Summary
Nearly all animals reproduce sexually through the production and fusion of sperm and egg cells, yet little is known about the ancestry of animal sexual reproduction. Moreover, the sexual cycle of the closest living relatives of animals, the choanoflagellates [1, 2], remains completely unknown. The choanoflagellate Monosiga brevicollis, possesses a “meiotic toolkit” of genes [3], but the lack of polymorphisms detected during genome sequencing precluded inferences about its ploidy or sexual cycle [1]. Here we report that a related choanoflagellate, Salpingoeca rosetta [4, 5], has a sexual life cycle and transitions between haploid and diploid states. Haploid cultures of S. rosetta became diploid in response to nutrient limitation. This ploidy shift coincided with anisogamous mating, during which small, flagellated cells fused with larger flagellated cells. Distributions of polymorphisms in laboratory strains of S. rosetta provided independent evidence of historical recombination and mating. The ability of S. rosetta to produce morphologically differentiated gametes and to engage in sexual reproduction has implications both for reconstructing the evolution of sex in the progenitors of animals and for establishing classical genetics in choanoflagellates.
Results and Discussion
Discovery of cryptic differences in ploidy among laboratory cultures of S. rosetta
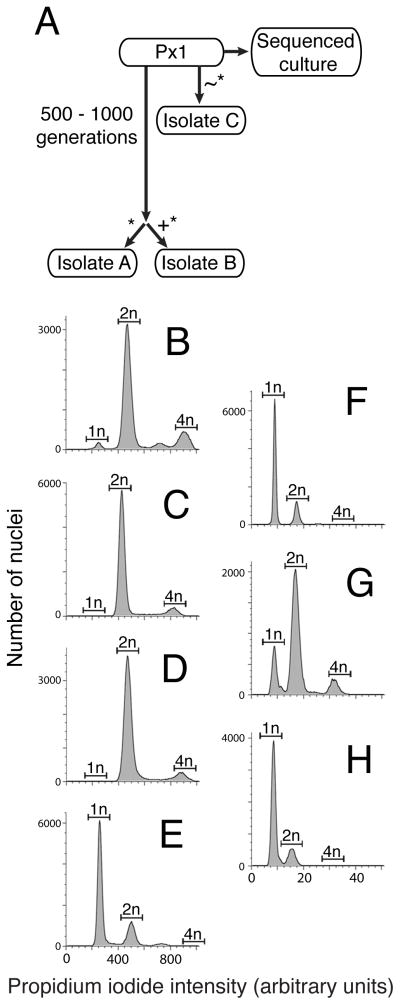
We investigated the ploidy of a laboratory culture of S. rosetta called Px1 [6, 7] and three derived laboratory strains, isolates A – C (Fig. 1A), using flow cytometry on propidium iodide-stained S. rosetta nuclei [8]. S. rosetta isolates A and B were diploid (Fig. 1C and D, Fig. S1A – D), with 2n and 4n peaks representing cells in the G1 and G2 stages of the cell cycle, respectively. In contrast, isolate C was haploid (Fig. 1E), and Px1 produced a mix of diploid- and haploid-sized peaks, in addition to a small 3n-sized peak (Fig. 1B). Because each of the tested isolates was derived from Px1 (Fig. 1A), we inferred that S. rosetta may be capable of switching ploidy under laboratory conditions, perhaps as part of a sexual life cycle. Nonetheless, after three months under standard culturing conditions, isolate C remained haploid and isolates A and B remained diploid (Fig. S1E – H).
Figure 1. S. rosetta cultures transition between haploid and diploid states in response to changing nutrient availability.
(A) The S. rosetta cultures used in this study were all derived from a Px1 culture [6] through clonal isolation (*), during which one cell was isolated and propagated to establish the new culture line. For isolate A, a Px1 culture was propagated for 500–1000 generations before the new culture was established through clonal isolation. For isolate B, clonal isolation occurred after EMS mutagenesis (+). For isolate C, clonal isolation occurred after replacing the feeder bacteria from Px1 with a new species, Echinicola pacifica (~). During preparation for genome sequencing, Px1 was passaged rapidly, which may have resulted in changes in ploidy (see Supplemental Experimental Methods). (B) – (E) S. rosetta laboratory cultures can be haploid, diploid, or have a mix of haploid and diploid cells, as revealed by flow cytometry of propidium iodide-stained nuclei from unsynchronized cultures of Px1 (B), isolate A (C), isolate B (D), and isolate C (E). The expected locations of the 1n, 2n, and 4n peaks were based on an internal standard of S. cerevisiae (Fig. S1A – D). (F) – (H) Nutrient limitation induces haploid cultures to become diploid. Flow cytometry showed that isolate C was haploid after growth in HN medium for six days (F). Following growth of isolate C in unenriched sea water for six days, most cells were diploid (G). Transferring diploid isolate C cells back to HN medium for three days resulted in a return to the haploid state (H). Haploid-to-diploid transitions under nutrient limitation occurred within clonal cultures and did not require mixing of multiple clones. See also Figure S1.
Altered nutrient availability triggers changes in S. rosetta ploidy
Because sex and meiosis in many unicellular eukaryotes are triggered by changes in nutrient availability [9–12], we next tested the influence of different culture media on S. rosetta ploidy. A haploid isolate C culture that had been maintained in high-nutrient (HN) media was divided in two, with one half continuing in HN media while the other half was transferred to unenriched sea water. After 6 days, isolate C cells grown in HN media remained haploid (Fig. 1F), whereas approximately 89% of isolate C cells cultured in unenriched sea water were diploid (Fig. 1G). The change in ploidy was reversible; isolate C diploid cells transferred from unenriched sea water back into HN media returned to a haploid state within 3 days (Fig. 1H, Experimental Procedures).
Cultures of S. rosetta fed only A. machipongonensis bacteria (i.e. isolate A, B, and Px1) grew poorly in HN media for unknown reasons. Still, we could induce isolate B to be largely haploid after daily passaging in cereal grass (CG) medium for 3 weeks, which provided a continual replenishment of nutrients in the culture (Fig. S1I). We hypothesize that the largely stable differences in ploidy between isolates A, B, and C were due to the relative levels of media richness and/or nutrient availability in the different cultures. Thus, we infer that nutrient limitation induces S. rosetta haploid populations to become diploid, perhaps through mating, and nutrient rich conditions promote a transition from diploidy to haploidy, potentially through meiosis.
Patterns of polymorphism reveal a history of sex and recombination
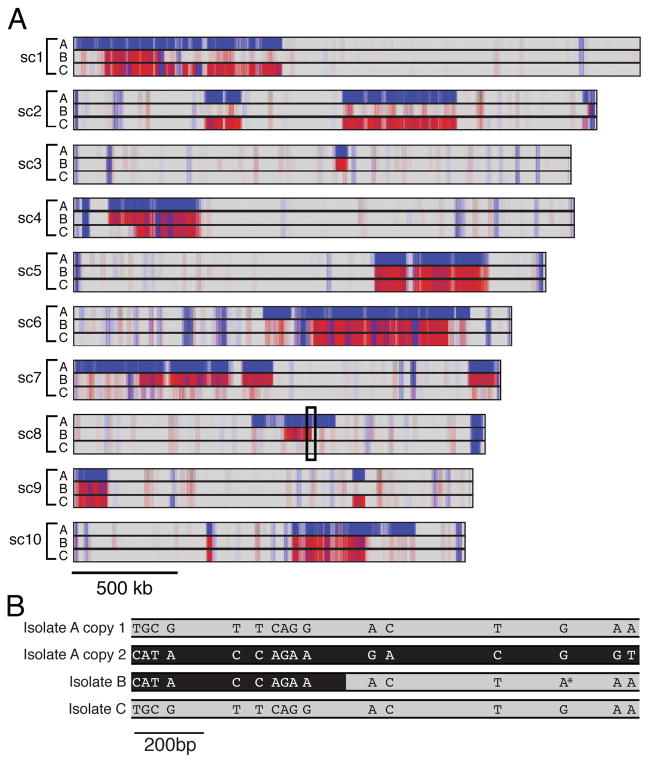
If S. rosetta has a sexual cycle, we would expect to find signatures of historical, meiotic recombination throughout the genome. In the absence of recombination, single nucleotide polymorphisms (SNPs) are inherited in haplotype blocks that span the length of each chromosome, whereas recombination can break up haplotype blocks into smaller chromosomal regions [13–16]. We found that SNPs in isolates A, B, and C were broken up into discrete haplotype blocks on 33 of the 40 largest supercontigs (which range in size from 0.4 to 2.6 Mb; Fig. 2A). The presence of haplotype blocks on the majority of large supercontigs suggested a history of genome-wide recombination generated during meiosis. The sharp boundaries at the edges of the haplotype blocks marked the chromosomal regions where we infer there has been genetic exchange between homologous chromosomes. Most SNPs were also shared among the isolates (Fig. 2A and B), suggesting that these polymorphisms are segregating in laboratory populations of S. rosetta. Furthermore, by interrogating the physical linkage of SNPs in these strains, we observed direct evidence of recent recombination in S. rosetta that has resulted in novel combinations of the segregating alleles (Fig. 2B, Fig S2A).
Figure 2. Patterns of polymorphism reveal a history of sex and recombination in S. rosetta.
(A) Mapping of SNP positions in isolates A, B, and C on the ten largest supercontigs suggests a history of recombination in S. rosetta. Sites that match the reference genome sequence are indicated in grey, homozygous SNPs in red and heterozygous SNPs in blue. Most SNPs are concentrated in large, contiguous haplotype blocks with sharp cut-offs that mark inferred sites of historical recombination. (B) SNPs in segregating haplotype blocks are shared among isolates. The boxed region of supercontig 8 indicated in (A) was cloned and sequenced from isolates A, B, and C to reveal the physical linkage among SNPs (see Fig. S2A for full alignment). Two distinct sequences were obtained from the heterozygous diploid isolate A; copy 1 (grey) matched isolate C and the reference genome, whereas copy 2 (black) contained many SNPs relative to the reference genome. In isolate B, the 5′ end of the region shares the copy 2 (black) haplotype, while the 3′ end is more similar to the reference sequence (grey). The transition from black to grey in isolate B indicates the inferred region of recombination. Isolate B also has a single SNP (*) that was not detected in any other sequenced sample and likely arose within isolate B. See also Figure S2.
In addition to offering evidence for historical recombination, the segregating polymorphisms in diploid isolates A and B yielded insights into S. rosetta sex. We found that 84% of the SNPs in isolate B were homozygous, suggesting that this strain became diploid either by self-fertilization or through auto-diploidization (Fig. 2A and B, Fig S2B). In isolate A, however, 93% of the SNPs were heterozygous and this strain had nonidentical homologous chromosomes, suggesting that isolate A arose through the mating (i.e. fusion) of two genetically distinct individuals. Thus, we propose that S. rosetta isolates A and B have experienced a history of sex and meiosis, which involved the generation of diploids (either outcrossed or selfed), recombination between genetically distinct chromosomes, and segregation of recombinant chromosomes into different cells.
Morphological evidence for sex during haploid-to-diploid transitions
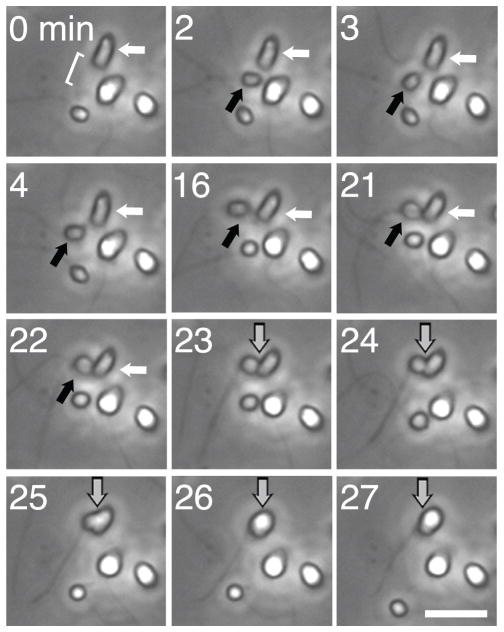
To investigate the cell morphology and behavior of putative S. rosetta gametes, we monitored cultures of S. rosetta during the haploid-to-diploid transition. Twenty-four hours after transferring haploid isolate C cells into unenriched sea water to induce the switch to diploidy, we monitored the culture using time lapse microscopy for 10 hours. We observed approximately 1,200 total cells and recorded 6 cell fusion events, indicative of mating (Fig. 3, Movie S1). All mating events involved a small, rounded, uniflagellated cell (provisionally termed the ‘male gamete’) fusing with and entering a larger, ovoid, uniflagellated cell (the ‘female gamete’). For approximately 10 to 30 minutes prior to fusion, the male gamete cell body contacted the microvillar collar of the female gamete, possibly as part of a recognition process. We also observed many examples of adhesion between cells that did not result in fusion, and adherent clumps of cells often persisted over a period of minutes to hours. Successful cell fusions were always initiated by contact between the basal end of the male gamete (opposite the flagellum) and the base of the collar of the female gamete. Each cell fusion event we observed lasted for 5 to 8 minutes and resulted in a slightly larger, more rounded cell.
Figure 3. Cell differentiation and cell fusion during haploid-to-diploid transitions.
Time-lapse microscopy shows the fusion of morphologically distinct cells in an isolate C culture following 24 hours of nutrient limitation. A small, rounded cell (the ‘male gamete’, black arrow) entered the field of view (minute 2) and adhered to a larger, ovoid cell (‘female gamete’, white arrow; minutes 3–22) before cell fusion (gray arrow; minutes 23–27). The bracket in the first panel indicates the location of the microvillar collar of the female gamete. Cells were imaged once per minute using 40x phase contrast microscopy. Scale bar is 10 μm. See also Figure S3 and Movie S1.
Immunofluorescence microscopy of cultures undergoing the haploid-to-diploid transition provided further support for a sexual cycle in S. rosetta. Isolate C cells grown rapidly in HN media (Fig. S3A), tended to have a rounded cell body and a doughnut-shaped nucleoplasm. In contrast, isolate C cells in unenriched sea water were often found in disorganized, adherent clumps consisting of both male and female gamete types (Fig. S3B). Contacts between adherent cells were through actin-based microvillar collars and/or filopodia. Strikingly, we also observed a bridge of DNA between the nuclei of adherent male and female gametes (Fig. S3B), consistent with a process of genetic transfer.
Choanoflagellates feed by phagocytosing bacterial prey at the base of their collars, but several pieces of evidence indicate that S. rosetta cell fusion is sex rather than cannibalism. First, the male gamete appears to take on a consistent orientation prior to fusion, which would not be expected if it were merely prey. Second, starvation induced changes in nuclear morphology that were suggestive of genetic transfer (Fig. S3B). Third, fusion was preceded by a long period of cell contact, and only a minority of male-female encounters resulted in cell fusion. This may indicate that signaling between the gametes or another form of recognition might be required for cell fusion, rather than indiscriminate engulfment. Finally, cell fusion occurred as haploid populations were becoming diploid, as would be expected in the case of sex.
Conclusions and implications
We conclude that the choanoflagellate S. rosetta has a sexual phase to its life cycle. Haploid-to-diploid transitions (i.e. sex) occurred in nutrient limiting conditions, while diploid-to-haploid transitions (presumably meiosis) occurred in nutrient rich conditions. Although this sexual response is the reverse of the meiotic response of S. cerevisiae to nutrient limitation [10], diverse other unicellular eukaryotes respond to nutrient limitation through sexual differentiation and fusion [9, 11]. During the S. rosetta haploid-to-diploid transition, we observed the fusion of previously undocumented, morphologically differentiated cell types that we infer are anisogamous gametes. As both cell fusion and the haploid-to-diploid transitions occurred within clonal cultures (without mixing different isolates together), we conclude that S. rosetta is homothallic (i.e. that it can self-fertilize). This is consistent with the finding that diploid isolate B has genome-wide homozygosity. However, diploid isolate A has genome-wide heterozygosity, implying that S. rosetta can also undergo outcrossed mating, which may be harnessed in the future for crosses and genetics in choanoflagellates.
One implication of the homothallism of S. rosetta is that a single genotype can produce both male and female gametes. This suggests that mating type is not permanent and instead that a sort of mating type switching may occur, possibly through sex chromosome loss or epigenetic mechanisms. Understanding how S. rosetta gamete identity is specified may help to illuminate the ancestry of animal gametogenesis.
Although sexual systems have diversified extensively within the fungal clade [17], sex in animals has remained relatively consistent [18] suggesting that choanoflagellates could shed light on ancestral sexual systems in Opisthokonta, the clade that includes animals, choanoflagellates, and fungi. Morphologically, the fusion of flagellated, anisogamous gametes in choanoflagellates bears a striking resemblance to the flagellated, anisogamous gametes of Allomyces macrogynus, a basal-branching fungal species [19, 20]. In addition to the morphological similarities of the gametes (uniflagellated female cells that are 2–4 times larger than the male cells), both species also appear to have a biflagellated intermediate that forms after cell fusion, with nuclear fusion occurring while the cell retains multiple flagella [19] (Fig. S3 C – D). If these features are shared among the gametes of other Opisthokont groups, the last common ancestor of fungi and animals may have had sex that resembled mating in S. rosetta. Finally, the discovery of sex in choanoflagellates represents a previously unreported mode of cell-cell recognition and adhesion in the closest relatives of animals. Mechanisms used for gamete recognition and fusion in the ancestors of animals may have been co-opted for somatic cell adhesion during the evolution of animal multicellularity [21–23].
Experimental procedures
Growth media
Unenriched sea water was prepared by adding 32.9 g Tropic Marin sea salts (Wartenberg, Germany) to 1 L water to a salinity of 32–37 parts per thousand. Cereal grass (CG) medium was made by infusing unenriched sea water with cereal grass pellets (Basic Science Supplies, Rochester NY) [24]. High nutrient (HN) medium (250 mg/L peptone, 150 mg/L yeast extract, 150 μL/L glycerol in unenriched sea water) is made by diluting Sea Water Complete Medium [25] to 5% (vol/vol) in unenriched sea water.
Establishment of S. rosetta isolates
All strains of Salpingoeca rosetta currently in culture were derived from an environmental isolate that was started from a single, rosette colony (ATCC 50818) [5]. The Px1 culture (ATCC PRA-366), which was derived from the environmental isolate through a combination of antibiotic treatments and serial dilution, includes a single feeder bacterium, Algoriphagus machipongonensis [5, 6, 26]. Px1 has been sequenced and its genome is publicly available [6]. The ploidy of the Px1 population from which genomic DNA was isolated for genome sequencing was unknown. However, given the data presented here, we infer in retrospect that the clone that founded Px1 was a heterozygous diploid (similar to isolate A) and that, during the rapid expansion of the culture prior to genome sequencing, a subset of these cells underwent meiosis (see Supplemental Experimental Procedures).
Isolates A, B, and C were derived from Px1 through multiple clonal isolation steps (Fig. 1A). To generate isolates A and B, a culture of Px1 was passaged in CG medium every 3–4 days for approximately 9 months. Using an approximation of 1 generation per 10 hours (derived from the growth curve in [4]), we estimate that this corresponds to 500–1000 generations. Subsequently, this long-term culture was mutagenized with EMS prior to clonal isolation. Isolate A was derived from the control sample and isolate B was derived from the mutagenized sample. Clonal isolation was achieved using 2 serial dilution-to-extinction steps, similar to [27].
In isolate C, A. machipongonensis was replaced with a new feeder species, Echinicola pacifica (DSM 19836) [28] through repeated antibiotic treatment and dilution-to-extinction. These antibiotic treatment and dilution procedures were repeated until E. pacifica was the only remaining bacterial species in the culture, as monitored by plating and 16S RFLP analysis [5]. After this time, antibiotic treatment was discontinued. This strain is publicly available as ATCC PRA-390 under the name SrEpac, for S. rosetta grown with E. pacifica bacteria.
Culturing conditions
Isolate C was propagated in HN medium, yielding primarily haploid cultures of chains and slow swimmer cells [5]. Px1 and isolates A and B were propagated in CG medium, which yielded primarily diploid cultures of the cate cells. All cultures were propagated by diluting 1 mL of culture into 9 mL fresh medium every 2–3 days.
To induce sex under nutrient poor conditions, a stationary phase culture of isolate C was pelleted and resuspended in an equivalent volume of unenriched sea water. These conditions did not provide nutrients to support bacterial growth, and therefore the choanoflagellates grazed down the bacteria and themselves became starved within 3–6 days.
To return diploid isolate C cells to a haploid state, 1 mL of isolate C culture in unenriched sea water (prepared as above) was added to 9 mL HN medium. Ploidy was assessed 3 days later. To induce isolate B cells to become haploid, 2 mL isolate B culture was added to 8 mL CG medium daily for 3 weeks.
Quantification of DNA content
We used flow cytometry to measure the DNA content of isolated, propidium iodide-stained S. rosetta nuclei. Three to four million choanoflagellate cells and their prey bacteria were concentrated to 250 μL and centrifuged through 2mL Percoll/sorbitol/sea water mixture to enrich for S. rosetta cells and reduce the concentration of bacteria before gentle osmotic lysis was applied to isolate individual S. rosetta nuclei. Nuclei were stained with propidium iodide and fluorescence was quantified by flow cytometry, with yeast cells included as an internal standard in each sample.
Genomic DNA sequencing and SNP analysis
We sequenced the genomes of isolates A, B, and C to detect polymorphisms. For each isolate, S. rosetta genomic DNA was prepared by phenol chloroform extraction and physically separated from bacterial DNA on a CsCl gradient [29]. Samples were sequenced with 100 bp Illumina paired end reads to a median read depth of 186x, 161x, and 37x respectively for isolates A, B, and C.
Raw reads were trimmed using TrimmomaticPE [30] to remove Illumina adapters and low quality base calls. Trimmed reads were mapped to the S. rosetta reference genome [6] using BWA [31]. After removing duplicates using PicardTools (http://picard.sourceforge.net), variants were called using samtools and bcftools with default parameters [32]. All subsequent analyses used only high-quality SNPs with a quality score > 100. The frequency of SNP-containing reads was calculated as the number of reads at a locus that contained the SNP divided by the total number of reads at the position. To generate the map of SNPs across the genome, we binned SNPs into 20 kb sliding windows and plotted their positions semi-transparently, so that higher concentrations of SNPs were displayed as a darker color.
Immunofluorescence and time lapse video microscopy
To generate time lapse movies, isolate C cells were cultivated in unenriched sea water for 24 hours before transfer to a 35 mm Fluorodish glass bottomed dish (World Precision Instruments, Sarasota FL). After allowing 5–10 minutes for cells to settle to the bottom of the dish, multiple fields of view was imaged once per minute with 40x phase contrast microscopy for a period of 10 hours. Movies were prepared from still images using the Animation setting in ImageJ 1.43 [33] at frame rate of 7 frames per second, which corresponds to a speed of 420x real time. For immunofluorescence staining, cells cultured under the same conditions were allowed to settle live on poly-L-lysine coated coverslips (BD Biosciences) for 30 minutes. After a 6% acetone/4% formaldehyde fixation, cells were stained as in [34] with E7 anti-β-tubulin primary antibody (Developmental Studies Hybridoma Bank, Iowa City, IA), Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibody (Molecular Probes, Carlsbad, CA), 6 U/mL rhodamine phalloidin (Molecular Probes, Carlsbad, CA), and 0.5 μg/mL DAPI (Invitrogen) before mounting in 4 μL roLong Gold antifade reagent with DAPI (Molecular Probes, Carlsbad, CA). All images were taken using a Leica DMI6000 B inverted compound microscope and Leica DFC350 FX camera. Immunofluorescence images were obtained using a 100x oil immersion objective.
Supplementary Material
Highlights.
Choanoflagellates can reproduce asexually as both haploids and diploids
Altered nutrient levels trigger the choanoflagellate sexual life cycle
Patterns of polymorphism provide evidence of historical recombination
Flagellated male and female gamete cells fuse during haploid-to-diploid transitions
Acknowledgments
We thank Z. Cande, J. Rine, D. Richter, and members of the King laboratory for helpful discussions and comments on the manuscript, the Drubin laboratory for yeast strains, and H. Nolla for flow cytometry expertise. This work was supported by funding from the National Institutes of Health (NIH GM089977-01). T. L. was supported by a Graduate Research Fellowship from the National Science Foundation (DGE 1106400). N. K. is a Senior Scholar in the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carr M, Leadbeater BSC, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci USA. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr M, Leadbeater BSC, Baldauf SL. Conserved Meiotic Genes Point to Sex in the Choanoflagellates. Journal of Eukaryotic Microbiology. 2010;57:56–62. doi: 10.1111/j.1550-7408.2009.00450.x. [DOI] [PubMed] [Google Scholar]
- 4.Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr Biol. 2010;20:R875–6. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife. 2012:1. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousin A, Heel K, Cowling WA, Nelson MN. An efficient high-throughput flow cytometric method for estimating DNA ploidy level in plants. Cytometry. 2009;75A:1015–1019. doi: 10.1002/cyto.a.20816. [DOI] [PubMed] [Google Scholar]
- 9.Sager R, Granick S. Nutritional control of sexuality in Chlamydomonas reinhardi. J Gen Physiol. 1954;37:729–742. doi: 10.1085/jgp.37.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JJ. The metabolism of yeast sporulation. II. Stimulation and inhibition by monosaccharides. Can J Microbiol. 1957;3:81–90. doi: 10.1139/m57-010. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn SI, Hallegraeff GM, Bolch CJ. Vegetative reproduction and sexual life cycle of the toxic dinoflagellate Gymnodinium catenatum from Tasmania, Australia. Journal of Phycology. 1989;25:577–590. [Google Scholar]
- 12.O’Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. Available at: http://www.nature.com/doifinder/10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 13.Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, et al. Probing Meiotic Recombination and Aneuploidy of Single Sperm Cells by Whole-Genome Sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper MA, Adam RD, Worobey M, Sterling CR. Population Genetics Provides Evidence for Recombination in Giardia. Current Biology. 2007;17:1984–1988. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Marshall WL, Berbee ML. Population-Level Analyses Indirectly Reveal Cryptic Sex and Life History Traits of Pseudoperkinsus tapetis (Ichthyosporea, Opisthokonta): A Unicellular Relative of the Animals. Molecular Biology and Evolution. 2010;27:2014–2026. doi: 10.1093/molbev/msq078. [DOI] [PubMed] [Google Scholar]
- 16.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang Y-M, Su C-H, Bennett RJ, Wang Y, et al. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature. 2013:1–6. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SC, Ni M, Li W, Shertz C, Heitman J. The Evolution of Sex: a Perspective from the Fungal Kingdom. Microbiology and Molecular Biology Reviews. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schatten H, AC . Fertilization in Invertebrates. In: Tarin J, Cano A, editors. Fertilization in Protozoa and Metazoan Animals: Cellular and Molecular Aspects. Berlin: Springer; 2000. pp. 27–73. [Google Scholar]
- 19.Pommerville J, Fuller MS. The cytology of the gametes and fertilization of Allomyces macrogynus. Archives of microbiology. 1976;109:21–30. doi: 10.1007/BF00425108. [DOI] [PubMed] [Google Scholar]
- 20.Pommerville J. Morphology and physiology of gamete mating and gamete fusion in the fungus Allomyces. Journal of Cell Science. 1982;53:193–209. [Google Scholar]
- 21.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 22.King N. The Unicellular Ancestry of Animal Development. Developmental Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Koschwanez JH, Foster KR, Murray AW. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife. 2013;2:e00367–e00367. doi: 10.7554/eLife.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King N, Young SL, Abedin M, Carr M, Leadbeater BSC. Starting and maintaining Monosiga brevicollis cultures. Cold Spring Harbor Protocols. 2009;2009 doi: 10.1101/pdb.prot5148. pdb.prot5148. [DOI] [PubMed] [Google Scholar]
- 25.Atlas RM. Handbook of Microbiological Media. 3. CRC Press; 2004. [Google Scholar]
- 26.Alegado RA, Grabenstatter JD, Zuzow R, Morris A, Huang SY, Summons RE, King N. Algoriphagus machipongonensis sp. nov., co-isolated with a colonial choanoflagellate. International Journal of Systematic and Evolutionary Microbiology. 2013;63:163–168. doi: 10.1099/ijs.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King N, Young SL, Abedin M, Carr M, Leadbeater BSC. Isolation of Single Choanoflagellate Cells from Field Samples and Establishment of Clonal Cultures. Cold Spring Harbor Protocols. 2009;2009 doi: 10.1101/pdb.prot5147. pdb.prot5147 pdb.prot5147. [DOI] [PubMed] [Google Scholar]
- 28.Nedashkovskaya OI, Kim SB, Vancanneyt M, Lysenko AM, Shin DS, Park MS, Lee KH, Jung WJ, Kalinovskaya NI, Mikhailov VV, et al. Echinicola pacifica gen. nov., sp. nov., a novel flexibacterium isolated from the sea urchin Strongylocentrotus intermedius. International Journal of Systematic and Evolutionary Microbiology. 2006;56:953–958. doi: 10.1099/ijs.0.64156-0. [DOI] [PubMed] [Google Scholar]
- 29.King N, Young SL, Abedin M, Carr M, Leadbeater BSC. Separation of Choanoflagellate and Bacterial Genomic DNA. Cold Spring Harbor Protocols. 2009;2009 doi: 10.1101/pdb.prot5154. pdb.prot5154–pdb.prot5154. [DOI] [PubMed] [Google Scholar]
- 30.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Research. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King N, Young SL, Abedin M, Carr M, Leadbeater BSC. Visualizing the Subcellular Localization of Actin, Beta-tubulin, and DNA in Monosiga brevicollis. Cold Spring Harbor Protocols. 2009;2009 doi: 10.1101/pdb.prot5150. pdb.prot5150–pdb.prot5150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.