits are formed. To test this model, we used proteomic approaches to assay changes in the levels of ribosomal proteins and assembly factors in preribosomes when RPLs functioning in early, middle, and late steps of pre-60S assembly are depleted. Our results demonstrate that structural domains of eukaryotic 60S ribosomal subunits are formed in a hierarchical fashion. Assembly begins at the convex solvent side, followed by the polypeptide exit tunnel, the intersubunit side, and finally the central protuberance. This model provides an initial paradigm for the sequential assembly of eukaryotic 60S subunits. Our results reveal striking differences and similarities between assembly of bacterial and eukaryotic large ribosomal subunits, providing insights into how these RNA–protein particles evolved.
Cells invest a major percentage of their energy to construct ribosomes necessary for growth and proliferation. These protein-synthesizing machines contain a complex lattice of ribosomal RNAs (rRNAs) stabilized by ribosomal proteins (RPs). How these components are assembled together to produce functional ribosomes is a fundamental question in ribonucleoprotein (RNP) biology. Pioneering experiments to reconstitute bacterial subunits in vitro revealed that assembly is hierarchical and cooperative (Held et al. 1974; Rohl and Nierhaus 1982). Folding of rRNAs creates binding sites for primary RPs. Their binding induces conformational changes in rRNA, which enables the subsequent assembly of secondary and tertiary binding RPs (Shajani et al. 2011; Woodson 2011). More recent studies have shown that assembly in vitro can proceed via parallel pathways (Adilakshmi et al. 2008; Mulder et al. 2010) in which individual RPs contact different rRNA elements in multiple stages throughout assembly.
Although extremely efficient, ribosome biogenesis is much more complicated inside cells, especially in eukaryotes, where nascent ribosomal subunits transit from the nucleolus to the cytoplasm. In vivo, RPs are incorporated coincident with transcription of precursor rRNAs (pre-rRNAs), which undergo folding, modification, and processing to remove spacer sequences (Supplemental Fig. 1). These events are facilitated in yeast by >300 trans-acting assembly factors (AFs) (Maxwell and Fournier 1995; Karbstein 2011; Kressler et al. 2012). These dynamically intertwined events are subject to rigorous quality control mechanisms so that assembly is rapid yet accurate in order to meet the cellular demand to produce more than a million ribosomes per cell cycle (Grummt 1999). Misassembled ribosomes in humans are linked to developmental abnormalities and many forms of cancer (Freed et al. 2010; Ellis and Gleizes 2011; McCann and Baserga 2013).
While the binding sites for only a handful of yeast AFs in nascent ribosomes have been identified (Ulbrich et al. 2009; Granneman et al. 2010, 2011; Sengupta et al. 2010; Strunk et al. 2011; Bradatsch et al. 2012; Greber et al. 2012; Matsuo et al. 2013), recently solved crystal structures of mature yeast ribosomes show in exquisite detail where each individual RP interacts with rRNA (Ben-Shem et al. 2011). Knowing these final endpoints of RP localization provides a strategic platform to investigate the connection between spatial and temporal aspects of ribosome biogenesis. Most RPs assemble with preribosomes early in the biogenesis pathway (Kruiswijk et al. 1978; Ferreira-Cerca et al. 2007; Babiano and de la Cruz 2010; Babiano et al. 2012; Gamalinda et al. 2013; Ohmayer et al. 2013). As the nascent subunits mature, the association of RPs with pre-rRNA is strengthened by dynamic rearrangement of initial protein–RNA encounter complexes (Ferreira-Cerca et al. 2007; Adilakshmi et al. 2008; Ohmayer et al. 2013).
RP depletions in yeast and siRNA-mediated knockdowns in cultured mammalian cells showed that most RPs are essential for ribosome biogenesis (Ferreira-Cerca et al. 2005; Robledo et al. 2008; Poll et al. 2009; Babiano and de la Cruz 2010; O'Donohue et al. 2010; Jakovljevic et al. 2012; Gamalinda et al. 2013). However, phenotypic analyses of these mutants were primarily limited to assaying effects on pre-rRNA processing and nucleo–cytoplasmic export of preribosomes. Nevertheless, investigations of small subunit RPs (RPSs) suggested that the two discernable structural domains of the 40S subunit are sequentially assembled (Ferreira-Cerca et al. 2007; O'Donohue et al. 2010). The “body” substructure (containing the 5′ and central domains of 18S rRNA) is formed first, followed by the “head” substructure (containing the 3′ major domain of 18S rRNA). This bipartite assembly of eukaryotic 40S subunits in vivo parallels observations from thermodynamic and kinetic studies of bacterial 30S subunit assembly in vitro and in vivo (Held et al. 1974; Mulder et al. 2010; Chen and Williamson 2013).
In contrast, structural principles governing assembly of large ribosomal subunits are less clear. The segregation of large subunits into distinct structural domains is not as apparent as for small subunits (Ban et al. 2000); the six rRNA domains are more intertwined in large subunits (Holbrook 2008). Hence, it was initially striking to find that depletion of individual yeast large subunit RPs (RPLs) led to specific rather than global pre-rRNA processing defects (Ferreira-Cerca et al. 2005; Hofer et al. 2007; Robledo et al. 2008; Poll et al. 2009; Babiano and de la Cruz 2010; O'Donohue et al. 2010; Babiano et al. 2012; Fernandez-Pevida et al. 2012; Jakovljevic et al. 2012; Gamalinda et al. 2013), which are grouped into early, middle, and late classes (Table 1). From these results, we observed a pattern previously not described: RPLs belonging to these phenotypic classes are remarkably clustered into distinct structural neighborhoods of the yeast 60S subunit (Fig. 1; Ben-Shem et al. 2011). Recent studies also demonstrate that RPLs influence the assembly of other RPLs and AFs into preribosomes, although information derived from these studies is fragmentary. To our knowledge, no global picture of the hierarchy of 60S subunit assembly from the beginning to the end has been described thus far. We reasoned that the correlation between the location and function of RPLs might reflect sequential construction of stable pre-60S assembly intermediates, analogous to the biogenesis of small ribosomal subunits (Held et al. 1974; Ferreira-Cerca et al. 2007; Mulder et al. 2010; O'Donohue et al. 2010).
Table 1.
Summary of effects of depleting RPLs on processing of pre-60S rRNAs
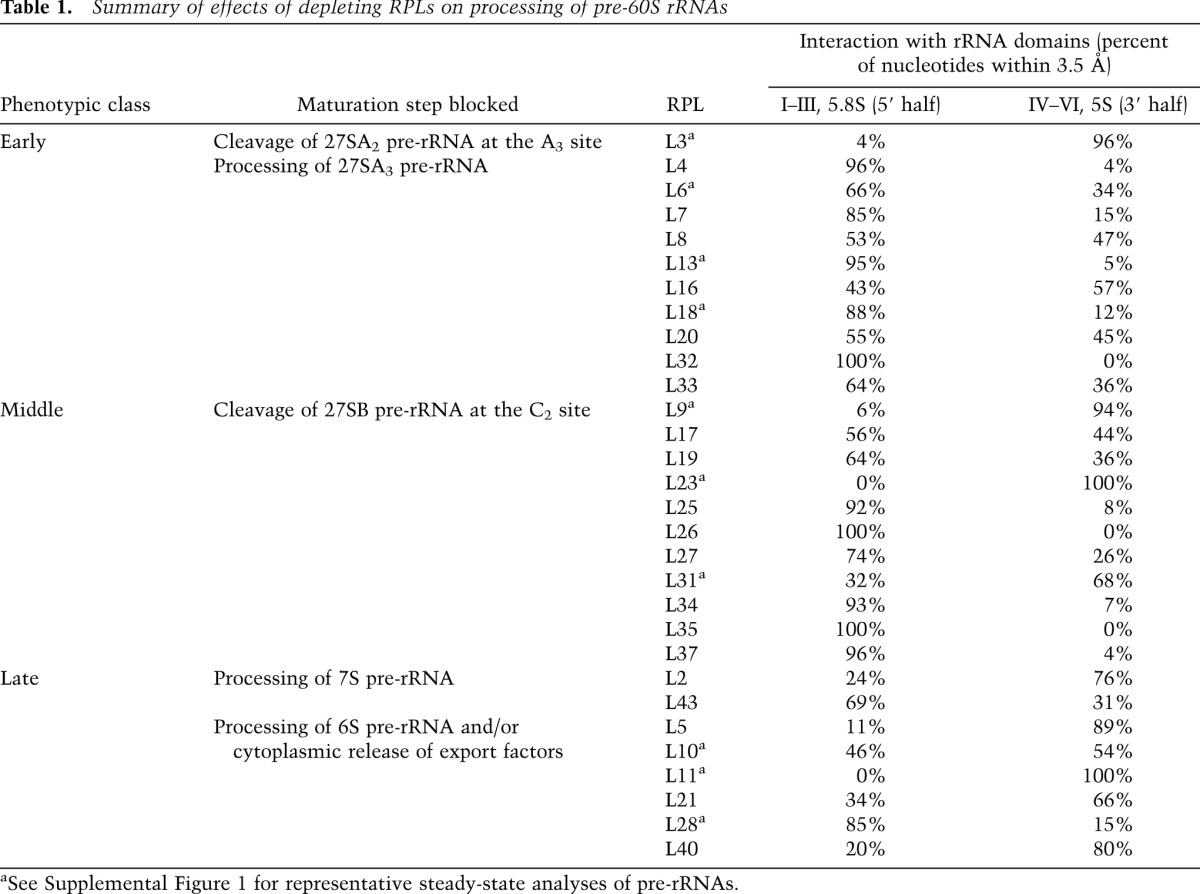
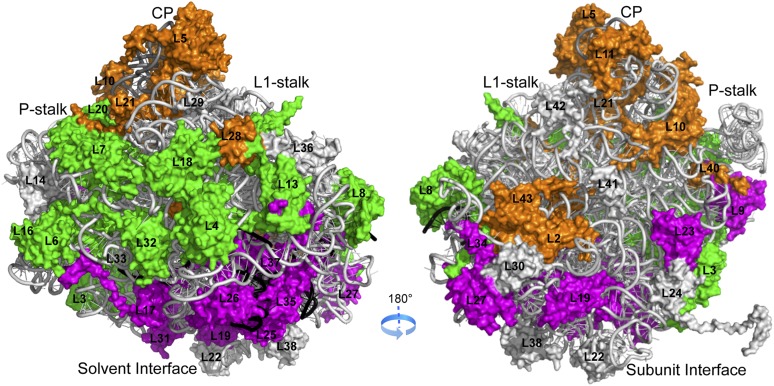
Figure 1.
Correlation between function and localization of RPLs. The location of early-acting (green), intermediate-acting (magenta), and late-acting (orange) RPLs is mapped onto the crystal structure of the yeast 60S subunit (Protein Data Bank [PDB] accession nos. 3U5D and 3U5E). rRNAs and RPLs are shown in cartoon and surface representations, respectively. 5.8S, 25S, and 5S rRNAs are colored black, light gray, and dark gray, respectively.
To test this model, we systematically characterized effects on preribosomes upon depleting RPLs functioning in early, middle, and late steps of yeast 60S subunit assembly. We dissected how binding of RPLs influences association of other RPLs and AFs with preribosomes in order to form stable pre-rRNPs capable of undergoing successive pre-rRNA processing events. Our global survey supports a sequential model for 60S subunit assembly. The convex solvent side is formed first, followed by the polypeptide exit tunnel, the flat subunit interface, and finally the central protuberance. This hierarchy serves as an initial framework to further understand 60S assembly in vivo. Our results also support the hypothesis that the formation of the 3′ end of 27S pre-rRNA is important for early steps of 60S assembly occurring near the 5′ end of pre-rRNA. Finally, we discuss similar principles underlying construction of bacterial and eukaryotic large subunits.
Results
As shown in Figure 1, RPLs classified according to their requirement for successive steps in maturation of pre-rRNAs cluster together in the structural model of the yeast large subunit. Within the early-acting group (Fig. 1, green), L3 is required for the first step of 27SA2 pre-rRNA processing and is bound to domain VI. However, RPLs required for the next early step, processing of 27SA3 pre-rRNA, are localized on the convex side of the solvent interface, containing domains I and II of 25S rRNA (Supplemental Table 1). The middle-acting RPLs (Fig. 1, magenta), required for 27SB pre-rRNA processing, largely cluster around the rim of the polypeptide exit tunnel, outlined by domains I and III of 25S/5.8S rRNA. Within the late-acting group (Fig. 1, orange), RPLs functioning in 7S pre-rRNA maturation are located on the intersubunit surface, while those RPLs important for 6S pre-rRNA processing and cytoplasmic maturation cluster around the central protuberance, where 5S rRNA sits in between domains II and V.
To investigate whether these clusters reflect a hierarchical construction of structural neighborhoods of RPLs and AFs, we analyzed components of mutant preribosomes that are present when each RPL is depleted. Preribosomes were purified using different TAP-tagged AFs associated with pre-60S complexes (see the Materials and Methods). Preribosomes from 28 different RPL depletion mutants were assayed by SDS-PAGE and silver staining, and 11 were followed up by Western blotting. Among these, eight representative mutants were subjected to more detailed analysis by iTRAQ mass spectrometry. These representatives were chosen such that when combined with other RPL mutants that we previously analyzed by mass spectrometry, we were able to investigate the roles of RPLs from throughout the entire 60S subunit structure to derive a general pattern of assembly. We previously individually characterized early-acting RPLs L7 (domain II) and L8 (domain I) on the solvent interface (Jakovljevic et al. 2012), middle-acting L25 (domain III) around the exit tunnel (Ohmayer et al. 2013), and late-acting RPLs L2 and L43 (domains IV and V) on the flat subunit interface as well as L21 (domain V) around the central protuberance (Ohmayer et al. 2013). In this study, we included early-acting L18 (domain II) on the solvent side and L3 (domain VI) at the periphery of the solvent interface; middle-acting L17 (5.8S/25S domain I) around the exit tunnel opposite L25 as well as L9 and L23 (domains IV–V), which are middle-acting RPLs farther away from the rim of the exit tunnel; and late-acting RPLs L10, L11, and L28 on or around the central protuberance.
Binding of RPLs is coupled with association of AFs with preribosomes
Consistent with an early block in assembly, preribosomes depleted of early-acting RPLs, but not middle- or late-acting RPLs, are enriched for AFs present in early pre-60S complexes, as assayed by silver staining and Western blotting (Fig. 2A,B; Supplemental Figs. 3, 4). In addition, levels of six interdependent AFs required for 27SA3 pre-rRNA processing (A3 factors) and at least one AF required for 27SB pre-rRNA processing (B factors) are reduced in preribosomes, consistent with an early processing defect.
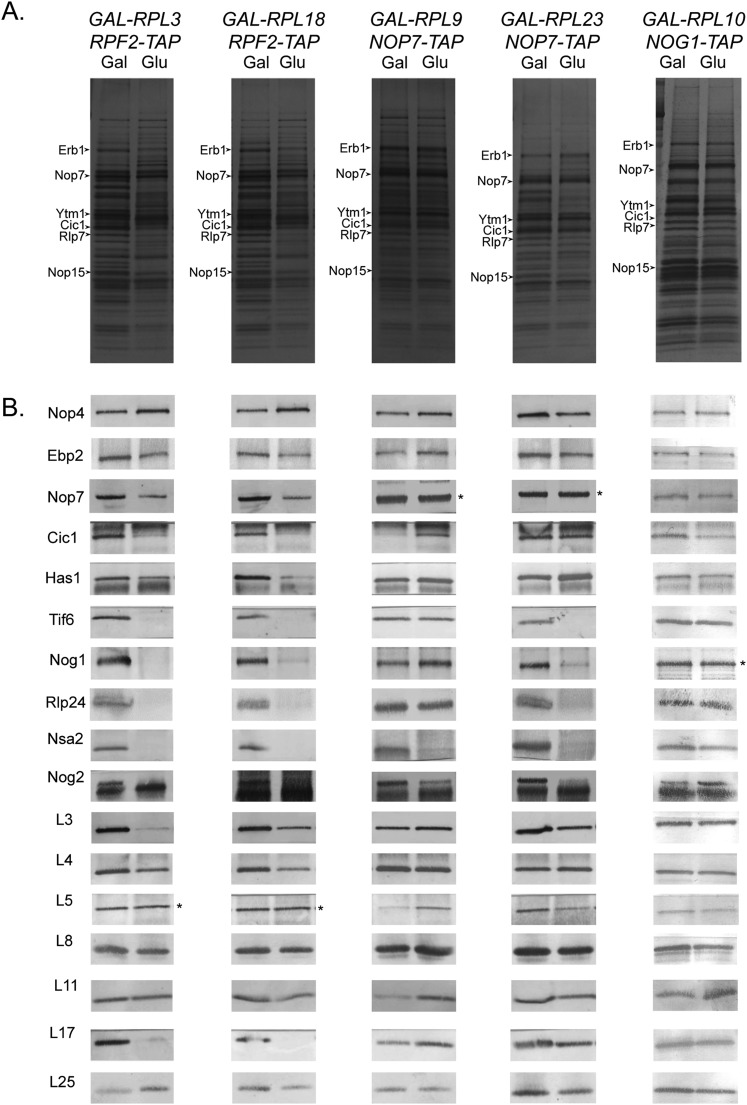
Figure 2.
Effects of depleting each RPL on preribosome composition. TAP-tagged AFs Rpf2, Nop7, or Nog1 were used to affinity-purify pre-60S complexes from conditional strains where each RPL is either expressed or depleted for 16 h. (A) Protein constituents of affinity-purified preribosomes were separated by SDS-PAGE and visualized by silver staining. Proteins in these SDS-PAGE profiles are labeled based on comigration with polypeptide bands previously identified by mass spectrometry, shift in gel mobility of epitope-tagged versions of corresponding proteins, and decrease in protein band intensity when expression of the corresponding gene is turned off. (B) Western blotting was carried out using available antibodies against specific RPLs or biogenesis factors as well as against HA, Myc, or TAP in cases where proteins are epitope-tagged. (*) Loading controls.
Semiquantitative mass spectrometry confirmed and extended these results (Fig. 3; Supplemental Table 2). The effects observed when early-acting RPL L3 or L18 is depleted are consistent with previous results obtained upon depleting other early-acting RPLs L7 and L8 (Jakovljevic et al. 2012). Early preribosome components accumulate (i.e., SSU processome constituents, RPSs, Nop6, Nop4, Noc1, Nop13, and Rrp5). A3 factors (Nop7, Erb1, Ytm1, Rlp7, Cic1, Nop15, and Has1) and B factors (Tif6, Nog1, Rlp24, Dbp10, and Spb4) are reduced. AFs that associate later with downstream 27SB/7S pre-rRNAs are also reduced (Noc3, Nsa2, Nog2, Rsa4, Nug1, Mrt4, Arx1, and, when detected, its partner, Alb1). These latter AFs are important for final steps of processing of ITS2 and subsequent nuclear export of preribosomes. In addition to these changes, depletion of L3 also leads to the reduction of early AFs Ebp2 and Brx1, which function upstream of 27SA3 pre-rRNA processing. This is consistent with the role of L3 in this earliest step in 27S pre-rRNA processing—cleavage at the A3 site of 27SA2 pre-rRNA.
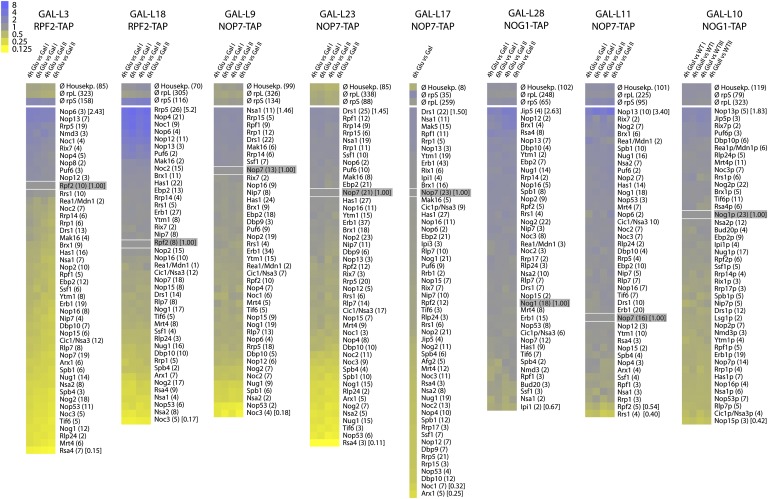
Figure 3.
Comparison of changes in levels of AFs in preribosomes by semiquantitative analysis after depleting each RPL. Resulting pairwise comparisons of the iTRAQ ratios of all identified large ribosomal subunit AFs were normalized to the iTRAQ ratio of the bait protein, log2-transformed, and displayed as a heat map. The average number of peptides by which each protein was identified is indicated in parentheses. The average iTRAQ ratios of all identified housekeeping proteins and SSU r-proteins are also included. Bait proteins used for affinity purification are highlighted, and the time of RPL depletion (in hours) is indicated. The normalized iTRAQ ratios of selected proteins (with the highest or lowest values) are given in brackets.
In contrast to the early group of RPLs, depletion of middle-acting RPLs located around the exit tunnel, such as L25 (domain III) (Ohmayer et al. 2013) and L17 (domain I), as well as those bound to the 3′ half of 25S rRNA (L9 and L23) did not lead to the reduction of A3 and most B factors (Fig. 3; Supplemental Table 2). However, some B factors (Spb4 and/or Dbp10) and late-associating AFs (i.e., Noc3, Nsa2, Nog2, Rsa4, Nop53, and Arx1) are diminished from preribosomes (Fig. 3; Supplemental Table 2; Ohmayer et al. 2013). Nog2 and Rsa4 bind to the subunit interface and at the base of the central protuberance, respectively (Ulbrich et al. 2009; Matsuo et al. 2013). Their reduction in preribosomes is likely because these structural neighborhoods are not properly assembled when middle-acting RPLs are depleted (see “Assembly of 60S Ribosomal Subunit Domains Is Hierarchical,” below). Arx1 binds to RPLs and rRNA sequences around the exit tunnel (Bradatsch et al. 2012; Greber et al. 2012). Thus, its reduction may reflect proximal effects of RPL assembly on binding of factors; assembly of the exit tunnel might be required to establish proper contacts with Arx1. Another proximal effect that we observed was reduction of Tif6, Rlp24, and Nog1 in preribosomes after depleting L23 (Fig. 3; Supplemental Table 2). Previously, Tif6 was found to bind to L23 (Gartmann et al. 2010), Rlp24 was suggested to act as a placeholder for L24 that is adjacent to L23 (Saveanu et al. 2003), and Nog1 was hypothesized to localize to the subunit close to L23 and L24 (Bradatsch et al. 2012). Hence, binding of Tif6, Rlp24, and Nog1 may depend on L23-induced structural rearrangements in this region.
Finally, the effects of depleting late-acting RPLs L10, L11, and L28 (Fig. 3; Supplemental Table 2) are consistent with previous observations upon depleting other late-acting RPLs L2, L43, and L21 (Ohmayer et al. 2013). Preribosomes isolated from L10, L11, and L28 depletion mutants still contain many early AFs. However, some early AFs are underrepresented in preribosomes when L10 (the A3 factors Nop7, Erb1, Ytm1, Cic1, Rlp7, and Has1) or L28 (e.g., Ssf1, Nsa1, and Ipi1) is depleted, consistent with previous observations that these factors are normally released from preribosomes before the late steps blocked in these mutants. The specific reduction of Rpf2 and Rrs1 when L11 is depleted reflects their presence in a subcomplex together with L5 (Zhang et al. 2007).
These collective results demonstrate a logical coupling between binding of RPLs, their role in pre-rRNA processing, and the stable association of AFs required for different steps in assembly. This may in part explain how the absence of different RPLs can affect distinct steps of pre-rRNA maturation, although most RPLs are present in the earliest assembly intermediates before any of these steps occur. Based on the known and suggested binding sites of AFs relative to the depleted RPLs, some of these effects on AF assembly (e.g., Arx1, Tif6, Nog1, and Rlp24) may reflect local effects.
Assembly of 60S ribosomal subunit domains is hierarchical
L3 is required for the earliest step of 27S pre-rRNA processing—cleavage at the A3 site of 27SA2 pre-rRNA. However, in contrast to all other early-acting RPLs, L3 largely binds to domain VI at the 3′ end of 25S rRNA instead of domains I or II at the 5′ end (Supplemental Table 1). Depletion of L3 results in an overall reduction in levels of most RPLs in preribosomes (Fig. 4; Supplemental Table 3), suggesting global instability of L3-depleted particles, consistent with its very early role in assembly. One might argue that this destabilization in the absence of L3 may be simply due to its size. L3 is the largest yeast RPL (∼44 kDa) and has multiple contacts with rRNA (Ben-Shem et al. 2011). However, such is not the case; depletion of L4, the second largest RPL (∼40 kDa), does not lead to such an extreme early defect (Poll et al. 2009), although L4 exhibits equally extensive interactions with rRNA (Ben-Shem et al. 2011). Rather, we posit that the location of L3 in proximity to both the 5′ and 3′ ends of 25S/5.8S rRNA (Supplemental Fig. 6C) is responsible for its pivotal role in assembly (see the Discussion).
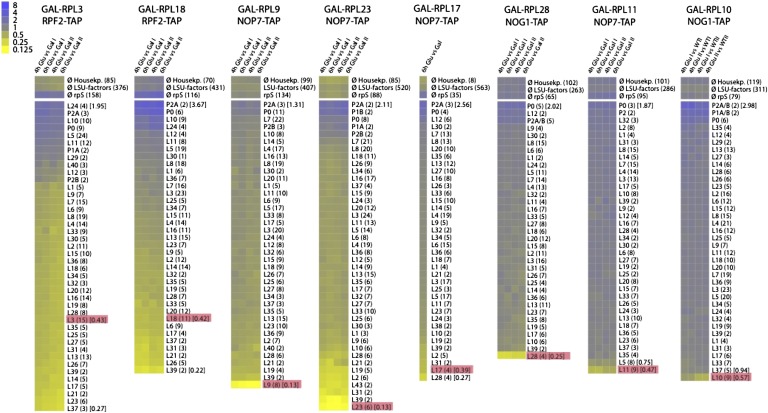
Figure 4.
Comparison of changes in levels of other RPLs in preribosomes by iTRAQ after depleting each RPL. Resulting pairwise comparisons of the iTRAQ ratios of all identified RPLs were normalized to the iTRAQ ratio of the median value of all RPL ratios, log2-transformed, and displayed as a heat map. The average iTRAQ ratios of all identified housekeeping proteins and SSU r-proteins are also included. RPLs conditionally depleted are highlighted, and the time of depletion (in hours) is indicated. Heat maps are labeled as in Figure 3. The enrichment of RPLs that associate with pre-60S complexes in the cytoplasm might be due to the their ability to alternatively assemble in the nucleus (Francisco-Velilla et al. 2013).
A smaller, more specific subset of RPLs is diminished when L18 (domain II) is depleted (Fig. 4; Supplemental Table 3), reminiscent of both proximal and distal effects observed upon depleting two other early-acting RPLs, L7 (domain II) and L8 (domain I) (Jakovljevic et al. 2012). Early-acting domain II RPLs adjacent to L18 are reduced (e.g., L6, L14, L20, and L33), whereas early-acting RPLs bound to domain I remain present at wild-type levels. Middle-acting RPLs bound to domains I and III of 25S/5.8S rRNA around the exit tunnel (e.g., L17, L19, L26, L31, L35, L37, and L39) as well as late-acting RPLs on the subunit interface (e.g., L2 and L43) and around the central protuberance (e.g., L21 and L28) are also reduced. All of these diminished RPLs assemble with early pre-rRNAs. Previous work has shown that assembly of RPLs is stabilized as particles mature (Ohmayer et al. 2013). Hence, these reductions of early-, middle-, and late-acting RPLs in preribosomes upon depleting an individual early-acting RPL may reflect sequential effects on the transition to more stable association of RPLs with preribosomes. This strengthened binding is likely mediated via both local and distal effects on pre-rRNP structure.
The majority of middle-acting RPLs cluster around the exit tunnel (Fig. 1), where domains I and III of 25S/5.8S rRNA come together. The exceptions are L9 and L23, which are bound to the 3′ half of 25S rRNA. Nevertheless, the effects of depleting of L9 or L23 are very similar to those upon depleting other middle-acting RPLs, including L25 (domain III) (Ohmayer et al. 2013) or L17 (domain I) (Fig. 4; Supplemental Table 3). None of the early-acting RPLs bound to the solvent interface are affected; they remain present at wild-type levels. However, some middle-acting RPLs in proximity to the exit tunnel are diminished (e.g., L19, L31, and L39), suggesting moderate perturbation of this immediate RNP neighborhood. L2 and L43 on the subunit interface are also diminished. The association of these two RPLs becomes tighter after processing of 27SB pre-rRNA (Ohmayer et al. 2013); hence, their reduction in middle-acting RPL mutants is likely an indirect effect of blocking 27SB pre-rRNA processing. The remaining RPLs reduced when L9 or L23 is depleted (e.g., L10, L21, L28, L36, and L40) are located at the base of the central protuberance (Fig. 4; Supplemental Table 3), consistent with their reported reduction when late-acting RPLs L2 and L43 are depleted (Ohmayer et al. 2013).
Last, late-acting RPLs can be subdivided into two classes: those that are necessary for processing 7S pre-rRNA and those more important for events downstream from this processing step (Table 1). L2 and L43 belong to the former and are located on the subunit interface. While early- and middle-acting RPLs remain present in preribosomes when L2 or L43 is depleted, late-acting RPLs at the base of the central protuberance are diminished, including L10 and L21 (Ohmayer et al. 2013). Among the three late-acting RPL depletion mutants that we tested in this study, L11 and L28 are necessary for 6S pre-rRNA processing, similar to L21, which we previously characterized (Ohmayer et al. 2013), whereas L10 is required for final cytoplasmic 60S subunit maturation (Hofer et al. 2007). Depletion of L11 only affects stable association of L5 (Fig. 4; Supplemental Table 3), which is likely due to their presence together in a subcomplex (Zhang et al. 2007). Upon depletion of L10 or L28, all of the other early-, middle-, and late-acting RPLs are present at wild-type levels. Consistent with these defects in association of RPLs around the central protuberance when late-acting RPLs are depleted, cryo-electron microscopy (cryo-EM) of late pre-60S complexes indicates that this rRNP region has not yet formed the structure found in mature ribosomes (Bradatsch et al. 2012).
The general patterns that emerged from our characterization of effects on RPL depletion are the following: Depletion of early-acting RPLs on the solvent interface affects stable association of a subset of early-, middle-, and late-acting RPLs with preribosomes; depletion of middle-acting RPLs around the exit tunnel affects the stable binding of middle- and late-acting RPLs with preribosomes; and depletion of late-acting RPLs on the subunit interface and around the central protuberance affects the binding of only late-acting RPLs. Furthermore, we also know that most RPLs are found in the earliest pre-60S particles, yet their association with preribosomes is thought to become more stable as assembly proceeds, by creating more contacts with pre-rRNA (Ferreira-Cerca et al. 2007; Adilakshmi et al. 2008; Ohmayer et al. 2013). Thus, analyzing the RPL content of preribosomes depleted of early-, middle-, and late-acting RPLs enabled us to arrive at a conclusion that assembly of the 60S ribosomal subunit is hierarchical. L3, which binds domain VI and is close to the 5′ and 3′ ends of 5.8S/25S rRNAs, is critical to initiate early assembly steps. Binding of the other 10 early-acting RPLs to domains I and II promotes the construction of the polypeptide exit tunnel RNP neighborhood around 25S/5.8S rRNA domains I and III. After construction of the exit tunnel, binding of RPLs on the subunit interface is stabilized, and then, finally, assembly of the RNP region around the central protuberance is completed.
Preribosomes become more stable as assembly proceeds
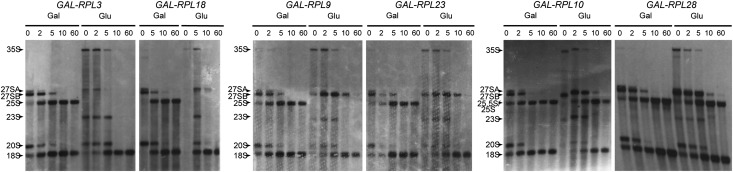
We previously found that abortive assembly intermediates are turned over after depleting RPLs functioning in early and middle steps of pre-60S assembly (Jakovljevic et al. 2012; Gamalinda et al. 2013). Preribosomes blocked early in 60S assembly are more rapidly degraded than preribosomes blocked at middle steps, indicating that pre-60S complexes are progressively stabilized throughout successive maturation steps. To expand on these observations, we assayed for the synthesis and turnover of pre-rRNA intermediates in the absence of RPLs functioning in late assembly steps and also investigated the effects upon depleting additional RPLs required for early and middle steps or pre-rRNA processing (Fig. 5). Consistent with previous observations, pre-60S ribosomes containing unprocessed 27SA pre-rRNAs are rapidly turned over when early-acting RPL L3 or L18 is depleted. In cells depleted of middle-acting RPLs L9 and L23, aberrant 27SB pre-rRNAs are also eventually targeted for degradation but more slowly than when early-acting RPLs are depleted. In contrast, after depletion of late-acting RPLs L10 and L28, 27S pre-rRNAs do not undergo significant turnover and are processed into downstream rRNA species, although more slowly than in wild-type cells (Poll et al. 2009). This suggests that final rearrangements in preribosomes occur in a less efficient fashion when L10 or L28 is absent. Consistent with these observed differences in kinetics of pre-rRNA turnover, more changes in preribosome composition were detected upon depletion of early-acting RPLs than when middle- and late-acting RPLs were depleted. We conclude that preribosomes become more stable as they assemble. Rather than participating directly in pre-rRNA processing, RPLs may function to create stable assembly intermediates that are in the correct conformation to undergo pre-rRNA processing. These processing events may then enable subsequent stabilization of a downstream assembly neighborhood.
Figure 5.
Synthesis and turnover of pre-rRNA intermediates. The kinetics of pre-rRNA processing was assayed by metabolic pulse-chase experiments with [3H-methyl]-methionine. Precursors and mature rRNAs are labeled accordingly.
Discussion
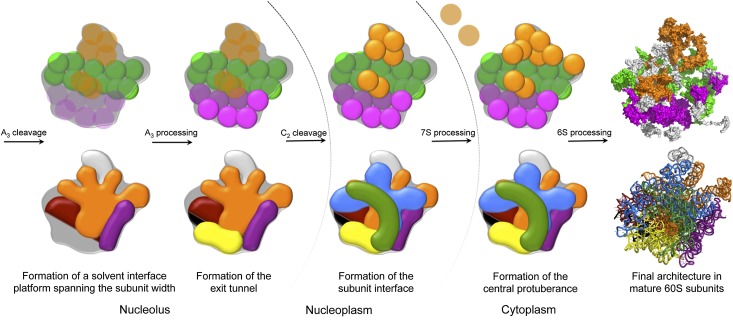
It has become increasingly apparent that the hierarchical assembly of small ribosomal subunit domains is largely conserved from bacteria to eukaryotes (Held et al. 1974; Ferreira-Cerca et al. 2005, 2007; Mulder et al. 2010; O'Donohue et al. 2010; Chen and Williamson 2013; Clatterbuck Soper et al. 2013). What remained unclear is whether the same is true for large ribosomal subunits. In this study, we systematically depleted individual RPLs that function in early, middle, and late steps of 60S assembly and assayed the effects on preribosome composition. This global analysis has enabled us to generate a new model for hierarchical assembly of 60S ribosomal subunits and how these steps are coupled with pre-rRNA processing (Fig. 6). Binding of RPLs to domains I and II creates a stable preribosome conformation able to undergo processing of 27SA3 pre-rRNA. These early assembly steps are coupled with the construction of the polypeptide exit tunnel and initiation of processing of 27SB pre-rRNA. Cleavage at the C2 site in ITS2 of 27SB pre-rRNA stabilizes the intersubunit domain, after which 7S pre-rRNA can be processed. Finally, the rRNP neighborhood around the central protuberance is constructed. Binding of RPLs is tightly connected to the folding of rRNA, and this linkage dictates the order by which structural domains of the 60S ribosomal subunits are assembled. This hierarchy serves as an initial paradigm for a more detailed structural understanding of eukaryotic large subunit assembly.
Figure 6.
Model for hierarchical assembly of yeast 60S ribosomal subunits. General principles of large subunit assembly. A representation of the 60S ribosomal subunit is illustrated, as viewed from the subunit interface. The 60S subunit is outlined in gray. The top panel shows sequential tightening of association of RPLs (circles). RPL groups are colored as in Figure 2, and stabilized association is indicated by a shift from transparent to solid circles. The bottom panel shows sequential stabilization of rRNA domains. 5.8S rRNA and domains I, II, III, IV, V, and VI are colored black, red, orange, yellow, green, blue, and purple, respectively.
In the 40S ribosomal subunit, each RPS interacts almost exclusively with one 18S rRNA domain; primary binders interact with 5′ rRNA sequences, whereas tertiary binders contact 3′ rRNA sequences. In contrast, in the 60S subunit, most RPLs bind to at least three domains of rRNA (Supplemental Table 1). These scattered interactions of members of early-, intermediate-, and late-acting RPLs with rRNA do not immediately suggest a 5′–3′ directionality of assembly. However, some contacts might be preferentially established earlier, while some interactions occur later, explaining the progressive stabilization of RPL binding (Ferreira-Cerca et al. 2007; Sahasranaman et al. 2011; Ohmayer et al. 2013). Interestingly, with the exception of L3 in domain VI, early-acting RPLs have more interactions with the 5′ half of 25S rRNA in domains I and II, while late-acting RPLs have more interactions with the 3′ half of 25S rRNA in domains IV and V (Supplemental Table 1). This is consistent with observations in bacteria, where domains I, II, and VI are stabilized early in assembly, whereas domains IV and V are stabilized during later steps (Li et al. 2013). This correlation between the timing of function of RPLs in subunit assembly and their primary binding sites suggests that, with the exception of domain VI, the 5′–3′ directionality of stable RPL binding influences large ribosomal subunit maturation events in an ordered fashion.
Structures of large ribosomal subunits suggest that assembly involves the formation of a compact intermediate where the root helices of all six rRNA domains (Supplemental Fig. 6A), including those containing the 5′ and 3′ ends of 5.8S/25S rRNAs, are brought together to facilitate subsequent structuring events. The beginning and end of each rRNA secondary structure domain in yeast and bacteria are clustered together in the three-dimensional structure of mature large subunits (Supplemental Fig. 6B). In support of this model, the formation of a compact intermediate that possesses native tertiary interactions (i.e., correctly oriented helices) is a key step during folding of large RNAs such as ribozymes (Bokinsky et al. 2003; Buchmueller and Weeks 2003; Pyle et al. 2007; Behrouzi et al. 2012). The role of L3 very early in 60S subunit assembly is consistent with this compaction. L3 is aptly positioned close to the 5′ end of 5.8S rRNA as well as the 3′ end of 25S rRNA of mature 60S subunits (Ben-Shem et al. 2011). Hence, in the earliest pre-60S particle, L3 is possibly also close to the 5′ and 3′ ends of 27SA2 pre-rRNA. (The 5′ end of 27SA2 pre-rRNA is ∼150 nucleotides [nt] upstream of what becomes the 5′ end of mature 5.8S rRNA, while the 3′ end of 27SA2 pre-rRNA is identical to the 3′ end of 25S rRNA.) These sequences might have to be juxtaposed during an early compaction event to commence 60S assembly, similar to bacterial large subunit assembly where sequences flanking 23S rRNA form a helix recognized by RNase III (Shajani et al. 2011). The bacterial homolog of L3 occupies a similar position close to the ends of 23S rRNA and is required to initiate 50S assembly (Nowotny and Nierhaus 1982).
Effects observed upon depleting L3 also support the previous hypothesis that proper transcription and structuring of the 3′ rRNA sequences are crucial to signal cleavage at the A3 site close to the 5′ end of 27SA2 pre-rRNA (Hitchen et al. 1997; Allmang and Tollervey 1998). Specific 3′ ETS mutations were found to not only affect processing of this transcribed spacer but also inhibit cleavage at the A3 site in ITS1, >4000 nt upstream. In contrast to the other early-acting RPLs that bind domains I and II, L3 binds to a significant fraction of domain VI at the 3′ end of 25S rRNA and is the RPL that makes the most contacts with domain VI (Supplemental Table 1; Ben-Shem et al. 2011). Depletion of L3 elicits the earliest block in 27S pre-rRNA processing: cleavage at the A3 site in 27SA2 pre-rRNA (Supplemental Fig. 2A,C), similar to that seen when depleting the N-terminal half of Rrp5 (Eppens et al. 1999) that cross-links immediately downstream from the A3 site (Lebaron et al. 2013). Thus, while cleavage at the A2 site to create the 5′ end of 27S pre-rRNA occurs cotranscriptionally (Kos and Tollervey 2010), subsequent processing at the A3 site of 27SA2 pre-rRNA appears to occur post-transcriptionally, after proper formation of the 3′ end of 27S pre-rRNA. Recently, Tollervey and colleagues (Lebaron et al. 2013) reported that the AF Rrp5 potentially links processes at the 5′ and 3′ ends of 27SA2 pre-rRNA and facilitates packaging of preribosomes into a compact intermediate. Our results suggest that L3 may function together with Rrp5 to carry out these events. Interestingly, Rrp5 remains present in preribosomes in the absence of L3 (Fig. 3), possibly suggesting a more direct role for L3 in these events.
RPLs predominantly interacting with domains I and II are required for processing of 27SA3 pre-rRNA during early steps of pre-60S assembly (Fig. 1, green). These two domains span the width of the large subunit, with domain II occupying almost the entire solvent interface (Ben-Shem et al. 2011). Strikingly, several helices of domain II resemble finger-like projections oriented toward the subunit interface, interacting with domains IV and V (Supplemental Fig. 7A). This feature is conserved between bacteria and eukaryotes (Supplemental Fig. 7B; Ban et al. 2000). We therefore propose a scenario where binding of RPLs to domain II may help form this rRNA digitation, providing a stable rRNP platform for folding of all other rRNA domains. The binding of early-acting RPLs to 5′ rRNA regions highlights the importance of cooperativity during assembly of large RNPs. As observed for ribozymes, cooperativity in rRNA folding is established early in the process, which facilitates the formation of productive versus antagonistic conformers (Behrouzi et al. 2012).
Landmark in vitro studies on Escherichia coli 50S subunits established the thermodynamic binding interdependencies for assembly of bacterial RPLs (Rohl and Nierhaus 1982). More recently, a refined in vivo assembly map was put forward by Chen and Williamson (2013) that largely corresponds to the Nierhaus map (Rohl and Nierhaus 1982). Our collective studies on yeast RPLs (Babiano et al. 2012; Jakovljevic et al. 2012; Gamalinda et al. 2013; Ohmayer et al. 2013) show the following differences from bacterial large subunit assembly: First, while incorporation of bacterial RPLs occurs via distinct assembly groups, most yeast RPLs are present in early assembly intermediates but differ in the step of assembly for which they are required and become more stably assembled in a sequential fashion. Second, several assembly relationships observed in bacterial RPLs are not found for their eukaryotic homologs (Supplemental Fig. 8A). Bacterial homologs of early-acting yeast RPLs are not necessarily primary binders in vitro and do not significantly influence binding of other RPLs. Conversely, the bacterial homologs of the majority of middle- and late-acting yeast RPLs are mostly primary binders in vitro and affect binding of other bacterial RPLs to a greater extent. Nevertheless, our study also revealed a noteworthy similarity between bacterial and eukaryotic large ribosomal subunits. Assembly of both large subunits begins at the convex solvent side opposite the peptidyl transferase center and concludes with the formation of the central protuberance (Supplemental Fig. 8B; Chen and Williamson 2013; Li et al. 2013). The middle step of bacterial large subunit assembly is dispersed around the 50S structure (Chen and Williamson 2013), whereas for 60S subunits, it involves the formation of the polypeptide exit tunnel. These observations suggest that the principles underlying the biosynthesis of large ribosomal subunits appear to be evolutionarily conserved despite the more numerous steps of pre-rRNA processing, additional rRNA and protein elements, and the added complexity of intranuclear trafficking and nucleocytoplasmic export in eukaryotes. It is interesting to note that the final steps of large ribosomal subunit assembly occur around regions important for ribosome function, including the central protuberance that participates in subunit joining, the peptidyl transferase center, and the GTPase activation center. This provides further credence to an emerging theme in ribosomal subunit maturation in which translational capacities of both subunits are evaluated before joining to form 80S ribosomes (Bussiere et al. 2012; Strunk et al. 2012; Karbstein 2013). Biogenesis of these catalytic RNPs seems to have evolved to save for the finale the most crucial quality control system to inspect their prime reason for existence.
In conclusion, our current and previous functional characterization of pre-60S ribosomes reveals how the assembly of eukaryotic large ribosomal subunits is linked to stable association of RPLs. The connection between cotranscriptional binding of RPLs and folding of nascent pre-rRNA influences post-transcriptional stabilization of 60S subunit structural neighborhoods. This complex interplay dictates the largely 5′–3′ order by which structural domains of the 60S subunit are formed: starting from the convex solvent side, followed by the polypeptide exit tunnel, and finishing with the flat intersubunit side and then the central protuberance. Our results also reveal that the stable formation of these neighborhoods is linked with pre-rRNA processing steps and the AFs that facilitate these events, suggesting plausible checkpoint mechanisms for each step in the maturation of 60S ribosomal subunits, from the nucleolus to the cytoplasm. This study demonstrates how the fundamental events of ribosome assembly—namely, pre-rRNA folding and processing and RP binding—are dynamically intertwined.
Materials and methods
Construction of yeast strains and depletion of RPLs in vivo
The yeast strains used in this study and their corresponding genotypes are listed in Supplemental Table 4. Conditional RPL mutant strains were engineered as previously described (Ferreira-Cerca et al. 2005; Jakovljevic et al. 2012). To construct strains conditional for expression of the RPs encoded by two genes in the haploid genome, we took two approaches. In some cases, both copies of the gene were deleted, and a plasmid bearing a GAL1/10 promoter fusion of one of the genes was present. In other cases, we deleted one copy of the gene and replaced the other copy with a version driven by the GAL1/10 promoter in the genome. Chromosomal gene fusions with 3xHA, 13-Myc, and TAP tag cassettes were generated as described (Longtine et al. 1998). The PCR oligonucleotides and plasmids used in this study are available on request. Yeast strains bearing RPL genes fused to the GAL1/10 promoter were cultivated at 30°C in rich medium containing either 2% galactose or glucose to express or repress RPL gene expression, respectively.
Analyses of affinity-purified preribosomes by SDS-PAGE, Western blotting, or iTRAQ mass spectrometry
Pre-60S complexes associated with TAP-tagged proteins were purified via a single-step affinity purification technique using IgG-conjugated magnetic beads (Sahasranaman et al. 2011). Proteins from purified pre-rRNPs were separated by SDS-PAGE and analyzed by silver staining or Western blotting as previously described (Sahasranaman et al. 2011). Semiquantitative analysis of protein samples using iTRAQ labeling and liquid chromatography/mass spectrometry as well as subsequent pairwise comparisons of the wild-type and mutant preribosomes were carried out as previously described (Ohmayer et al. 2013). For each of the resulting pairwise comparisons, the iTRAQ ratios of all identified RPLs or AFs were normalized to the median value of all RPL ratios or to the iTRAQ ratio of the bait protein, respectively. iTRAQ ratios were log2-transformed and displayed as heat maps using Java Treeview (http://www.eisenlab.org/eisen/?page_id=42).
For early-acting RPL mutants, TAP-tagged Rpf2 was used to purify preribosomes because this AF is present in 90S particles as well as in each of the pre-60S particles. Importantly, Rpf2 does not function in 27SA2 or 27SA3 pre-rRNA processing (Zhang et al. 2007), and its assembly is unaffected when these early steps are blocked (Sahasranaman et al. 2011). For middle- and late-acting RPLs, the AF Nop7 was used for purification of preribosomes for similar reasons. Nop7 is present in 90S and pre-60S complexes, is not required for 27SB or 7S pre-rRNA processing, and does not depend on proteins required for these processing steps for association with preribosomes. TAP-tagged AF Nog1 was used to purify preribosomes from some late-acting RPLs (especially L10, which assembles in the cytoplasm). Nog1 is present in 90S and all pre-60S particles, stably assembles with preribosomes in the absence of L10, and is released in the cytoplasm. Importantly, the population of pre-rRNAs with which TAP-tagged Rpf2, Nop7, or Nog1 is associated does not change significantly in the absence of RPLs (Supplemental Fig. 2C; Jakovljevic et al. 2012) or AFs (Talkish et al. 2012) required for the corresponding step that is blocked. Thus, the changes that we observed in our depletion mutants are most likely not due to the TAP-tagged proteins failing to assemble with preribosomes or being associated with a different population of assembly intermediates.
Steady-state and kinetic analyses of pre-rRNA processing
Steady-state analyses of mature rRNAs and their precursors by primer extension and Northern hybridization and metabolic pulse-chase analysis were carried out as previously described (Schillewaert et al. 2012; Gamalinda et al. 2013; Ohmayer et al. 2013).
Analysis of ribosome structure
Tertiary structural models for E. coli (Protein Data Bank [PDF] accession no. 2AW4) (Schuwirth et al. 2005) and Saccharomyces cerevisiae large ribosomal subunits (PDB accession nos. 3U5D and 3U5E) (Ben-Shem et al. 2011) were downloaded from the PDB (http://www.rcsb.org). PyMol (http://www.pymol.org) was used to visualize structures and generate images. Secondary structures of yeast 5.8S/25S rRNAs were retrieved from The Comparative RNA Web site (http://www.rna.ccbb.utexas.edu).
Acknowledgments
We are very grateful to J. de la Cruz and J. McManus as well as the members of the Woolford laboratory for critically reading this manuscript. We are also grateful to P. Milkereit for the supervision and valuable discussions about the project. We also thank S. Melnikov for providing the coordinates of RPL contacts with rRNA sequences within 3.5 Å. We also thank the following people for generous gifts of antibodies and plasmids: M. Fromont-Racine, F. Lacroute, P. Linder, D. Goldfarb, J. Maddock, M. McAlear, E. Tosta, J. Brodsky, J. Warner, L. Lindahl, A. Johnson, S. Rospert, K. Siegers, E. Hurt, M. N. Hall, and A. Jacobson. This work was supported by National Science Foundation grant MCB0818534 (to. J.L.W.), the Deutsche Forschungsgemeinschaft in the research unit SFB 960 (to U.O.), and the de Vries Fellowship (to M.G.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.228825.113.
References
- Adilakshmi T, Bellur DL, Woodson SA 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Tollervey D 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J Mol Biol 278: 67–78 [DOI] [PubMed] [Google Scholar]
- Babiano R, de la Cruz J 2010. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38: 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Gamalinda M, Woolford JL Jr, de la Cruz J 2012. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol Cell Biol 32: 3228–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Behrouzi R, Roh JH, Kilburn D, Briber RM, Woodson SA 2012. Cooperative tertiary interaction network guides RNA folding. Cell 149: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X 2003. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci 100: 9302–9307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnadig M, Tollervey D, Bottcher B, Beckmann R, Hurt E 2012. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmueller KL, Weeks KM 2003. Near native structure in an RNA collapsed state. Biochemistry 42: 13869–13878 [DOI] [PubMed] [Google Scholar]
- Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW 2012. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol 197: 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Williamson JR 2013. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol 425: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA 2013. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol Cell 52: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Gleizes PE 2011. Diamond Blackfan anemia: Ribosomal proteins going rogue. Semin Hematol 48: 89–96 [DOI] [PubMed] [Google Scholar]
- Eppens NA, Rensen S, Granneman S, Raue HA, Venema J 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5: 779–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pevida A, Rodriguez-Galan O, Diaz-Quintana A, Kressler D, de la Cruz J 2012. Yeast ribosomal protein L40 assembles late into precursor 60 S ribosomes and is required for their cytoplasmic maturation. J Biol Chem 287: 38390–38407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20: 263–275 [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P 2007. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell 28: 446–457 [DOI] [PubMed] [Google Scholar]
- Francisco-Velilla R, Remacha M, Ballesta JP 2013. Carboxy terminal modifications of the P0 protein reveal alternative mechanisms of nuclear ribosomal stalk assembly. Nucleic Acids Res 41: 8628–8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Bleichert F, Dutca LM, Baserga SJ 2010. When ribosomes go bad: Diseases of ribosome biogenesis. Mol Biosyst 6: 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL Jr 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R 2010. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Swiatkowska A, Tollervey D 2010. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J 29: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N 2012. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Grummt I 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol 62: 109–154 [DOI] [PubMed] [Google Scholar]
- Held WA, Ballou B, Mizushima S, Nomura M 1974. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem 249: 3103–3111 [PubMed] [Google Scholar]
- Hitchen J, Ivakine E, Melekhovets YF, Lalev A, Nazar RN 1997. Structural features in the 3′ external transcribed spacer affecting intragenic processing of yeast rRNA. J Mol Biol 274: 481–490 [DOI] [PubMed] [Google Scholar]
- Hofer A, Bussiere C, Johnson AW 2007. Mutational analysis of the ribosomal protein Rpl10 from yeast. J Biol Chem 282: 32630–32639 [DOI] [PubMed] [Google Scholar]
- Holbrook SR 2008. Structural principles from large RNAs. Annu Rev Biophys 37: 445–464 [DOI] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K 2011. Inside the 40S ribosome assembly machinery. Curr Opin Chem Biol 15: 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K 2013. Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J 2012. The power of AAA-ATPases on the road of pre-60S ribosome maturation—molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Krop JM 1978. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta 517: 378–389 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe A, French SL, Dudnakova T, de Lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D 2013. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol Cell 52: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Chen Y, Guo Q, Zhang Y, Yuan Y, Ma C, Deng H, Lei J, Gao N 2013. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res 41: 7073–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, Hurt E 2013. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 10.1038/nature12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ 1995. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934 [DOI] [PubMed] [Google Scholar]
- McCann KL, Baserga SJ 2013. Genetics. Mysterious ribosomopathies. Science 341: 849–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR 2010. Visualizing ribosome biogenesis: Parallel assembly pathways for the 30S subunit. Science 330: 673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny V, Nierhaus KH 1982. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc Natl Acad Sci 79: 7238–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Poll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, et al. 2013. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisiae. PLoS ONE 8: e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL Jr, Tschochner H, Milkereit P 2009. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 4: e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM, Fedorova O, Waldsich C 2007. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci 32: 138–145 [DOI] [PubMed] [Google Scholar]
- Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M 2008. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl R, Nierhaus KH 1982. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci 79: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: Role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M 2003. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillewaert S, Wacheul L, Lhomme F, Lafontaine DL 2012. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol Cell Biol 32: 430–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J 2010. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL III, Karbstein K, Skiniotis G 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K 2012. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr 2012. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 40: 8646–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138: 911–922 [DOI] [PubMed] [Google Scholar]
- Woodson SA 2011. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res 44: 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes & Dev 21: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]