Cellular senescence can be described as a state of stable cell cycle arrest in response to diverse stresses. Senescence is a collective phenotype of multiple effectors, and their intensity and combination can be different depending on triggers and cell types. In this review, Salama et al. summarize effector mechanisms and highlight some key components of the collective phenotype of senescence.
Keywords: senescence, aging, DNA damage, oncogenes, tumor suppressors, inflammation, metabolism
Abstract
Cellular senescence is a stress response that accompanies stable exit from the cell cycle. Classically, senescence, particularly in human cells, involves the p53 and p16/Rb pathways, and often both of these tumor suppressor pathways need to be abrogated to bypass senescence. In parallel, a number of effector mechanisms of senescence have been identified and characterized. These studies suggest that senescence is a collective phenotype of these multiple effectors, and their intensity and combination can be different depending on triggers and cell types, conferring a complex and diverse nature to senescence. Series of studies on senescence-associated secretory phenotype (SASP) in particular have revealed various layers of functionality of senescent cells in vivo. Here we discuss some key features of senescence effectors and attempt to functionally link them when it is possible.
Cellular senescence was originally identified as a state of “permanent” cell cycle arrest resulting from the limited replicative capacity of normal human diploid fibroblasts (HDFs) in culture (called replicative senescence) (Hayflick 1965; Shay and Wright 2000). While replicative senescence was later associated with telomere shortening or dysfunction, different cellular stressors have also been demonstrated to induce a similar phenotype. Thus, more generally, senescence can be described as a state of stable cell cycle arrest in response to diverse stresses (Campisi and d'Adda di Fagagna 2007; Collado et al. 2007; Kuilman et al. 2010). Recently, however, this rather bland description has been considerably revised due to our increased understanding of its non-cell-autonomous activities, which are mediated by the senescence-associated secretory phenotype (SASP). The non-cell-autonomous activities of senescence provide an understanding of the functional relevance of senescent cells in pathophysiology, particularly diverse roles in the tumor microenvironment, and the suggestion that they may contribute to declining organ function with aging. Thus, senescence is not a singular state but rather a heterogeneous phenotype driven by diverse inputs leading to diverse outputs. In addition to the SASP, senescence is associated with other effector mechanisms, providing additional complexity to the phenotype. This view is consistent with the idea that, in contrast to the apoptotic program, senescence is a collective phenotype of multiple effector programs, which form functional networks of senescence (Young et al. 2013). Furthermore, recent studies have revealed “nonpathological” roles for senescence during embryonic development. Senescence, much like apoptosis, contributes to embryonic patterning (Muñoz-Espín et al. 2013; Storer et al. 2013). In addition, ERVWE1-mediated cell fusion, which is required for the placental syncytiotrophoblast formation, induces senescence, although precise roles for senescence in the development and function of the placenta remain to be elucidated (Chuprin et al. 2013). These studies reinforce the concept of a diversity of senescence phenotypes and suggest that senescence might be a more widespread phenomenon than it previously appeared to be.
A number of senescence markers have been characterized in the prototypical HDF system, but many of these findings have been validated in other cell and in vivo systems (Table 1). These markers often are associated with the effector mechanisms of senescence, as discussed below. For example, some markers of the DNA damage response (DDR) (d'Adda di Fagagna 2008) and components of the SASP (Kuilman and Peeper 2009; Coppé et al. 2010) have been used as in vitro or in vivo markers of senescence. Even senescence-associated β-galactosidase (SA-β-Gal) activity, the de facto hallmark of senescence (Dimri et al. 1995), may reflect increased activity of autophagy, at least in some cases (Gerland et al. 2003; Narita et al. 2011). Since the effector mechanisms of senescence are not necessarily specific for senescence or present in all forms of senescence, interpretation of senescence markers needs to be context-dependent; more importantly, multiple markers need to be considered in combination. It is conceivable that the different combinations of effectors and markers result from differences in the “quality” of the phenotype. As discussed later, such a “spectrum” of senescence is consistent with the recent idea of an actively maintained state of “deep” or “late” senescence, which seems to be associated with specific (or additional) markers and effectors of senescence (Passos et al. 2010; De Cecco et al. 2013; Ivanov et al. 2013). The recent detailed characterization of the SASP, where the paracrine transmission of senescence to neighboring cells wanes during serial transmission, also provides evidence for a qualitative variation in the senescent phenotype (Acosta et al. 2013). Here we summarize effector mechanisms and highlight some key components of the collective phenotype of senescence (Fig. 1).
Table 1.
Selected list of senescence markers
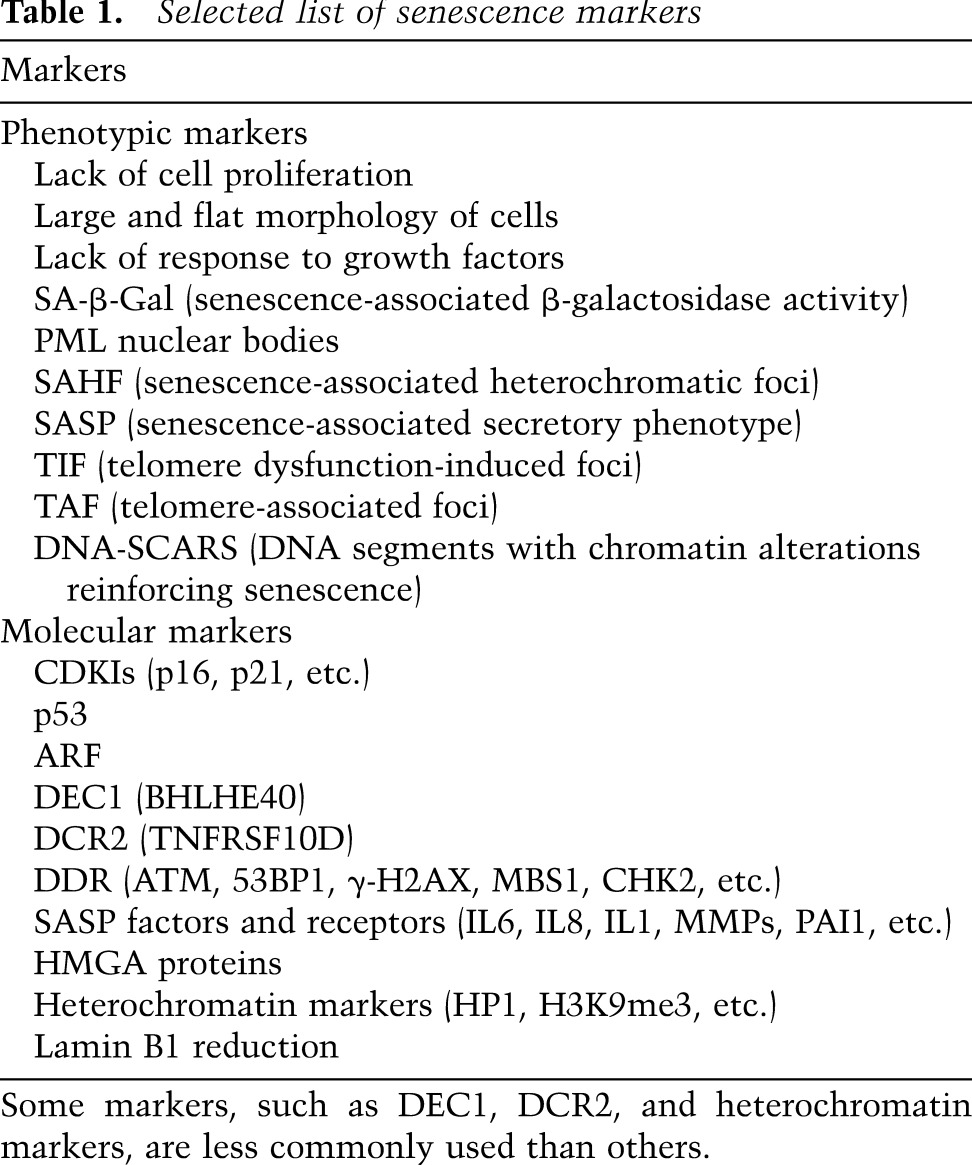
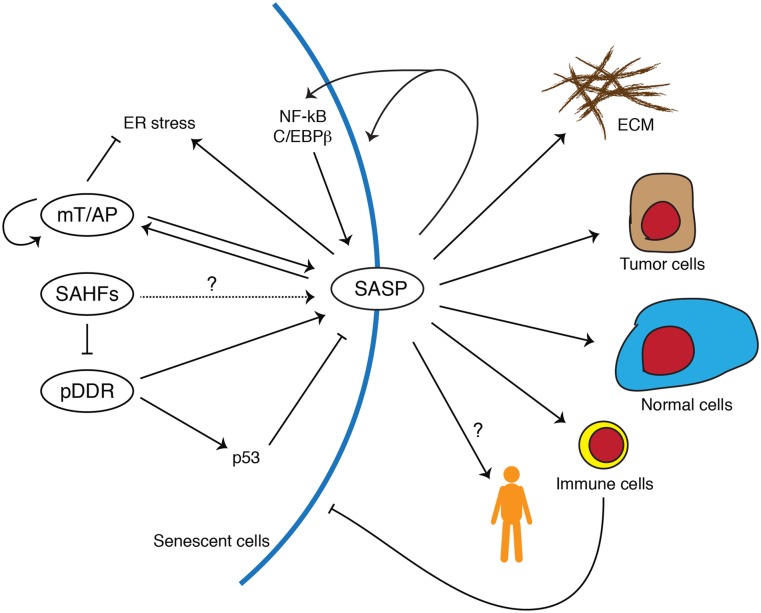
Figure 1.
Schematic model of senescence and its biological functions. Representative intrinsic effectors of senescence and their relationship are shown on the left. Spatial and functional associations between mTOR (mT) and autophagy (AP) augment each other and facilitate SASP. The SASP provokes ER stress and, as a countermeasure, autophagy. SAHFs might contribute to maintenance of constitutively active gene expression, such as SASP components. SAHFs also restrain DNA damage. Persistent DDR (pDDR) positively regulates many SASP components, whereas p53 negatively regulates or modulates the SASP. Representative impacts of the SASP are shown on the right. The SASP reinforces senescence through autocrine activities. The SASP can be amplified through positive feedback loops between proinflammatory cytokines and the transcription factors (NF-kB and C/EBPβ). Targets of paracrine activities of the SASP include the extracellular matrix (ECM), tumor cells (protumorigenic), normal cells (prosenescent), and immune cells. Senescent cells can be eliminated through the immune cells. It is also conceivable that the SASP causes systemic effects on individual fitness, facilitating aging.
DDR
Senescence can be induced by diverse stimuli, and activation of the DDR pathways is involved in both the induction and maintenance of senescence in many cases. The DDR is composed of DNA damage sensing, amplification of activities of upstream kinases ATM and ATR, the signal transduction through downstream kinases CHK2 and CHK1, and effector proteins such as p53 and CDC25 (d'Adda di Fagagna 2008). Persistent DDR in particular, which is represented by telomere dysfunction-induced foci (TIF) (Takai et al. 2003; Herbig et al. 2006) and DNA segments with chromatin alterations reinforcing senescence (DNA-SCAR) (Rodier et al. 2011), has been associated with the maintenance of senescence primarily through p53 activation (Rodier et al. 2009; Fumagalli et al. 2012).
It has been shown that the DDR kinases are activated during replicative senescence, which is associated with the gradual shortening of telomeres (Harley et al. 1990), and pairwise inhibition of ATM/ATR or CHK2/CHK1 delays the onset of replicative senescence (d'Adda di Fagagna et al. 2003; Herbig et al. 2004). Telomeres are nucleoprotein complexes composed of specialized tandem repeat sequences (5′-TTAGGG-3′) and an associated multiprotein complex called “shelterin” (Palm and de Lange 2008). Telomeres are located at the end of every chromosome, protecting DNA ends from a number of enzymes involved in DNA metabolism. The very end of the telomere is single-stranded, and this can loop back, forming a t-loop and invading between the telomeric dsDNA to form a D-loop structure (Palm and de Lange 2008). It has been shown that the shelterin protein TRF2 protects chromosomal ends (van Steensel et al. 1998) but also limits DNA repair when damage occurs within telomeres (Fumagalli et al. 2012).
Although the exact threshold of telomere length or the number of dysfunctional telomeres within a cell that can trigger senescence is still unclear (d'Adda di Fagagna 2008), a current structural model describes three states of telomeres: closed state (t-loop formed), intermediate state, and uncapped state (Cesare and Karlseder 2012). In this model, telomere shortening induces exposure of chromosomal ends, which provides substrates for the DDR, thus triggering cellular senescence (the intermediate state). Further shortening of telomeres (the uncapped state) leads to loss of TRF2, leading to interchromosomal or intrachromosomal fusions and a state called “crisis,” which is associated with a marked increase in genomic instability and cell death. The DDR at deprotected telomeres can be visualized by DDR marker proteins, such as 53BP1 and γ-H2AX (H2AX phosphorylated at Ser139) (d'Adda di Fagagna et al. 2003; Takai et al. 2003; Herbig et al. 2004). In the intermediate state, however, DDR-positive telomeres retain TRF2, which inhibits both chromosome end fusion and DNA repair. Thus, this state can result in a persistent DDR. In addition, the TRF2-dependent suppression of DNA repair within telomeres can yield DDR signaling from telomere-associated damage resulting from causes other than telomere shortening (Karlseder et al. 2002; Fumagalli et al. 2012). If DNA damage is dispersed throughout the genome (e.g., by γ-irradiation), damage within telomeres is irreparable and thus stabilized (Fumagalli et al. 2012; Hewitt et al. 2012; Suram et al. 2012). Interestingly, a recent study showed that damage at certain nontelomeric sites can also be persistent, resulting in DNA-SCAR formation (Rodier et al. 2011). Mechanisms for the suppression of DNA damage repair at nontelomeric sites remain to be elucidated.
In addition to replicative exhaustion and DNA damage-inducing agents, the DDR has also been shown to be a critical effector of oncogene-induced senescence (OIS) (d'Adda di Fagagna 2008). Strong activation of some oncogenes (such as Ras or Raf) paradoxically induces senescence (Serrano et al. 1997), and this process appears to be telomere-independent (Jones et al. 2000). The initial response of normal cells to constitutively active oncogenic stimuli is typically hyperproliferation, which appears to be “sensed” by cells, triggering a senescence phenotype (Lin et al. 1998). It has been shown that one mechanism for the link between the initial mitotic reaction and senescence induction is the S-phase-associated DDR: The oncogenic stress drives increased DNA replication origins, leading to accumulation of incomplete replication intermediates, resulting in DNA damage and activation of the DDR kinases (ATM, ATR, CHK2, and CHK1) (Bartkova et al. 2006; Di Micco et al. 2006; Mallette et al. 2007). These studies also showed that, depending on oncogenes or cell type, experimental inactivation of key DDR factors can inhibit OIS. Furthermore, consistent with the finding that telomeric DNA damage is irreparable, it has been reported that OIS is also associated with persistent DDR at telomeres (Suram et al. 2012).
Thus, the DDR appears to be a mechanism that underlies senescence, triggered by diverse stimuli. Of note, DDR-independent senescence has also been reported in some experimental conditions where p16, p38MAPK, or ATR is constitutively activated or tumor suppressor Pten is completely lost (Toledo et al. 2008; Rodier et al. 2009; Alimonti et al. 2010; Freund et al. 2011). In addition, two recent studies have identified “programmed” cellular senescence, which contributes to tissue remodeling during mammalian embryonic development at multiple regions (Muñoz-Espín et al. 2013; Storer et al. 2013). They showed that this developmental senescence typically shares some markers with OIS (including SA-β-Gal, p21, p15, and heterochromatin marks) but not the DDR. The up-regulation of p21 (a major p53 target) is critical for this phenotype, but, consistent with the lack of the DDR, the activation of p21 is p53-independent. Interestingly, while developmental senescence also exhibits up-regulation of some SASP components, major DDR targets (see below) like IL6 and IL8 do not appear to be up-regulated (Storer et al. 2013). Thus, these studies provide physiological contexts for DDR-independent senescence.
SASP
The key contribution of secretome alterations to the senescent phenotype has long been recognized. Early studies showed that late passage human fibroblasts or fibroblasts derived from subjects with premature aging syndromes produce secretory proteins, including plasminogen activator inhibitor type-1 (PAI-1), which later became a functional marker of senescence (Murano et al. 1991; Goldstein et al. 1994; Kortlever et al. 2006).
The secretome of senescent cells is complex, consisting of a range of cytokines, chemokines, and proteases, among others (Campisi 2005; Kuilman and Peeper 2009). This SASP or senescence messaging secretome (SMS) (Kuilman and Peeper 2009) reflects the non-cell-autonomous functionality of senescent cells and may underpin their in vivo role in the pathophysiology of aging and age-related disorders. However, the range of functions ascribed to members of the SASP is extremely diverse and includes both autocrine and paracrine signaling, protumorigenic and tumor-suppressive effects, and pro- and anti-inflammatory signaling.
Regulation of the SASP
Control of the secretome in senescent cells is achieved at many levels, from transcriptional regulation to autocrine feedback loops, but persistent DDR appears to be critical for regulation of the SASP (Coppé et al. 2010). Senescent cells induced by either genotoxic stress, replicative exhaustion, or oncogenic stress secrete myriad factors associated with inflammation, proliferation, and modulation of the extracellular matrix (ECM) (Acosta et al. 2008; Kuilman et al. 2008; Rodier et al. 2009).
Loss of ataxia telangiectasia-mutated (ATM) or other factors responding to DNA damage (such as NBS1 and CHK2) leads to reductions in some SASP factors, such as IL6 and IL8, central components of SASP, during not only DDR-induced senescence but also OIS (Rodier et al. 2009). OIS is also mediated by the DDR (Bartkova et al. 2006; Di Micco et al. 2006; Mallette et al. 2007). In addition, expression of the CDK inhibitors p16 or p21 leads to the induction of senescence without the development of a DDR; this senescent phenotype lacks a proinflammatory SASP (Rodier et al. 2009). Thus, the development of the SASP is largely dependent on a persistent DDR associated with senescence stimuli (Coppé et al. 2010). Interestingly, Coppé et al. (2008) showed that loss of p53 from HDFs rather augments persistent DDR and IL6 secretion, which plays a key role in protumorigenic paracrine activities of the SASP (see below). Thus, they propose an intriguing concept that p53 acts as a “cell-nonautonomous tumor suppressor.” This idea was reinforced by a recent study of chemically induced hepatocellular carcinoma (HCC) in mice, in which the p53-mediated SASP in hepatic stellate cells (HSCs) suppresses development of HCC in part through triggering the activation of anti-tumoral M1 macrophages (Lujambio et al. 2013).
Many SASP components are up-regulated at a transcriptional level (Shelton et al. 1999; Kuilman et al. 2008), and two transcription factors, NF-κB and C/EBPβ, have been shown to be up-regulated, activated, and bound to chromatin during OIS and to cooperatively regulate the inflammatory components of the SASP (Acosta et al. 2008; Kuilman et al. 2008; Chien et al. 2011; Jing et al. 2011). Using a transcriptomic approach, Kuilman et al. (2008) have shown that depletion of IL6, which they confirmed to be a direct transcriptional target of C/EBPβ, results in collapse of the inflammatory SASP network and senescence bypass. More recently, IL1 signaling was shown to be an upstream effector of both NF-κB and C/EBPβ and therefore IL6 and IL8 induction (Orjalo et al. 2009; Hubackova et al. 2012; Acosta et al. 2013). Interestingly, Acosta et al. (2013) identified inflammasomes, multimolecular innate immune complexes, as a critical regulator of this process. Analogous to the apoptosome, which activates caspase-9 and the subsequent cascade of the classical apoptosis pathway, inflammasomes activate caspase-1 (formerly known as IL1 converting enzyme), which is required for the initiation and amplification of IL1 signaling (Strowig et al. 2012; Hoare and Narita 2013). In addition, recent studies have shown that IL1 (and TGFβ, another SASP component) can mediate senescence through activating oxidative stress and DNA damage, providing another level of positive feedback loop (Hubackova et al. 2012; Acosta et al. 2013). These studies collectively suggest that the inflammatory SASP consists of a complex hierarchical network, which involves robust signal amplification.
Functions of the SASP
There are diverse downstream effects of the SASP that are dependent on the context and signal-receiving cell situation. These effects include protumorigenesis (paracrine), immunomodulation (paracrine), senescence reinforcement (paracrine and autocrine), and modulation of the tissue microenvironment (paracrine) (Hoare and Narita 2013). In addition to the cell-autonomous tumor suppressor function of senescence, which could be reinforced by the SASP through an autocrine feedback loop, the paradoxical protumorigenic effects of the SASP have been noted (Coppé et al. 2010). It has been shown that secreted proteins from senescent HDFs promote proliferation and malignant transformation of premalignant epithelial cells (Krtolica et al. 2001; Bavik et al. 2006). For example, these paracrine effects include promotion of epithelial–mesenchyme transition and invasion, tumor vascularization, and abnormal tissue morphology, which are mediated by the proinflammatory cytokines IL6 and IL8, VEGF, and the metalloprotease MMP3 (Parrinello et al. 2005; Coppé et al. 2006, 2008; Kuilman et al. 2008). Interestingly, however, the paracrine protumorigenic activity of SASP appears to be more relevant to cells that are already primed toward malignant transformation.
How, then, does the SASP affect normal cells? In their seminal works, the Peeper and Gil groups (Acosta et al. 2008; Kuilman et al. 2008) showed that IL6 and IL8, key components of the SASP, ‘”reinforce” senescence rather than “spread” senescent phenotypes to the healthy neighboring cells. This requirement for some commitment of target cells toward either malignant transformation or senescence might explain to some extent the pleotropic functions of these cytokines. However, recent studies have shown that some SASP factors, including IL1β, can induce senescence in normal cells. Thus, senescence can be transmitted to untransformed neighboring cells through the paracrine activity of the SASP (Hubackova et al. 2012; Acosta et al. 2013). IL1β needs to be cleaved and modified by inflammasome-activated caspase-1, and therefore the inflammasome is involved in local propagation of not only inflammation but also senescence in the tissue microenvironment. Interestingly, the propagation of paracrine senescence appears to be under control, since transmissibility of the SASP wanes during serial transmission of senescence (Acosta et al. 2013). The significance of the inflammasome–IL1β signal as an upstream effector of senescence was also observed in murine HSC senescence. This can be induced by obesity-associated gut microbial production of the bile acid metabolite deoxycholic acid (DCA), although, in this case, the paracrine effect of the HSC SASP appears to be protumorigenic on hepatocytes in the presence of the carcinogen DMBA (Yoshimoto et al. 2013).
Paracrine effects of the SASP also provoke anti-tumor immunity (Serrano 2011; Hoenicke and Zender 2012). Apoptotic cells physiologically are rapidly eliminated through phagocytosis to avoid harmful necrotic inflammation. Xue et al. (2007) first showed that senescent cells can also be eliminated in vivo. They used a H-ras-driven mouse liver cancer model in which the endogenous level of p53 could be modulated by inducible RNAi. They showed that reactivation of the p53 pathway results in ras-driven tumor regression. Strikingly, they found no sign of apoptosis but instead induction of senescence; the SASP triggers infiltration of natural killer (NK) cells and other innate effector cells to eliminate tumor cells. Recently, using a similar model, Iannello et al. (2013) showed that the recruitment of NK cells into tumors in which p53 had been reactivated requires the chemokine CCL2 and that the elimination of those tumor cells by NK cells is dependent on the NK surface receptor NKG2D, known to be involved in anti-tumoral immunity. However, both studies used recipient mice that lacked mature T-lymphocytic responses. In contexts where all components of the immune system are intact, antigen-specific CD4+ T-lymphocyte responses against activated oncogenes were demonstrated to be crucial in the clearance of senescent cells (Kang et al. 2011b). “Senescence surveillance” by both innate and adaptive immune responses, triggered by paracrine SASP signaling, has been further demonstrated in several other model systems (Krizhanovsky et al. 2008; Acosta et al. 2013; Lujambio et al. 2013).
Interestingly, it has been shown that nonmalignant cells, including infiltrating immune cells, surrounding early neoplastic lesions exhibit focal p16 activation (Burd et al. 2013). The data suggest that paracrine activities of the SASP not only recruit immune cells but also transmit senescence to them. However, as the investigators comment, the possibility cannot be excluded that immune cells become senescent elsewhere and then are preferentially recruited to the emerging neoplastic region (Burd et al. 2013). In any case, the specific roles of the senescence program in these immune cells need further validation.
Beyond cell-to-cell communication, recent evidence also links senescence with tissue architectural integrity. Chronic toxic liver injury in mice leads to the development of hepatic fibrosis that mimics human chronic liver injury. Upon liver injury, HSCs initially activate, proliferate, and develop a profibrotic secretome. These mice exhibit accumulation of senescent cells, which are primarily derived from HSCs (Krizhanovsky et al. 2008). However, when activated HSCs subsequently become senescent, the profibrotic secretome switches to a more typical SASP-like secretome, which contains both proinflammatory cytokines and matrix-degrading enzymes; this is generally felt to be “fibrolytic,” thereby limiting liver fibrosis. In mice lacking p53, HSCs fail to undergo senescence, continue to proliferate, and secrete profibrotic factors, leading to exaggerated hepatic fibrosis. Furthermore, senescent activated HSCs are eventually eliminated through senescence surveillance, by either NK cells (Krizhanovsky et al. 2008) or macrophages (Lujambio et al. 2013). Thus, the proper timing of senescence induction and subsequent immune-mediated clearance of HSCs appears to be essential for the physiological process of tissue repair in the liver.
How senescence is triggered in HSCs remains unclear. However a recent study has shown an interesting interplay between damaged hepatocytes and activated HSCs: Expression of the matricellular protein CCN1 is up-regulated and secreted from hepatocytes upon liver damage; CCN1 triggers senescence in HSCs through binding α6β1 integrin, a receptor of CCN1 (Kim et al. 2013). An earlier study from the same group revealed that CCN1 also plays an essential role in skin wound healing, where CCN1 triggers senescence of local myofibroblasts. Similar to the senescent HSCs within the injured liver, CCN1-induced senescent myofibroblasts limit excessive fibrosis (Jun and Lau 2010). The level of CCN1 also appears to be increased in other senescence-associated tissues, such as benign prostatic hyperplasia (BPH) (Castro et al. 2003; Sakamoto et al. 2004). Thus, it will be interesting to know whether the involvement of CCN1 in both senescence and tissue repair is more generalized, perhaps even in the context of the tumor microenvironment.
These different SASP effects are not mutually exclusive, and, within a given microenvironment, the complex cell-to-cell or cell-to-ECM communications through a cocktail of SASP components have diverse effects depending on cell types and cell conditions. Understanding and manipulating the net effects of such a complex signaling network as the SASP appears to be very challenging but would extend our opportunities for modulating senescence-related disorders. In addition, the recent study from the Schmitt laboratory (Dörr et al. 2013) provided insight into another aspect of the SASP unrelated to its downstream effects, but rather the process of protein synthesis, that provokes endoplasmic reticulum (ER) stress. This point is discussed in the context of senescence-associated metabolic changes below.
Nucleus, chromatin, and gene regulation
The nuclear phenotype and patterns of gene expression are drastically altered during senescence, with down-regulation of cell cycle genes, up-regulation of senescence marker genes, and senescence-associated alteration of the secretome (Shelton et al. 1999). Thus, it is possible that senescence is associated with some unique gene regulatory mechanisms. For example, p16, a functional biomarker of senescence, is negatively regulated by histone H3 Lys 27 trimethylation (H3K27me3) and its docking proteins of the polycomb group (Jacobs et al. 1999), which can be countered by JMJD3, the H3K27 demethylase (Agger et al. 2009; Barradas et al. 2009).
Another example for senescence-associated gene regulation can be seen in the recent study reported by Chicas et al. (2010). Cell cycle genes are generally repressed when cells are arrested; thus, their repression is not specific to senescence. Among the Retinoblastoma protein family (Rb, p107, and p130), the contribution of Rb to repressing a subset of cell cycle genes (namely, genes involved in DNA replication) is unique to senescence, whereas all Rb family members show redundant activity in repressing those gene during quiescence (Chicas et al. 2010). This study provides a potential explanation for why, of the Rb family, Rb itself is almost exclusively mutated in cancer and highlights the tumor-suppressive role of Rb and senescence. Several mechanisms have been proposed for Rb's involvement in senescence-associated gene regulation. For example, the tumor suppressor promyelocytic leukemia (PML), a component of PML nuclear bodies (PML NBs, also known as promyelocytic oncogenic domains [PODs]), is a marker of senescence: During senescence, PML is up-regulated, the size and number of PML NBs are increased (Table 1), and PML contributes to senescence through regulation of p53 and Rb status (Ferbeyre et al. 2000; Pearson et al. 2000; Bischof et al. 2002, 2005). Vernier et al. (2011) recently showed that Rb–E2F complexes are recruited to PML NBs, where heterochromatin protein 1 (HP1) (Zhang et al. 2005) and the Rb phosphatase PP1α (protein phosphatase 1α) are also located during senescence. They showed that PML overexpression represses some E2F targets, whose promoters can be enriched for E2F as well as PML; thus, the investigators propose that PML NBs are the sites for nucleation of heterochromatin at E2F targets (Vernier et al. 2011). It has also been shown that Rb is involved in silencing of E2F target genes during senescence through its physical association with AGO2 (a key component of RNA-induced silencing complex [RISC]) and microRNA (e.g., let-7) (Benhamed et al. 2012).
Senescence is also often associated with global changes in chromatin structure and altered expression of proteins that affect chromatin structure (Adams 2007). For example, upon the induction of senescence in HDFs, senescence-associated heterochromatic foci (SAHFs) become readily visible as distinct DAPI-dense foci, the formation of which is typically dependent on the Rb pathway (Narita et al. 2003). In addition, a subset of genes repressed in senescent cells shows an increase of repressive histone marks in the genic regions (Narita et al. 2003; Di Micco et al. 2011; Sadaie et al. 2013). These studies suggest some correlation between alterations in global chromatin structure and gene expression, but how the higher-order chromatin structure affects specific gene expression remains to be elucidated.
SAHFs
SAHFs were originally described in OIS and, to a lesser extent, in replicative and DNA damage-induced senescence HDFs (Narita et al. 2003). SAHFs are enriched for repressive epigenetic marks, such as methylated H3K9, HP1, and macroH2A, whereas euchromatic marks are excluded from SAHFs (Narita et al. 2003; Zhang et al. 2005). A series of studies from the Adams laboratory (Zhang et al. 2005, 2007; Ye et al. 2007a,b) have shown that, as an early event in senescence, translocation of the HIRA histone chaperone complex to PML NBs is required for SAHF formation. They also found that the relocalization of HIRA to PML bodies is driven by down-regulation of the canonical Wnt signaling pathway (Ye et al. 2007a). SAHFs are also characterized by alteration of chromatin architectural proteins, including accumulation of HMGA proteins and loss of linker histone H1 (Funayama et al. 2006; Narita et al. 2006). Thus, a number of factors are either functionally or physically involved in the process.
Interestingly, it has been proposed that SAHFs consist of individual chromosomes (Funayama et al. 2006; Zhang et al. 2007), and each focus exhibits a multilayered structure, which consists of a H3K9me3 constitutive heterochromatic core surrounded by H3K27me3 facultative heterochromatic layer (Chandra et al. 2012). Strikingly, our genome-wide analyses of these two repressive histone marks revealed that the SAHF structure arises through a spatial rearrangement of pre-existing repressively marked chromatin.
How does the global structure of senescent chromatin affect expression of individual genes? One might speculate that such a process, which clearly segregates constitutive heterochromatin, facultative heterochromatin, and active euchromatin into exclusive layers, would create an environment in which not only gene repression but also gene activation can be very efficiently coordinated and maintained (Chandra and Narita 2013). Thus, it is possible that “local” genic chromatin modifications can be separately regulated, and the “global” structural chromatin alterations facilitate the expression of genes depending on their chromatin states and/or positions. Di Micco et al. (2011) showed that SAHF-like structures can be retained when OIS bypass is achieved by p53 or ATM depletion. Therefore, SAHF-like structures may also confer context-dependent downstream effects, much like SASP, which also can remain active when senescent cells resume proliferating by p53 depletion in a low p16 background (Coppé et al. 2008). Di Micco et al. (2011) also showed that SAHFs restrain the DDR, suggesting that the role of SAHFs is not limited to gene regulation.
Lamin B1
The nuclear lamina, a filamentous structure beneath the inner nuclear membrane, has been implicated in the integrity of nuclear structure, chromatin positioning, and gene expression. The nuclear lamina in somatic cells contains two major proteins: A-type and B-type lamins (Dechat et al. 2010). Recently, Lamin B1 (LMNB1) was reported to be specifically down-regulated during senescence in HDFs (Shimi et al. 2011). Since then, several studies have confirmed the reduction of LMNB1 in various senescent cell types (Barascu et al. 2012; Freund et al. 2012; Dreesen et al. 2013; Sadaie et al. 2013; Shah et al. 2013). However, the functional relationship between altered LMNB1 levels and senescence phenotypes is still unclear.
The extent to which the senescent phenotype is affected by modulation of LMNB1 levels varies between studies. However, RNAi-mediated depletion of LMNB1 appears to be associated with reduced cell proliferation to various degrees in most (Shimi et al. 2011; Dreesen et al. 2013; Sadaie et al. 2013; Shah et al. 2013). Shimi et al. (2011) showed that depletion of LMNB1 is sufficient to induce senescence through modulation of reactive oxygen species (ROS) production. Perhaps paradoxically, they showed that depletion of LMNB1 leads to a reduction of ROS in a p53-depenent manner. Similarly, Barascu et al. (2012) showed that increased levels of LMNB1 are associated with ROS up-regulation and senescence. Generally, ROS is thought to be a cause of cellular damage, and it has been shown that ROS is increased during senescence. Furthermore, high levels of exogenous oxidative stress can induce senescence (Passos et al. 2007). However, increasing evidence suggests that modest levels of ROS can also contribute to cellular proliferation and survival (Sena and Chandel 2012). Thus, alterations in LMNB1 might contribute to a fine-tuning of ROS levels.
Shah et al. (2013) also showed induction of senescence by LMNB1 depletion in HDFs. They found large-scale chromatin domains enriched for H3K4me3 and H3K27me3 (called “mesas”) and depleted for H3K27me3 (called “canyons”) in senescent HDFs; these were correlated with the expression of key senescence-associated genes. Interestingly, LMNB1 depletion triggers these mesas and canyons. We also observed a reduction of LMNB1 in senescent HDFs and melanocytes (Sadaie et al. 2013). Interestingly, despite the global reduction of LMNB1, we observed redistribution of LMNB1 along the genome; LMNB1 is preferentially reduced from regions enriched for H3K9me3 rather than H3K27me3, but there are small regions (only 2% of the genome) where LMNB1 accumulates. Genes within these LMNB1-increased regions, which include some cell cycle genes, tend to be repressed during senescence, with increased H3K27me3 across the gene bodies. In addition, we showed a correlation between LMNB1 reduction (particularly from H3K9me3 regions) and spatial repositioning of perinuclear heterochromatin (H3K9me3-enriched) and SAHF formation. These studies suggest that alteration of LMNB1 might also contribute to senescence through gene regulation in some specific regions.
Compared with LMNB1 depletion, phenotypes induced by ectopic LMNB1 were more heterogeneous between studies: Overexpression of LMNB1 either delays (Shimi et al. 2011) or enhances senescence (Barascu et al. 2012; Dreesen et al. 2013) or exhibits a minimal effect on proliferation (Sadaie et al. 2013). Precise reasons behind these differences are not clear. In addition to the aforementioned dual role of ROS in proliferation depending on its levels, the fine balance between LMNB1 and LMNA may also be differentially perturbed depending on the system (Hutchison 2012; Dreesen et al. 2013). It is important to note that the senescence-associated complex regional alterations of genomic LMNB1 binding, dependent on local chromatin states, might be masked by enforced increase of global LMNB1 levels.
Other chromatin alterations associated with senescence
While SAHFs are characterized microscopically, De Cecco et al.(2013) recently identified another type of senescence-associated chromatin alteration using a biochemical approach: formaldehyde-assisted isolation of regulatory elements (FAIRE), a method for genome-wide mapping of open chromatin regions. They showed, in replicative senescent HDFs, an overall condensation of chromatin in euchromatic regions, with the exception of some specific genes. On the other hand, chromatin of repetitive sequences, including major classes of retrotransposon as well as pericentromeric regions, which are highly condensed in normal cells, tend to become open in senescent cells. The decondensation of these heterochromatic regions is associated with expression of transposable elements (such as LINE1 elements) and their transposition, particularly at later time points after the induction of senescence. They also showed the decondensation of pericentromeric heterochromatin in late senescent cells.
Ivanov et al. (2013) have recently shown another unexpected observation: the loss of chromatin during senescence. They found that reduction of LMNB1 and loss of nuclear envelope integrity in senescent cells are associated with the appearance of cytoplasmic chromatin fragments (CCFs). These are targeted to autophagy for degradation, resulting in low histone content. A reduction of total histone content was seen within the deeper (or “mature”) portions of nevi, a model for in vivo OIS (Ivanov et al. 2013). Thus, similar to the redistribution of the relative condensation of chromatin mentioned above, CCFs also appear to be a late event during senescence. The physiological significance of these chromatin alterations that are associated with late or deep senescence remains to be elucidated, or they might be considered “degenerative” alterations as the investigators suggest (De Cecco et al. 2013; Ivanov et al. 2013).
Senescence and changes in cellular metabolism
Senescence is currently defined by the lack of cell cycle progression or proliferation. However, senescent cells are often enlarged, suggesting that they retain the capacity to “hypertrophy.” Thus, there appears to be an uncoupling between cellular proliferation and growth during senescence. In addition, the functional activities of senescent cells along with the en masse secretion of proteins further suggest that senescent cells are metabolically highly active. This has been confirmed by recent studies that have measured various aspects of metabolism during senescence (Fig. 2).
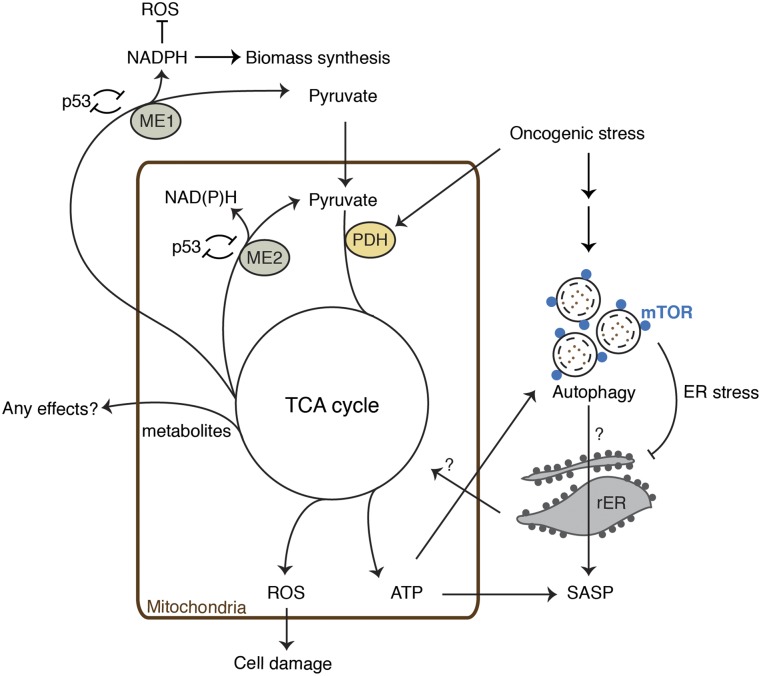
Figure 2.
Metabolic pathways involved in senescence—a collective view of recent studies. Recent studies suggest that mitochondrial metabolism is highly active during OIS and TIS. In addition to the TCA cycle, TIS was also associated with enhanced glycolysis and fatty acid catabolism (Dörr et al. 2013). It was proposed that enhanced ATP production facilitates energy-consuming effectors, such as SASP and autophagy. Production of the SASP evokes ER stress, which is mitigated by autophagy. We showed that spatial coupling of autolysosomes and mTOR (TASCC) facilitates IL6/8 synthesis in OIS cells. We hypothesize that the rapid protein turnover drives nascent synthesis of secretory proteins, but the precise mechanism for the functional relationship between the TASCC and SASP remains to be elucidated (Narita et al. 2011). ME1 and ME2 have been shown to be p53-repressive targets, and their down-regulation activates p53, triggering and/or reinforcing senescence. MEs are important for the production of NADPH, a reducing agent, which is also necessary for biomass synthesis. Some metabolites derived from the TCA cycle or shift in their balance may also have cellular effects. This is a collective interpretation of data from multiple studies; thus, components do not necessarily link to each other in the same context. For example, the significance of the reciprocal regulation of MEs and p53 is unclear in OIS conditions, and the mechanism for PDH activation appears to be different depending on oncogenes. Note that earlier studies suggest that senescent cells are associated with dysfunctional mitochondria (Moiseeva et al. 2009). In all cases, ROS production appears to be increased.
Protein degradation pathways
Macroautophagy (referred to as autophagy in this review) is a an evolutionarily conserved catabolic process, which involves isolation of cytoplasmic organelles and macromolecules by double membranous autophagosomes and their en masse degradation through fusion with lysosomes (forming autolysosomes) (Mizushima and Komatsu 2011). Autophagy is a bulk nonspecific cytoplasmic degradation mechanism. Some level of specificity of the cargo for degradation is directed through autophagic receptors, including p62 (also known as sequestosome-1), NBR1, and Nix (Johansen and Lamark 2011). The basal level of autophagy plays an important role for quality control of macromolecules and energy homeostasis, but it can also be induced in response to various cytotoxic stresses (Hoare et al. 2011). Increasing data indicate a significant association between senescence and autophagy in various cell systems (Gerland et al. 2003; Gamerdinger et al. 2009; Sasaki et al. 2010; Capparelli et al. 2012; Dörr et al. 2013). Some studies have touched on the functional relevance of autophagy in senescence, where its role appears to vary depending on cell type and senescence triggers (Patschan et al. 2007; Young et al. 2009; Leidal et al. 2012; Mosieniak et al. 2012; Singh et al. 2012; Dörr et al. 2013). Gamerdinger et al. (2009) showed a gradual shift in the degradation pathway for polyubiquitinated proteins from the proteasome pathway to autophagy during replicative senescence in HDFs. Other studies also imply the significance of another function of autophagy as a cell survival mechanism, providing some balance between senescence and cell death upon the onset of cellular stress (Patschan et al. 2007). Indeed, some studies have shown that inhibition of autophagy facilitates senescence (Kang et al. 2011a; Wang et al. 2012), but this might be due in part to the loss of cell-protective roles of autophagy, which facilitates senescence effectors under some stressful conditions, where cells are not prone to apoptosis.
We recently showed that autophagy is activated during the dynamic phase of OIS in HDFs, which was accompanied by a concerted up-regulation of genes involved in both autophagy and lysosomal biogenesis (Young et al. 2009). The kinetics of these gene expression changes were similar to that of SASP components, implying some level of correlation between autophagy and the SASP. Indeed, inhibition of autophagy suppressed the production of both IL6 and IL8 and delayed the establishment of senescence. Our data suggest a functional link between two effector programs, autophagy and SASP, involved in senescence, although we do not exclude a more direct functional link of autophagy with OIS. How do autophagy, the bulk protein degradation process, and the SASP, which represents a process of massive protein synthesis, occur together? Interestingly, a number of studies have shown that mTOR, a master regulator of protein synthesis, also positively regulates senescence in different systems (Demidenko et al. 2009; Blagosklonny 2012; Kolesnichenko et al. 2012; Pospelova et al. 2012; Garbers et al. 2013). We recently showed that the subcellular localization of autophagy and mTOR might play a key role for the simultaneous activation of anabolic and catabolic processes during senescence (Narita et al. 2011). mTOR is a nutrient-sensitive kinase that promotes global protein translation and ribosome biogenesis and inhibits the ULK1 complex, which initiates autophagosome formation (Mizushima and Komatsu 2011). On the other hand, it has been shown that amino acid stimulation triggers mTOR recruitment to the surface of lysosomes, where mTOR is activated, in a Rag GTPase-dependent manner (Sancak et al. 2008, 2010; Bar-Peled and Sabatini 2012). Thus, mTOR inhibits the initial step of autophagy, whereas the last stage of autophagy (autolysosomes) can facilitate mTOR activation. Interestingly, OIS is associated with accumulation of autolysosome compartments, where a Rag GTPase-dependent sequestration of mTOR correlates with enhanced autophagosome formation as well as cytokine production (Narita et al. 2011). Although whether the actual kinase activity of mTOR colocalized with autolysosomes during senescence has not been confirmed, the spatial proximity of autolysosomes and mTOR (Tor autophagy spatial coupling compartment [TASCC]) may provide a mechanism for the simultaneous activation of protein degradation and “local protein synthesis” in certain contexts. Lysosomal clusters have also been associated with senescence in different contexts (Gerland et al. 2003; Dörr et al. 2013).
In addition to autophagy, the second major protein-degradative pathway—the proteasome—also has functional relevance to the phenotype. It was recently shown that aberrant Ras/ERK signaling leads to senescence-associated protein degradation (SAPD), whose targets include proteins required for cell cycle progression, cell migration, mitochondrial functions, RNA metabolism, and cell signaling (Deschênes-Simard et al. 2013). Inactivation of individual SAPD targets is sufficient for senescence induction. Thus, in addition to autophagic “bulk” degradation, “selective” proteasomal protein degradation also plays important roles in senescence.
Further evidence links the rapid protein turnover and production of secretory proteins during senescence. In a chemotherapy-induced senescence model, SASP production was associated with increased energy requirements, ER stress, an unfolded protein response, and increased autophagy activity (Dörr et al. 2013). The investigators proposed that the highly active SASP overwhelms the cellular capacity for synthesis and the processing of secretory proteins, thus leading to proteotoxic stress, which can be attenuated by simultaneous activation of autophagy in an energy-dependent manner. The data also suggest that the increase of oxygen consumption (and thus perhaps ATP production) during senescence is autophagy-dependent. Thus, therapy-induced senescent (TIS) cells are more susceptible to interruption of exogenous energy supplies or autophagy than normal cells or nonsecretory senescent cells. This study provides an additional view of the functional relationship between anabolic and catabolic processes, which are simultaneously activated during senescence. This implies that the SASP and the energy required for its production confer a potential mechanism for clearance of senescent cells independent of downstream functions of each secretory protein.
Mitochondria metabolism
In addition to the active protein metabolism described above, recent reports have shown that mitochondrial oxidative metabolism is up-regulated in senescent cells to support the metabolic demands of these cells (Dörr et al. 2013; Kaplon et al. 2013). The collaborative study from the Peeper and Gottlieb groups (Kaplon et al. 2013) has recently investigated the metabolic profile of senescent HDFs caused by oncogenic BRAFV600E. This revealed that pyruvate dehydrogenase (PDH) plays a key role in OIS. Pyruvate is the last product of glycolysis, and PDH converts pyruvate to acetyl-CoA, which enters the TCA cycle for cellular respiration. Thus, PDH lies at the junction between glycolysis (the anaerobic reaction in the cytoplasm) and the TCA cycle (the aerobic reaction within the mitochondria). PDH activity is fine-tuned by two enzymes, PDH kinases (PDKs) and PDH phosphatases (PDPs), which inhibit and activate PDH, respectively. During OIS caused by BRAFV600E, PDH is activated through suppression of PDK1 and induction of PDP2, resulting in increased TCA cycle activity and concomitant high oxygen consumption. The similar increase in the PDH activity and oxygen consumption was observed during RasG12V-induced senescence, although in this context, levels of PDK1 and PDP2 appear to be unaltered. PDH activation is not simply a consequence of OIS, since activation of PDH by the PDK1 depletion is sufficient for senescence induction in HDFs, and ectopic PDK1 expression inhibits OIS. Interestingly, PDH activation appears to be toxic even in BRAFV600E-driven melanoma cells, which have already bypassed OIS, suggesting that the PDK1–PDP2–PDH axis is not only crucial in tumor suppression but also a potential target for tumor treatment.
On the other hand, there has been a link between senescence and dysfunction of mitochondrial metabolism (Passos et al. 2007). Curiously, it has been shown that cellular levels of ATP are decreased through mitochondrial dysfunction in OIS HDFs (Moiseeva et al. 2009). In addition, in replicative senescent HDFs, a partial uncoupling of oxidative phosphorylation in mitochondria was reported; thus, even if the oxygen consumption is increased, ATP production may not be efficient (Hütter et al. 2004). Although, the Peeper and Gottlieb groups (Kaplon et al. 2013) did not report the actual levels of ATP production in OIS cells, another recent study by Dörr et al. (2013) has shown an increase of ATP production in both TIS lymphomas and OIS HDFs. They showed that glycolysis as well as the oxygen consumption rate are enhanced in these conditions and that the enhanced energy production is critical for the fitness of senescent cells, as discussed earlier in this review. The reason for the apparent discrepancy between recent and earlier studies on mitochondrial capacity for energy production is not clear. Changes in metabolic profiles in response to oncogenic or genotoxic stress might be very sensitive to the intensity and/or duration of that stress. Thus, subtle differences in experimental systems might affect single time point snapshots of the profile. In any case, all studies commonly detect up-regulation of ROS, which has been implicated as a critical mediator of senescence (Lee et al. 1999; Lu and Finkel 2008; Moiseeva et al. 2009).
How do the hyperactive TCA cycle and energy production contribute to senescence? It is conceivable that increased ROS facilitates the process. In addition, as Dörr et al. (2013) proposed, increasing cellular energy production appears to be a strategy for handling the energy consuming senescence effectors, including SASP and autophagy. Moreover, glycolysis (an anaerobic process) and the TCA cycle (an aerobic process) favor biomass synthesis and energy generation, respectively, and their balance is critical for cell proliferation. The hyperactive TCA cycle not only enhances ROS production but also compromises biomass synthesis (required for cell proliferation), such as production of lipids, nucleotides, and amino acids (Vander Heiden et al. 2009). In addition, considering recent evidence that metabolites derived from the TCA cycle, such as succinate, fumarate, and α-ketoglutarate, have profound effects on transcriptional regulation (Kaelin and McKnight 2013), it is also tempting to speculate that those metabolites or a shift in their balance may affect the phenotype through their cellular effects.
A recent study by Jiang et al. (2013) has shown that perturbation of malic enzymes (MEs), which are associated with the TCA cycle, modulates senescence. MEs are important for NADPH production, which is required for biomass synthesis, such as lipid production. They showed that MEs are repressive targets of p53 and that depletion of MEs reciprocally activates p53, thus inducing p53-dependent senescence in HDFs, while their ectopic expression delays replicative senescence. Although the consequences of ME depletion appear to be cell type-dependent (Jiang et al. 2013), it is possible that depletion of MEs diverts malate metabolism to producing NADH, which is the main electron donor for the electron transport chain that drives ATP production in mitochondria. In addition, NADPH is a reducing agent; thus, depletion of MEs would also enhance oxidative stress. Indeed, Jiang et al. (2013) showed an increased ROS production upon knockdown of MEs (ME2 in particular) in HDFs. It would be interesting to know whether the hyperactive mitochondrial respiration and reciprocal regulation of p53 and ME are functionally connected (Fig. 2).
Senescence stability
Senescence has previously been defined using terms such as “permanent arrest” or “irreversible cell cycle exit.” However, a frequently asked question about senescence is: “Is senescence really irreversible?” In culture, quiescent cells can readily resume their cell cycle upon release from inhibitory effects, such as low serum or contact inhibition; senescent cells are refractory to similar treatments (Seshadri and Campisi 1989, 1990; Serrano et al. 1997). The lack of response to growth factor stimulation in particular is reinforced by the stable arrest of OIS cells, where signaling downstream from growth factor stimuli is constitutively active. This appears to be true in vivo, exemplified by melanocytic nevi that frequently harbor the oncogenic BRAFV600E mutation.
However, efforts to rigorously investigate the stability of senescence have revealed that the stability of cell cycle arrest also varies depending on the cellular context. It has been shown that replicative senescence in BJ skin fibroblasts can be reversed upon p53 depletion (Beauséjour et al. 2003). This is consistent with the view that persistent DDR contributes to the maintenance of the senescent phenotype primarily through the p53 pathway (d'Adda di Fagagna 2008). Compared with senescent BJ cells, which tend to have relatively low expression of p16, attempts to reverse senescence in WI38 lung embryonic fibroblasts in which the p16–Rb pathway is intact are less successful (Beauséjour et al. 2003). Interestingly, perhaps due to the different activity in the p16–Rb pathway, WI38 cells are also more SAHF-competent. As mentioned earlier, SAHF formation is largely dependent on the p16–Rb pathway but not on the p53 pathway in OIS HDFs (Narita et al. 2003). If the speculation above is correct that the SAHF-like segregation of chromatin, due to its epigenetic marks, is associated with efficient maintenance of the gene expression profile (for both constitutively expressed and repressed), SAHFs, at least in part, might contribute to the refractoriness of senescence arrest to the removal of the persistent p53 signal.
Another insight into the stability of senescence arrest was derived from the field of cell reprogramming. In 2009, six studies showed that the p53 and INK4A pathways are activated during generation of induced pluripotent stem (iPS) cells using the classical four factors OCT4, SOX2, KLF4, and c-MYC (or three factors without c-MYC). Collectively, they suggest that senescence can be a barrier to the reprogramming to generate iPS cells from fibroblasts (Banito et al. 2009; Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marión et al. 2009; Utikal et al. 2009). More recently, however, iPS cells were successfully generated from replicatively senescent HDFs at a rate comparable with their proliferative counterparts (Lapasset et al. 2011). For this, two other key regulators of embryonic stem cell identity, NANOG and LIN28, were added to the classical four; therefore, senescent cells can be reprogrammed if conditions are optimal. Interestingly, the investigators claim that, during the reprogramming of senescent cells, the disappearance of SAHFs preceded the restoration of proliferation, supporting a role for SAHFs in the maintenance of senescence arrest.
Senescence in vivo
OIS has been intensively studied in culture systems since it was first described in detail (Serrano et al. 1997). In 2005, a series of studies identified OIS and its related phenotype, loss of tumor suppresser-induced senescence (as exemplified by PTEN loss-induced cellular senescence [PICS]) (Alimonti et al. 2010) in several murine and human premalignant lesions (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005; Lazzerini Denchi et al. 2005; Michaloglou et al. 2005), reinforcing the tumor-suppressive role of senescence (Narita and Lowe 2005). However, as the diverse activities of the SASP illustrate, the role of senescence on tumorigenesis needs careful evaluation based on the net effect on the tumor microenvironment as a whole. The impact of senescent cells on the microenvironment has also been shown in the context of tissue injury and repair (see “Functions of the SASP”) (Krizhanovsky et al. 2008; Jun and Lau 2010; Kim et al. 2013).
Replicative senescence, the prototype of the phenotype, involves “time”; thus, it is often referred to as “cell aging.” It is not surprising that senescence has long been suspected to play a role in both organismal aging and declining organ function with age (Rudolph et al. 2000; Cristofalo et al. 2004). Indeed, senescent markers, including SA-β-Gal, DDR, telomere dysfunction, and p16, have been detected in various tissues from old individuals (Dimri et al. 1995; Krishnamurthy et al. 2004; Herbig et al. 2006; Jeyapalan et al. 2007; Flores et al. 2008) and age-related diseased organs in mammals (Minamino et al. 2002; Price et al. 2002). Such age-dependent up-regulation of p16 was also confirmed using p16 reporter mouse models (Yamakoshi et al. 2009; Burd et al. 2013). But does senescence play a causative role in aging? A series of studies in 2006 touched on this important question (Janzen et al. 2006; Krishnamurthy et al. 2006; Molofsky et al. 2006). They showed that p16 is up-regulated in tissue stem cell/progenitor compartments such as hematopoietic stem cells (HSCs), pancreatic islet cells, and neuronal stem cells in an age-dependent manner and thus constrains their regenerative capacity. p16 is a CDK inhibitor (CDKI) and thus inhibits cell cycle progression. Compared with other CDKIs, it is very specific for senescence and probably is one of the best “functional” biomarkers of senescence. Indeed, they showed that genetic ablation of p16 restores the decline of the regenerative capacity in those tissue stem cell/progenitor cells to some extent. These studies support the causative effect of senescence for aging, at least at an organ level.
Consistently, Baker et al. (2008) have shown that p16 deficiency attenuates aging phenotypes in a subset of tissues where p16 is activated in BubR1 hypomorphic progeroid mice. Instead of delaying senescence by depleting p16, the same group also took a different strategy through elimination of p16-positive senescent cells. They generated an elegant mouse model in which p16-positive senescent cells can be inducibly and selectively killed (Baker et al. 2011). In the same progerioid mice, lifelong or even late-life elimination of p16-expressing senescent cells delays age-associated phenotypes in those p16-sensitive tissues. Of note, Baker et al. (2008, 2011) showed that, in contrast to the case of p16 ablation, which accelerates lung tumor development in the BubR1 hypomorphic background, clearance of p16-expressing senescent cells does not appear to show obvious side effects. Thus, clearance of senescent cells represents a promising approach for anti-aging therapy. However, at the same time, these studies also revealed the existence of p16-independent age-associated phenotypes, at least in the BubR1 hypomorphic progeroid model.
The beneficial effect of elimination of p16-expressing senescent cells is somehow reminiscent of the phenotype of super-Arf/p53 (s-Arf/p53) mice, which harbor normally regulated single extra gene doses of Arf and p53 (Matheu et al. 2007). The average life span of s-Arf/p53 mice is significantly longer than control wild-type mice, although the maximum life span is similar. This anti-aging effect of additional Arf/p53 seems to be independent of their tumor suppressor effects. The precise mechanism for this anti-aging effect is not entirely clear. As the investigators propose, some p53 target genes under physiological conditions have a global anti-oxidant effect, which helps in part attenuating aging (Matheu et al. 2007). It is tempting to speculate that the extra doses of these tumor suppressors might also contribute to efficient clearance of senescent cells.
Concluding remarks
Recent advances in our understanding of senescence suggest that what once was thought to be an endpoint of stress response has a functional relevance in various pathophysiological contexts, including aging and age-associated diseases, inflammation, and tissue repair. In addition, identification and characterization of senescence effector programs such as the DDR, the mTOR/autophagy signal network, the protein secretory program, and epigenetic gene regulation, which are shared with other physiological contexts, have been promoting interdisciplinary approaches to tackle this complex phenotype. A challenge is to understand senescence as a high-order network of these different pathways and molecules. The diversity in the quality of the phenotype according to the combination of the effector mechanisms and biomarkers may seem to make the definition of senescence obscure. Unfortunately, there is no such thing as a “senescence score.” The recent approach using “high content analysis” to study senescence might improve our skill for diagnosing senescence, at least in vitro (Acosta et al. 2013). Further understanding of the “senescence network” will be required to obtain more systematic and quantitative tools for senescence diagnosis. Such advances in understanding senescence would also extend our opportunity to modulate this complex phenotype in various pathophysiological contexts.
Acknowledgments
We thank C. Frezza for critical reading and helpful discussions, and members of the Narita laboratory for general discussions. This work was supported by the University of Cambridge, Cancer Research UK, Hutchison Whampoa, the Human Frontier Science Program (M.N. and R.S.), and Cancer Research UK Clinician Scientist Fellowship (M.H.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.235184.113.
References
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018 [DOI] [PubMed] [Google Scholar]
- Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang T-W, Lasitschka F, Andrulis M, et al. 2013. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol 15: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD 2007. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene 397: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Cloos PAC, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K 2009. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23: 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, Cheng K, Varmeh S, Kozma SC, Thomas G, et al. 2010. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest 120: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederländer NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, et al. 2008. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barascu A, Le Chalony C, Pennarun GEL, Genet D, Imam N, Lopez B, Bertrand P 2012. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. EMBO J 31: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM 2012. SnapShot: mTORC1 signaling at the lysosomal surface. Cell 151: 1390–1390.e1 [DOI] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou M-M, Walsh MJ, et al. 2009. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23: 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou L-VF, Kolettas E, Niforou K, Zoumpourlis VC, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bavik C, Coleman I, Dean JP, Knudsen B, Plymate S, Nelson PS 2006. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res 66: 794–802 [DOI] [PubMed] [Google Scholar]
- Beauséjour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J 2003. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J 22: 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O 2012. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14: 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Kirsh O, Pearson M, Itahana K, Pelicci PG, Dejean A 2002. Deconstructing PML-induced premature senescence. EMBO J 21: 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Nacerddine K, Dejean A 2005. Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol Cell Biol 25: 1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blagosklonny MV 2012. Cell cycle arrest is not yet senescence, which is not just cell cycle arrest: Terminology for TOR-driven aging. Aging 4: 159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AHFM, Schlegelberger B, Stein H, Dörken B, Jenuwein T, Schmitt CA 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, Bardeesy N, Castrillon DH, Beach DH, Sharpless NE 2013. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell 152: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J 2005. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F 2007. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Capparelli C, Chiavarina B, Whitaker-Menezes D, Pestell TG, Pestell RG, Hulit J, Andò S, Howell A, Martinez-Outschoorn UE, Sotgia F, et al. 2012. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, ‘fueling’ tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle 11: 3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro P, Giri D, Lamb D, Ittmann M 2003. Cellular senescence in the pathogenesis of benign prostatic hyperplasia. Prostate 55: 30–38 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Karlseder J 2012. A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol 24: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Narita M 2013. High-order chromatin structure and the epigenome in SAHFs. Nucleus 4: 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret J-Y, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. 2012. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin H-K, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, Mccurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, et al. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25: 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuprin A, Gal H, Biron-Shental T, Biran A, Amiel A, Rozenblatt S, Krizhanovsky V 2013. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev 27: 2356–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: Senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M 2007. Cellular senescence in cancer and aging. Cell 130: 223–233 [DOI] [PubMed] [Google Scholar]
- Coppé J-P, Kauser K, Campisi J, Beauséjour CM 2006. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem 281: 29568–29574 [DOI] [PubMed] [Google Scholar]
- Coppé J, Patil C, Rodier F, Sun Y, Muñoz D, Goldstein J, Nelson P, Desprez P, Campisi J 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J-P, Desprez P-Y, Krtolica A, Campisi J 2010. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu Rev Pathol 5: 99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M 2004. Replicative senescence: A critical review. Mech Ageing Dev 125: 827–848 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F 2008. Living on a break: Cellular senescence as a DNA-damage response. Nat Rev Cancer 8: 512–522 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD 2010. Nuclear lamins. Cold Spring Harb Perspect Biol 2: a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV 2009. Rapamycin decelerates cellular senescence. Cell Cycle 8: 1888–1895 [DOI] [PubMed] [Google Scholar]
- Deschênes-Simard X, Gaumont-Leclerc M-F, Bourdeau V, Lessard F, Moiseeva O, Forest V, Igelmann S, Mallette FA, Saba-El-Leil MK, Meloche S, et al. 2013. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Genes Dev 27: 900–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, Zuffo RD, Matti V, d'Ario G, Montani E, et al. 2011. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol 13: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Däbritz JHM, Lisec J, Lenze D, Gerhardt A, Schleicher K, et al. 2013. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501: 421–425 [DOI] [PubMed] [Google Scholar]
- Dreesen O, Chojnowski A, Ong PF, Zhao TY, Common JE, Lunny D, Lane EB, Lee SJ, Vardy LA, Stewart CL, et al. 2013. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol 200: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev 14: 2015–2027 [PMC free article] [PubMed] [Google Scholar]
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA 2008. The longest telomeres: A general signature of adult stem cell compartments. Genes Dev 22: 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Patil CK, Campisi J 2011. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J 30: 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Laberge RM, Demaria M, Campisi J 2012. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell 23: 2066–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beauséjour CM, et al. 2012. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 14: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama R, Saito M, Tanobe H, Ishikawa F 2006. Loss of linker histone H1 in cellular senescence. J Cell Biol 175: 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C 2009. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers C, Kuck F, Aparicio-Siegmund S, Konzak K, Kessenbrock M, Sommerfeld A, Häussinger D, Lang PA, Brenner D, Mak TW, et al. 2013. Cellular senescence or EGFR signaling induces interleukin 6 (IL-6) receptor expression controlled by mammalian target of rapamycin (mTOR). Cell Cycle 12: 3421–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerland L-M, Peyrol S, Lallemand C, Branche R, Magaud J-P, Ffrench M 2003. Association of increased autophagic inclusions labeled for β-galactosidase with fibroblastic aging. Exp Gerontol 38: 887–895 [DOI] [PubMed] [Google Scholar]
- Goldstein S, Moerman EJ, Fujii S, Sobel BE 1994. Overexpression of plasminogen activator inhibitor type-1 in senescent fibroblasts from normal subjects and those with Werner syndrome. J Cell Physiol 161: 571–579 [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460 [DOI] [PubMed] [Google Scholar]
- Hayflick L 1965. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BPC, Chen DJ, Sedivy JM 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14: 501–513 [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM 2006. Cellular senescence in aging primates. Science 311: 1257. [DOI] [PubMed] [Google Scholar]
- Hewitt G, Jurk D, Marques FDM, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF 2012. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 3: 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare M, Narita M 2013. Transmitting senescence to the cell neighbourhood. Nat Cell Biol 15: 887–889 [DOI] [PubMed] [Google Scholar]
- Hoare M, Young ARJ, Narita M 2011. Autophagy in cancer: Having your cake and eating it. Semin Cancer Biol 21: 397–404 [DOI] [PubMed] [Google Scholar]
- Hoenicke L, Zender L 2012. Immune surveillance of senescent cells—biological significance in cancer- and non-cancer pathologies. Carcinogenesis 33: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubackova S, Krejcikova K, Bartek J, Hodny Z 2012. IL1- and TGFβ-Nox4 signaling, oxidative stress and DNA damage response are shared features of replicative, oncogene-induced, and drug-induced paracrine ‘Bystander senescence.’ Aging 4: 932–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CJ 2012. B-type lamins and their elusive roles in metazoan cell proliferation and senescence. EMBO J 31: 1058–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter E, Renner K, Pfister G, Stöckl P, Jansen-Dürr P, Gnaiger E 2004. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. Biochem J 380: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH 2013. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med 210: 2057–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443: 421–426 [DOI] [PubMed] [Google Scholar]
- Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U 2007. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev 128: 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X 2013. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Kase J, Dorr JR, Milanovic M, Lenze D, Grau M, Beuster G, Ji S, Reimann M, Lenz P, et al. 2011. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev 25: 2137–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Kipling D, Morris M, Hepburn P, Skinner J, Bounacer A, Wyllie FS, Ivan M, Bartek J, Wynford-Thomas D, et al. 2000. Evidence for a telomere-independent ‘clock’ limiting RAS oncogene-driven proliferation of human thyroid epithelial cells. Mol Cell Biol 20: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J-I, Lau LF 2010. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol 12: 676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, McKnight SL 2013. Influence of metabolism on epigenetics and disease. Cell 153: 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HT, Lee KB, Kim SY, Choi HR, Park S-C 2011a. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS ONE 6: e23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T-W, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. 2011b. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479: 547–551 [DOI] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EME, Cascante M, Shlomi T, et al. 2013. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498: 109–112 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T 2002. Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JCI 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-H, Chen C-C, Monzon RI, Lau LF 2013. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol Cell Biol 33: 2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnichenko M, Hong L, Liao R, Vogt PK, Sun P 2012. Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell Cycle 11: 2391–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernards R 2006. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol 8: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE 2004. Ink4a/Arf expression is a biomarker of aging. J Clin Invest 114: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE 2006. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443: 453–457 [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW 2008. Senescence of activated stellate cells limits liver fibrosis. Cell 134: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc Natl Acad Sci 98: 12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS 2009. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer 9: 81–94 [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LCW, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS 2010. The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez J-M, De Vos J, et al. 2011. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev 25: 2248–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Attwooll C, Pasini D, Helin K 2005. Deregulated E2F activity induces hyperplasia and senescence-like features in the mouse pituitary gland. Mol Cell Biol 25: 2660–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T 1999. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem 274: 7936–7940 [DOI] [PubMed] [Google Scholar]
- Leidal AM, Cyr DP, Hill RJ, Lee PWK, McCormick C 2012. Subversion of autophagy by Kaposi's sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe 11: 167–180 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Finkel T 2008. Free radicals and senescence. Exp Cell Res 314: 1918–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, et al. 2013. Non-cell-autonomous tumor suppression by p53. Cell 153: 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc M-F, Ferbeyre G 2007. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev 21: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Viña J, Blasco MA, Serrano M 2007. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448: 375–379 [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LCW, Soengas MS, Denoyelle C, Kuilman T, van der Horst CMAM, Majoor DM, Shay JW, Mooi WJ, Peeper DS 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724 [DOI] [PubMed] [Google Scholar]
- Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I 2002. Endothelial cell senescence in human atherosclerosis: Role of telomere in endothelial dysfunction. Circulation 105: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M 2011. Autophagy: Renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Moiseeva O, Bourdeau V, Roux A, Deschênes-Simard X, Ferbeyre G 2009. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol 29: 4495–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ 2006. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosieniak G, Adamowicz M, Alster O, Jaskowiak H, Szczepankiewicz AA, Wilczynski GM, Ciechomska IA, Sikora E 2012. Curcumin induces permanent growth arrest of human colon cancer cells: Link between senescence and autophagy. Mech Ageing Dev 133: 444–455 [DOI] [PubMed] [Google Scholar]
- Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, Rodríguez-Baeza A, Varela-Nieto I, Ruberte J, Collado M, et al. 2013. Programmed cell senescence during mammalian embryonic development. Cell 155: 1104–1118 [DOI] [PubMed] [Google Scholar]
- Murano S, Thweatt R, Shmookler Reis RJ, Jones RA, Moerman EJ, Goldstein S 1991. Diverse gene sequences are overexpressed in werner syndrome fibroblasts undergoing premature replicative senescence. Mol Cell Biol 11: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Lowe SW 2005. Senescence comes of age. Nat Med 11: 920–922 [DOI] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nũnez S, Chicas A, Hearn SA, Myers MP, Lowe SW 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126: 503–514 [DOI] [PubMed] [Google Scholar]
- Narita M, Young ARJ, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. 2011. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J 2009. Cell surface-bound IL-1α is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci 106: 17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Parrinello S, Coppé J-P, Krtolica A, Campisi J 2005. Stromal-epithelial interactions in aging and cancer: Senescent fibroblasts alter epithelial cell differentiation. J Cell Sci 118: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, von Zglinicki T 2007. DNA damage in telomeres and mitochondria during cellular senescence: Is there a connection? Nucleic Acids Res 35: 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, Miwa S, Olijslagers S, Hallinan J, Wipat A, et al. 2010. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol 6: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patschan S, Chen J, Polotskaia A, Mendelev N, Cheng J, Patschan D, Goligorsky MS 2007. Lipid mediators of autophagy in stress-induced premature senescence of endothelial cells. Am J Physiol Heart Circ Physiol 294: H1119–H1129 [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, et al. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207–210 [DOI] [PubMed] [Google Scholar]
- Pospelova TV, Leontieva OV, Bykova TV, Zubova SG, Pospelov VA, Blagosklonny MV 2012. Suppression of replicative senescence by rapamycin in rodent embryonic cells. Cell Cycle 11: 2402–2407 [DOI] [PubMed] [Google Scholar]
- Price JS, Waters JG, Darrah C, Pennington C, Edwards DR, Donell ST, Clark IM 2002. The role of chondrocyte senescence in osteoarthritis. Aging Cell 1: 57–65 [DOI] [PubMed] [Google Scholar]
- Rodier F, Coppé J-P, Patil CK, Hoeijmakers WAM, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J 2009. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 11: 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Muñoz DP, Teachenor R, Chu V, Le O, Bhaumik D, Coppé J-P, Campeau E, Beauséjour CM, Kim S-H, et al. 2011. DNA-SCARS: Distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci 124: 68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Millard M, Schreiber-Agus N, DePinho RA 2000. Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science 287: 1253–1258 [DOI] [PubMed] [Google Scholar]
- Sadaie M, Salama R, Carroll T, Tomimatsu K, Chandra T, Young ARJ, Narita M, Perez-Mancera PA, Bennett DC, Chong H, et al. 2013. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27: 1800–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Yokoyama M, Zhang X, Prakash K, Nagao K, Hatanaka T, Getzenberg RH, Kakehi Y 2004. Increased expression of CYR61, an extracellular matrix signaling protein, in human benign prostatic hyperplasia and its regulation by lysophosphatidic acid. Endocrinology 145: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y 2010. Autophagy mediates the process of cellular senescence characterizing bile duct damages in primary biliary cirrhosis. Lab Invest 90: 835–843 [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS 2012. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 48: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M 2011. Cancer: Final act of senescence. Nature 479: 481–482 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Seshadri T, Campisi J 1989. Growth-factor-inducible gene expression in senescent human fibroblasts. Exp Gerontol 24: 515–522 [DOI] [PubMed] [Google Scholar]
- Seshadri T, Campisi J 1990. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science 247: 205–209 [DOI] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE 2000. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol 1: 72–76 [DOI] [PubMed] [Google Scholar]
- Shelton DN, Chang E, Whittier PS, Choi D, Funk WD 1999. Microarray analysis of replicative senescence. Curr Biol 9: 939–945 [DOI] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev 25: 2579–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Matsuyama S, Drazba JA, Almasan A 2012. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy 8: 236–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, Yosef R, Pilpel N, Krizhanovsky V, Sharpe J, et al. 2013. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R 2012. Inflammasomes in health and disease. Nature 481: 278–286 [DOI] [PubMed] [Google Scholar]
- Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL, et al. 2012. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J 31: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T 2003. DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O 2008. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev 22: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G 2011. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev 25: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang XD, Lapi E, Sullivan A, Jia W, He Y-W, Ratnayaka I, Zhong S, Goldin RD, Goemans CG, et al. 2012. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci 109: 13325–13330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi K, Takahashi A, Hirota F, Nakayama R, Ishimaru N, Kubo Y, Mann DJ, Ohmura M, Hirao A, Saya H, et al. 2009. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J Cell Biol 186: 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Kennedy A, Banumathy G, Zhang R, Adams PD 2007a. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell 27: 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD 2007b. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499: 97–101 [DOI] [PubMed] [Google Scholar]
- Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JFJ, Tavaré S, Arakawa S, Shimizu S, Watt FM, et al. 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23: 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Narita M, Narita M 2013. Cell senescence as both a dynamic and a static phenotype. Methods Mol Biol 965: 1–13 [DOI] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30 [DOI] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]