Abstract
Recent findings suggest that both host and pathogen manipulate copper content in infected host niches during infections. In this review, we summarize recent developments that implicate copper resistance as an important determinant of bacterial fitness at the host-pathogen interface. An essential mammalian nutrient, copper cycles between copper (I) (Cu+) in its reduced form and copper (II) (Cu2+) in its oxidized form under physiologic conditions. Cu+ is significantly more bactericidal than Cu2+ due to its ability to freely penetrate bacterial membranes and inactivate intracellular iron-sulfur clusters. Copper ions can also catalyze reactive oxygen species (ROS) generation, which may further contribute to their toxicity. Transporters, chaperones, redox proteins, receptors and transcription factors and even siderophores affect copper accumulation and distribution in both pathogenic microbes and their human hosts. This review will briefly cover evidence for copper as a mammalian antibacterial effector, the possible reasons for this toxicity, and pathogenic resistance mechanisms directed against it.
Keywords: copper, pathogenesis, yersiniabactin, copper tolerance, metal biology, copper resistance
Introduction
Copper is both an essential mammalian micronutrient and a potent antibacterial agent. The Smith Papyrus, an ancient Egyptian medical text dated at 2400 BC, is the earliest medicinal archive to recommend copper sulfate to sterilize water and treat infections (Dollwet and Sorenson, 1985). Mesoamerican and Hellenistic civilizations used copper and copper salts to treat a broad variety of physical ailments, including microbial and parasitic infections. In 400 BC, Hippocrates prescribed copper salts to treat leg ulcers. In the nineteenth century, Victor Burq observed that copper workers in Paris appeared immune to recurrent cholera epidemics (Burq, 1867). He also noted that while neighboring towns were ravaged by frequent cholera epidemics, the pottery-making town of Aubagne was protected from these outbreaks. He attributed this protection to “… a rampart of copper dust” generated by copper-rich clay used by the city's potters. These observations led to rapid developments in the field of metallotherapy, and medically employed copper salts, amulets, and belts were widely used to treat dermatologic, gastrointestinal, and tubercular infections (Dollwet and Sorenson, 1985; Borkow, 2005) until the advent of commercially available antibiotics in 1932.
Human and animal studies now suggest a parallel between ancient medicinal copper use and antibacterial immune function. In this review, we summarize copper homeostasis mechanisms in the human host, and the means by which the host deploys the metal to combat infections. We describe the chemical and biochemical principles that define copper's toxicity, and how these toxic properties serve as potent leverage against invading pathogens. Finally, we discuss the pathogenic molecular, cellular, and biochemical responses that counter copper toxicity at host interface.
Copper as nutrient or toxin
With photosynthesis and dioxygen release in the atmosphere 2.7 billion years ago, the sulfides that sequestered copper were oxidized to sulfates, leading to increased copper bioavailability (Frausto da Silva and Williams, 1993). Copper-containing proteins appeared relatively late in an evolutionary timescale, likely in response to increasing need to use oxygen and oxygen containing molecules (Dupont et al., 2011; Nevitt et al., 2012). These enzymes are critical to cellular, biochemical and regulatory functions in the human host, leading to a nutritional requirement for sufficient copper levels. The most prominent examples include cytochrome c oxidase, the respiratory chain terminal electron acceptor, and Cu-Zn superoxide dismutase, required for defense against oxidative damage (Karlin, 1993). Putative copper binding proteins compose ~1% of the total eukaryotic proteome, suggesting that known cuproproteins represent only a minor fraction of the total (Andreini et al., 2008). Copper's role in host biology and defense is better understood by examining its chemistry.
Copper chemistry
Copper is the 26th most abundant in the earth's crust and exists as 2 stable and 9 radioactive isotopes. A transition metal, copper primarily exists as one of two stable oxidation states: Cu2+ in the oxidized cupric form, and Cu+ in the reduced cuprous form. Cu+ is a closed shell 3d10 transition metal ion with diamagnetic properties (Frausto da Silva and Williams, 1993). A soft Lewis acid, it favors tetrahedral coordination with soft bases such as hydrides, alkyl groups, cyanide, phosphines, and thiols from cysteine and thioether bonds with methionine (Crichton and Pierre, 2001). Cu2+ has a 3d9 configuration, is paramagnetic, and is an intermediate Lewis acid. In addition to ligands bound by Cu+, Cu2+ forms square planar complexes with sulphates, nitrates, nitrogen donors such as histidine, and oxygen donors like glutamate and aspartate (Bertini et al., 2007). Different ligand combinations, oxygenation levels, pH, organic matter, sulfates and carbonates, generate differential metal speciation and distinct metal coordination environments. Copper's value as a bioelement lies mainly in its unique electrochemical properties. The Cu+/Cu2+ couple has a high redox potential, which allows it to act as an electron donor/acceptor in redox reactions (Crichton and Pierre, 2001). Most copper enzymes span a range of +200 to +800 mV, enabling them to directly oxidize substrates such as ascorbate, catechol, and phenolates. The same electrochemical properties contribute to copper's toxic effects through several mechanisms, outlined below.
Copper as a Fenton reagent
Within superoxide and hydrogen peroxide-rich environments such as the phagosome, copper may propagate toxic hydroxyl radical formation by Fenton-like chemistry [Equation (1)] (Liochev, 1999).
| (1) |
Hydroxyl radicals are extremely reactive, cannot be scavenged by enzymatic reaction, and have a diffusion controlled half-life of ~10−9 s before reacting with organic molecules in vivo (Freinbichler et al., 2011), suggesting that hydroxyl radical damage would occur in close spatial proximity to copper ions. Extensive work has implicated reactive oxygen species (ROS) derived from metal-catalyzed oxidation in lipid, protein, and DNA oxidation (Yoshida et al., 1993; Liochev, 1999; Stadtman, 2006). Copper ions can also oxidize sulfhydryls such as cysteine or glutathione in a cycle between reactions [Equations (2), (3a,b) or (4a,b), followed by (5)]:
| (2) |
and
| (3a) |
| (3b) |
or
| (4a) |
| (4b) |
followed by
| (5) |
Hydrogen peroxide can in turn participate in reaction 1 and may further propagate radical formation.
Attempts to understand copper toxicity through classic copper-catalyzed Fenton chemistry to copper toxicity have produced contrary results. Macomber et al. exposed an Escherichia coli mutant with multiple copper efflux deficiencies to hydrogen peroxide (Macomber et al., 2007). Rather than exhibiting greater peroxide sensitivity [through Equation (1)], copper-loaded E. coli were instead more resistant to hydrogen peroxide. Furthermore, copper loading was associated with fewer, not more, oxidative DNA lesions. Lastly, EPR spectroscopy revealed no change in hydroxyl radical generation with copper addition. Most of the copper in overloaded strains was localized to the periplasm, where any hydroxyl radical generated would react locally before reaching DNA in the cytoplasm. This spatial compartmentalization may explain the lack of DNA damage. While there may exist circumstances in which copper propagates cytotoxic Fenton chemistry in vivo, this work suggests the existence of an alternative copper toxicity mechanism in E. coli.
Non-Fenton destruction of iron-sulfur complexes by copper
Recent evidence suggests a non-Fenton chemistry copper toxicity mechanism in which the reduced Cu+ ion is instrumental. Multiple investigators note that copper toxicity to bacteria is sustained or even enhanced in anoxic conditions (Beswick et al., 1976; Outten et al., 2001; Macomber and Imlay, 2009) where peroxide formation is minimal. Increased copper toxicity under anoxic conditions may reflect higher Cu+ prevalence. E. coli EPR spectroscopy indicates that considerable Cu2+ is converted to non-paramagnetic Cu+ under anoxic conditions (Beswick et al., 1976). Macomber et al. show that intracellular copper in overloaded E. coli is in the reduced Cu+ valence, likely due to cytosolic reduction and its ability to enter bacteria by traversing bacterial membranes (Macomber et al., 2007). Cu+ toxicity in the E. coli cytosol can be explained by its intense thiophilicity, which is sufficient to competitively disrupt key cytoplasmic iron-sulfur enzymes both in vitro and in vivo (Macomber and Imlay, 2009). Indeed, other “soft” thiophilic metal ions that do not act as Fenton reagents have been found to exert comparable toxicity (Jozefczak et al., 2012; Xu and Imlay, 2012). Together, these data provide compelling evidence linking copper toxicity to iron displacement from solvent-exposed dehydratase iron-sulfur clusters, resulting in metabolic disruption and branched chain amino acid auxotrophy.
Copper at the host-pathogen interface
Copper homeostasis is essential for human growth and development. Average daily human dietary copper intake varies from 0.6 to 1.6 mg/dL, with a free copper ion concentration of 10−13 M in human blood plasma (Linder and Hazegh-Azam, 1996). In mammalian cells, cytoplasmic metallothioneins, glutathione based redox maintenance, and the Cu/Zn superoxide dismutase mitigate copper toxicity (Fridovich, 1974; Babula et al., 2012; Hatori et al., 2012). This section reviews the basic characteristics of human copper transporters together with data that may speak to their functions during infection and inflammation.
Human copper physiology
Unlike antimicrobial peptides, proteolytic enzymes, or ROS, copper cannot be synthesized in situ during infections and so must be absorbed from the diet or mobilized from tissue depots for use by immune cells (see a more complete review Pena et al., 1999). Once dietary copper is absorbed from the intestinal lumen it is delivered to the liver, which exports it to the peripheral circulation or excretes it into the bile (Crampton et al., 1965; Vancampen and Mitchell, 1965). The liver incorporates copper into multiple proteins, including the secreted glycosylated multi-copper ferroxidase ceruloplasmin (Holmberg and Laurell, 1948). Ceruloplasmin-copper complexes bind Ctr1, an integral membrane protein that is structurally and functionally conserved from yeast to humans (Zhou and Gitschier, 1997). Ctr1 transports 60–70% of the total copper in flux. Ctr1 is responsive to copper levels: copper depletion increases Ctr1 expression at the plasma membrane through the recruitment from the intracellular pools, whereas elevated copper induces rapid transporter endocytosis from the plasma membrane to vesicles (Zhou and Gitschier, 1997; Petris et al., 2003; Guo et al., 2004). Following internalization by Ctr1, copper is shuttled to the trans-Golgi network by ATOX1/HAH1 in secretory compartments (Klomp et al., 1997). Atox1 gene deletion in mice results in perinatal lethality, reflecting its crucial role in normal cellular metabolism (Hamza et al., 2001). Copper is transferred directly from ATOX1 to the N-terminus of two homologous P1B-type ATPase Cu+ transporters, ATP7A (Chelly et al., 1993; Mercer et al., 1993; Vulpe et al., 1999) and ATP7B (Bull et al., 1993; Tanzi et al., 1993; Vulpe et al., 1993), located in the trans-Golgi network. Macrophages infected with Salmonella typhimurium exhibit increased Ctr1, ATP7A and ceruloplasmin gene expression, indicating that they play a role in restricting infection by professional intracellular pathogens (Achard et al., 2012).
Copper fills varied roles in mammalian biology, and it is notable that copper-deficiency is associated with numerous deficiencies in host defense (Kaim and Rall, 1996). Mutations in ATP7A result in a severe copper-deficiency known as Menkes disease (Kaler, 2011). Infants with Menkes' disease are more susceptible to Gram-negative infections, consistent with copper's role in restricting microbial growth (Menkes et al., 1962; Danks et al., 1972; Gunn et al., 1984). Conversely, Wilson's disease is characterized by excess copper accumulation in brain and liver tissues, resulting in cirrhosis and neurodegeneration that may manifest well after infancy. Other human copper deficiency studies reveal impaired phagocytic indices, decreased antibody response, impaired peripheral mononuclear cell proliferation, lower early T-cell activation and proliferation, and lower cytokine expression (Sullivan and Ochs, 1978; Prohaska and Lukasewycz, 1990). While these conditions suggest a specialized role for copper in antibacterial immunity, caution must be taken to differentiate this from a less specific, more general nutritional role in the host (Newberne et al., 1968; Sullivan and Ochs, 1978; Boyne and Arthur, 1981; Jones and Suttle, 1983; Koller et al., 1987; Prohaska and Lukasewycz, 1990; Crocker et al., 1992; Smith et al., 2008).
Copper physiology during infections
Although incompletely understood, there are indications that a coordinated physiologic response may increase both systemic and local copper availability during infections. Compared to normal controls, copper levels increase two- to ten-fold in the serum, livers and spleens of animals infected with a range of pathogens, including viruses, bacteria, and trypanosomes (Tufft et al., 1988; Crocker et al., 1992; Matousek De Abel De La Cruz et al., 1993; Ilback et al., 2003). Increased circulating copper may be selectively imported into infected sites, as indicated by two- to five-fold increase in copper-carrier proteins (Natesha et al., 1992; Chiarla et al., 2008). X-ray microprobe analyses indicate that copper's absolute atomic concentration in area density increases a hundred-fold to several hundred micromolar within granulomatous lesions of lungs infected with Mycobacterium tuberculosis, and high copper concentrations are selectively redistributed to the exudates of wounds and burns (Beveridge et al., 1985; Jones et al., 2001; Voruganti et al., 2005; Wagner et al., 2005). Whether this accumulation reflects uptake by myeloid cells alone or includes a tissue-wide response remains unclear.
Copper as a white blood cell antibacterial agent
In 2009, White et al. published findings from cultured macrophage-like RAW264.7 cells that are consistent with a copper-specific bactericidal system directed against phagocytosed E. coli (White et al., 2009). Phagosomal killing of K12 E. coli was greatly affected by copper content of the cell culture media. Microscopy and posttranscriptional silencing investigations linked this copper-dependent activity to ATP7A-mediated copper trafficking from the Golgi apparatus to E. coli-containing phagolysosomes. These studies suggest that in addition to its role in physiologic copper absorption, ATP7A fills a host defense function by transporting antibacterial quantities of copper ions to phagolysosomal compartments containing engulfed bacteria. Consistent with this finding, low-density lipoprotein (LDL) oxidation by macrophage-like THP-1 cells was found to be ATP7A-dependent, suggesting metal catalyzed oxidation by secreted copper ions (Qin et al., 2010). ATP7A is expressed in a broad range of both myeloid and non-myeloid cell types (La Fontaine et al., 2010; Wang et al., 2011), raising the possibility that a variety of cell types may similarly direct the copper payloads to kill internalized bacteria. These observations suggest a specific functional rationale for the array of mammalian copper transport genes upregulated by proinflammatory stimuli such as interferon-gamma and lipopolysaccharide and for the altered copper physiology noted above in section Copper as a Fenton Reagent. (Achard et al., 2012). Studies to identify macrophage lineages or even non-professional phagocytes that use copper-mediated antibacterial activity would be of great interest in the area of infection biology. To date, copper-dependent uropathogenic E. coli killing has been observed in both RAW264.7 cells and mouse peritoneal macrophages (Chaturvedi et al., 2013). Altogether, these findings suggest an intriguing parallel between ancient medicinal copper use and innate immune function.
Phagosomal copper may add to, and perhaps synergize with, the diverse cellular microbial killing strategies described since Elie Metchnikoff's pioneering work on phagocytosis (Gordon, 2008). These strategies are often functionally redundant and have been broadly grouped into oxidative killing mechanisms exemplified by the macrophage respiratory burst and non-oxidative killing mechanisms such as antimicrobial peptides and hydrolytic enzymes. Interactions between copper and more established antibacterial effectors within the phagosome's restricted space are likely. Membrane permeabilizing defenses may facilitate copper entry into bacteria, while high concentrations of respiratory burst-derived oxidants are likely to modulate redox active copper ions. These interactions may be spatially and temporally governed during and after the respiratory burst. One recent finding in E. coli suggests that copper's interactions with phagosomal superoxide may greatly impact intracellular bacterial survival (see section Superoxide Dismutation).
Copper-mediated killing by vertebrate immune systems would be expected exert selective pressure on copper resistance in pathogenic bacteria. Below, we review the virulence-associated copper resistance systems described in several human pathogens. The classic intracellular pathogen M. tuberculosis upregulates genes encoding copper efflux-associated P1B-type ATPases during macrophage infection (Ward et al., 2008; Rowland and Niederweis, 2012). Urinary E. coli isolates collected from patients with urinary tract infections (UTIs) exhibit higher growth than concomitant rectal isolates in a medium containing an inhibitory concentration of copper (Chaturvedi et al., 2012). Copper resistance genes are often observed in virulence-associated mobile genetic elements carried by E. coli as well as Legionella pneumophila, Klebsiella pneumoniae, and methicillin resistant Staphylococcus aureus (Sandegren et al., 2012; Shoeb et al., 2012; Gomez-Sanz et al., 2013; Trigui et al., 2013). E. coli and M. tuberculosis strains with engineered deficiencies in copper resistance genes exhibit impaired intracellular survival in phagocytic cells (White et al., 2009; Wolschendorf et al., 2011; Chaturvedi et al., 2013). To date, these observations suggest that resistance to copper-mediated killing among pathogens may be a virulence-associated property driven by host innate immunity.
Mechanisms of microbial copper tolerance
Copper's direct and indirect toxicity can alter enzyme specificity, disrupt cellular functions, and damage nucleic acid structure. Changes in copper concentrations during infection suggest that the host harnesses the metal's toxic properties to combat microbial growth. In response, pathogenic bacteria have evolved a series of protein- and small-molecule based defenses against copper toxicity. Unlike eukaryotic cells, most known bacterial cuproproteins are located within the cytoplasmic membrane or in the periplasmic space, perhaps to compartmentalize a potentially toxic metal species. Microbes use this copper sparingly in metabolism, and for electron transport in respiratory pathways. Given this, copper's cytoplasmic availability is tightly controlled, and data indicate that there are fewer than 104 free copper atoms per bacterial cell, reflecting cytoplasmic copper-responsive transcriptional regulators' high copper sensitivity (Outten and O'Halloran, 2001; Changela et al., 2003; Finney and O'Halloran, 2003).
Both Cu+ and Cu2+ can permeate the outer membrane of E. coli and enter the periplasm, but only Cu+ is able to cross the inner membrane and reaches the cytoplasm by a currently unknown mechanism. While no copper uptake genes have yet been identified in E. coli, the outer-membrane protein ComC (under transcriptional control of the TetR-like regulator ComR) may reduce the outer membrane's copper permeability (Mermod et al., 2012). It is speculated that cytoplasmic Cu+ is largely complexed by millimolar quantities of thiols such as glutathione. Interestingly, glutathione biosynthesis gene deletion has little effect on microbial copper response, indicating that its role in detoxifying copper in bacterial cells may either be limited or redundant (Helbig et al., 2008). In this regard, qualitative and quantitative analyses of cytosolic copper binding sites in bacteria would aid our understanding of copper toxicity.
Microbial copper-resistance systems span copper efflux (cue, cus, and extrachromosomal efflux systems), copper sequestration (CusF and siderophores), and copper oxidation (mixed copper oxidases and superoxide dismutase mimics). For the sake of brevity, the following sections primarily discuss Cu2+ detection and resistance proteins that have been described in E. coli (Figure 1). Their functional homologs in other microbial species are tabulated in Table 1 (see a more complete review Rademacher and Masepohl, 2012).
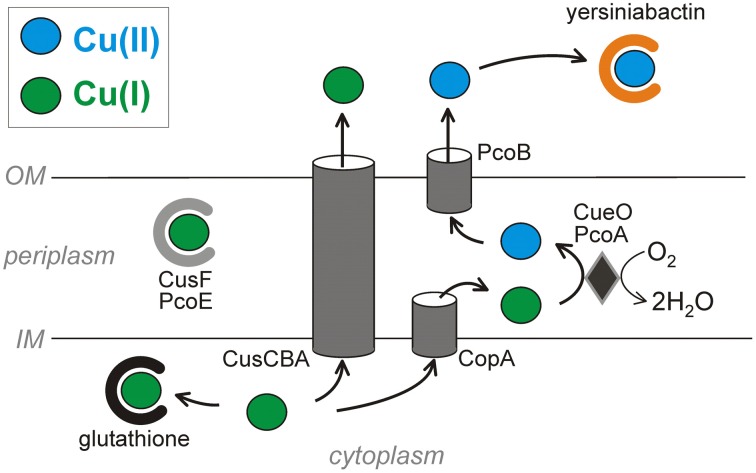
Figure 1.
Copper resistance strategies across pathogenic E. coli membranes. The virulence-associated siderophore yersiniabactin sequesters Cu2+ outside the cell and prevents its reduction to the more toxic Cu+. Copper ions that reach the cytosol are subject to chelation by glutathione and export by two ATPases. The CusCBA ATPase complex exports Cu+ from both the cytoplasm and the periplasm (via CusF) to the extracellular space. Alternatively, the CopA ATPase exports cytoplasmic copper across the inner membrane. Periplasmic Cu+ can bind the proteins CusF and PcoE or be oxidized by the mixed copper oxidases CueO or PcoA to less toxic Cu2+. PcoB has a putative function of exporting Cu2+ across the outer membrane. The systems are oriented to minimize free cytosolic copper ions by directing these to the periplasmic or extracellular spaces.
Table 1.
Species-wide distribution of copper-resistance proteins.
| Function | Protein | Species |
|---|---|---|
| Copper detection | SctR GolS* | S. typhimurium (Espariz et al., 2007; Pontel et al., 2007; Osman and Cavet, 2011) |
| RicR | M. tuberculosis (Festa et al., 2011) | |
| CueP | S. typhimurium (Pontel and Soncini, 2009) | |
| CrdRS | H. pylori (Waidner et al., 2005) | |
| CopY | Enterococcus hirae, Enterococcus faecium, Streptococcus mutans, Lactococcus lactis (Strausak and Solioz, 1997; Vats and Lee, 2001; Magnani et al., 2008; Reyes-Jara et al., 2010) | |
| Copper efflux | GolT | S. typhimurium (Espariz et al., 2007; Osman et al., 2010) |
| CtpV | M. tuberculosis (Rowland and Niederweis, 2012) | |
| CopA1 | P. aeruginosa (Thaden et al., 2010) | |
| CopA2 | P. aeruginosa (Gonzalez-Guerrero et al., 2010) | |
| CopA | Enterococcus hirae (Odermatt et al., 1994; Odermatt and Solioz, 1995) | |
| CopB | Enterococcus hirae (Odermatt et al., 1994; Odermatt and Solioz, 1995) | |
| Copper sequestration | CueP | S. typhimurium (Pontel and Soncini, 2009) |
| SmtA | Synechococcus (Shi et al., 1992) | |
| Copper oxidation | Rv0846c | M. tuberculosis (Rowland and Niederweis, 2013) |
| Copper homeostasis repression | YcnK | Bacillus subtilis (Chillappagari et al., 2009) |
| CstR | Staphylococcus aureus (Grossoehme et al., 2011) |
Copper efflux
The cue system. In E. coli, two chromosomal systems remove excess Cu+ from the cytosol (Outten et al., 2001). The cue system (for Cu efflux) transcriptionally activates both plasmid- and chromosomally-encoded copper homeostatic systems in response to intracellular Cu+ sensing through CueR, a MerR-family metalloregulatory transcriptional activator (Petersen and Moller, 2000; Stoyanov et al., 2001). CueR coordinates one Cu+ ion per monomer in an unusual and distinctive linear S–Cu+–S center encompassing two cysteine residues (C112 and C120) located at the dimer interface (Changela et al., 2003; Chen et al., 2003). Both holo- and apo-CueR bind to dyad-symmetric sequences at target promoters, but only holo-CueR activates transcription (Yamamoto and Ishihama, 2005; Andoy et al., 2009). A genome-wide transcriptional array study of the E. coli chromosome has identified 197 putative CueR-binding sites, which largely await experimental confirmation. Other bacteria that possess CueR-like copper-tolerance systems include Pseudomonas aeruginosa and S. typhimurium (Espariz et al., 2007; Pontel and Soncini, 2009; Thaden et al., 2010).
CueR is a copper-selective ortholog from multifunctional protein families that respond to a wide range of effector ligands (the MecI/BlaI-family repressors that mediate resistance to β-lactam antibiotics and the MerR family, respectively) (Brown et al., 2003; Portmann et al., 2006). While CueR is not widely distributed in bacterial genomes, Liu et al. describe one such copper-specific ubiquitous regulator (Liu et al., 2007). The intracellular copper sensor CsoR from M. tuberculosis is the founding member of what appears to be a large family of bacterial Cu+-responsive repressors, with greater than 170 projected members in archaeal, bacterial, and cyanobacterial genomes (Liu et al., 2007). Upon copper binding, CsoR is deactivated, leading to copper-resistance gene expression.
CueR upregulates copA and cueO gene expression (Outten et al., 2000; Stoyanov et al., 2001). These genes are associated with copper efflux and oxidation, respectively. CopA is a copper-exporting P1B-type ATPase active under high extracellular copper stress (Outten et al., 2000; Petersen and Moller, 2000; Fan and Rosen, 2002; Stoyanov et al., 2003). Mammalian and microbial P1B-type ATPases thus perform opposing functions that determine infection outcomes. Appropriate copper import and trafficking by mammalian ATPases is required to restrict microbial growth, while copper export by microbial ATPases is necessary to withstand this toxicity. CopA traverses the inner membrane and exports Cu+ from the cytosol in both oxic and anoxic conditions (Fan and Rosen, 2002; Kuhlbrandt, 2004; Arguello et al., 2007; Osman and Cavet, 2008). This efflux pump couples ATP hydrolysis to form an acylphosphate intermediate in the presence of Cu+ but not Cu2+. It is speculated that two amino-terminal metal binding domains with a CXXC motif confer metal binding specificity. copA mutants in E. coli, Streptococcus pneumoniae, and Neisseria gonorrhoeae all demonstrate impaired copper efflux, intracellular metal accumulation, and increased copper sensitivity in both oxic and anoxic conditions (Rensing et al., 2000; Outten et al., 2001; Shafeeq et al., 2011; Djoko et al., 2012).
The cus system. An independent copper efflux system, the cus (for Cus ensing) system confers copper-tolerance under moderate to high copper levels in oxic conditions (Outten et al., 2001). cusRSCFBA products are believed to form a multiunit transport complex that spans the periplasmic space and is anchored in both the inner and outer membranes (Mealman et al., 2012). While CopA exports excess Cu+ from the cytoplasm to the periplasm, CusRSCFBA effluxes Cu+ from the periplasm (Outten et al., 2001; Franke et al., 2003; Long et al., 2010).
CusRS is a two-component regulatory system that monitors copper stress in the cell envelope and is particularly active in anoxic copper stress conditions (Munson et al., 2000). In addition to CusRS, CpxRA, and YedWV are two other previously described copper-responsive E. coli two-component regulatory systems (Yamamoto and Ishihama, 2005, 2006). CusR and CusS exhibit homology with other plasmid-borne two-component systems that are also involved in metal responsive gene regulation. Membrane bound CusS senses periplasmic Cu+, which leads to protein autophosphorylation. CusS then donates the phosphoryl group to CusR, which activates the transcription of the cusCFBA and cusRS operons. CusA belongs to the resistance-nodulation-cell division (RND) proton antiporter family, CusB belongs to the membrane fusion protein family which anchor into the cytoplasmic membrane with a long periplasm-spanning domain, and CusC is an outermembrane protein with homology to the TolC-stress response protein (Franke et al., 2003; Delmar et al., 2013). CusF is a periplasmic metallochaperone that binds a single atom of Cu+ and participates in metal efflux by delivering the metal to CusC and CusB (Xue et al., 2008; Mealman et al., 2011).
Other prominent RND proton antiporters include the multidrug efflux systems AcrB and AcrF from E. coli, MexB from P. aeruginosa, and MtrD from N. gonorrhoeae (Nies and Silver, 1995; Paulsen et al., 1996). Interestingly, Cupriavidus metallidurans CH34 resistance to copper is attributed to RND protein expression (von Rozycki and Nies, 2009).
Extrachromosomally-encoded copper efflux systems. In environments where copper concentrations would overwhelm chromosomally encoded copper metabolic systems, microbes contain extrachromosomal loci that confer copper resistance. These loci are present in copper-resistant E. coli, Pseudomonas syringae, and Xanthomonas campestris pv. vesicatoria isolates (Tetaz and Luke, 1983; Bender and Cooksey, 1987; Brown et al., 1992; Voloudakis et al., 1993; Williams et al., 1993). All copper-resistant strains were isolated from agricultural areas characterized by repeated copper salt application as a feed additive, bactericidal agent, or antifungal agent. In these strains, the plasmid borne pco and cop operons confer copper resistance. These operons carry four related genes, pcoABCDRSE and copABCDRS, which are expressed from chromosomal copper-inducible promoters regulated by CusRS (Brown et al., 1995; Adaikkalam and Swarup, 2005). The genes copABCDRS are arranged in two operons, copABCD and copRS, respectively. This arrangement is also found in the pco determinant but with an additional gene, pcoE, further downstream. Extrachromosomal systems encode two-component regulators similar to CusRS, including PcoR and PcoS from the pco operon of E. coli; CopR and CopS from the cop operon, which provides copper resistance to P. syringae; and SilR and SilS from the sil locus, which provides silver ion resistance to Salmonella enterica serovar Typhimurium (Gupta et al., 1999). Similar to these copper efflux systems, extrachromosomal pco system encodes PcoB and PcoD, two copper pumps that are incorporated in the outer and inner membranes, respectively (Lee et al., 2002).
Extrachromosomal resistance systems are metal oxidation state selective. Recently published PcoC spectroscopic and crystallographic data and nuclear magnetic resonance (NMR) studies of the closely related P. syringae protein, CopC, reveal a biologically unprecedented thioether ligation (Arnesano et al., 2003a,b; Peariso et al., 2003). PcoC can bind both Cu2+ and Cu+: the protein exhibits a cupredoxin fold that binds Cu+ through two Met sulfur atoms and one nitrogen or oxygen ligand in a hydrophobic Metrich loop that is exposed to solvent on the protein surface. Cu2+ can bind a separate site in the same protein, where it coordinates water, as well as two histidine imidazoles and two other nitrogen or oxygen ligands. Following copper sensing, microbes respond to microenvironments that contain high concentrations of unligated copper by upregulating systems associated with copper efflux, oxidation, or sequestration.
Copper sequestration
In addition to copper oxidation and efflux systems, recent studies suggest that bacteria deploy both low molecular weight proteins and small molecules to bind and sequester intracellular copper. In E. coli, the periplasmic chaperone CusF binds copper, ultimately delivering it to CusCBA for export (Franke et al., 2003; Bagai et al., 2008; Xue et al., 2008; Mealman et al., 2012). Evidence indicates that PcoE acts as a soluble copper binder in the periplasm (Zimmermann et al., 2012). Across kingdoms, metallotheioneines sequester cytoplasmic copper (Leszczyszyn et al., 2011; Thirumoorthy et al., 2011; Gumulec et al., 2012). Recent work in M. tuberculosis shows that a five-locus regulon for copper resistance is upregulated during copper stress (Festa et al., 2011). This regulon includes MymT, a cytoplasmic metallothionein that binds Cu+ and attenuates copper toxicity (Gold et al., 2008). Although a native E. coli metallothionein has not yet been identified, data suggest that glutathione may exert similar cytoprotective effects by forming stable Cu+ complexes (Osterberg et al., 1979; Helbig et al., 2008; Macomber and Imlay, 2009).
Some microbial siderophores, low-molecular-weight iron chelating agents, sequester copper extracellularly and protect bacteria by minimizing intracellular copper penetration. There is precedent for this among environmental bacteria that express Cu+-binding compounds (those originally identified as copper binders are called chalkophores) such as methanobactin and phytochelatin (Cervantes and Gutierrez-Corona, 1994; Rauser, 1999; Kenney and Rosenzweig, 2012). In E. coli, chemically distinct siderophore types are observed to exert opposing copper phenotypes. Specifically, the catecholate siderophore enterobactin sensitizes E. coli to copper, likely through its ability to reduce cupric ion to the more toxic cuprous ion (Grass et al., 2004). Although known as a cuprous oxidase, CueO prevents this interaction by directly oxidizing catechols such as dihydroxybenzoic acid, an enterobactin biosynthetic precursor (Grass et al., 2004). Conversely, phenolate siderophores such as yersiniabactin bind Cu2+ in complexes that prevent reductive free Cu+ release (Chaturvedi et al., 2012). Uropathogenic E. coli strains that express yersiniabactin are protected from copper's toxic effects, suggesting that a strain's small molecule repertoire may affect its ability to survive and persist in a copper-rich environment. It is notable that yersiniabactin can protect bacteria with and without FyuA (the outer membrane ferric yersiniabactin importer) from copper toxicity, suggesting that yersiniabactin's iron uptake function does not contribute to this phenotype. Copper oxidation state selectivity among microbial small molecules is also observed in pyoverdin and pyochelin, two major siderophore types expressed by P. aeruginosa (Brandel et al., 2012). While both siderophores can bind Cu2+, Cu2+ supplementation upregulates genes involved in the synthesis of pyoverdin but downregulates those for pyochelin (Frangipani et al., 2008; Brandel et al., 2012). Data indicate that both siderophores prevent Cu2+ accumulation in the bacterial cell by 80% (Teitzel et al., 2006). Pyoverdin's selective expression indicates that it may play a direct role in copper tolerance, possibly by sequestering copper in reduction-resistant complexes like yersiniabactin. The chemical basis of pyoverdin's transcriptional selectivity is unclear, and response regulation is unknown. It is possible that ferric- and cupric siderophore complexes govern differential transcriptional responses.
It also remains unclear whether siderophore transport systems can discriminate between different metal bound forms. While sequestration by siderophores can attenuate copper toxicity, bacterial proteins that import siderophore-metal complexes may also play a role. The siderophore schizokinen eliminates copper's toxic effects on Anabaena (Clarke et al., 1987) but exacerbates copper toxicity in Bacillus megaterium (Arceneaux et al., 1984). It is possible that these differences arise from fundamental differences in metabolic and transport machinery between the two organisms. Copper schizokinen-mediated toxicity in Bacillus can be alleviated by the exogenous desferrioxamine, raising the possibility that cells transport iron to repair copper-mediated damage. This observation could be further explained by differences in each organism's ability to use its iron-uptake machinery to discriminate between cupric- and ferric-siderophore complexes. It is possible that copper indirectly affects siderophore expression by competitively inhibiting iron import or liberating intracellular iron, altering intracellular metal accumulation, and affecting a downstream biosynthetic feedback loop.
Copper oxidation
Mixed copper oxidases (MCO). Cu+ is more toxic than Cu2+ when applied under anoxic conditions, as demonstrated by Macomber and Imlay (2009). Consistent with this observation, E. coli cultures treated with both Cu2+ and reductants such as ascorbate or catechols demonstrate lower viability than those treated with Cu2+ alone (Chaturvedi et al., 2012). To detoxify extracytoplasmic Cu+, E. coli use the CueR-regulated multi-copper oxidase CueO to oxidize toxic cuprous copper to its less toxic cupric form (Grass and Rensing, 2001; Roberts et al., 2002; Singh et al., 2004). E. coli and S. typhimurium mutants lacking CueO exhibit extreme copper sensitivity in oxic conditions. CueO contributes to S. typhimurium virulence in a systemic murine infection model (Achard et al., 2010). A second, plasmid-borne 605 amino acid MCO called PcoA has also been described in E. coli. Periplasmic extracts containing PcoA exhibit copper-inducible oxidase activity, indicating that PcoA might similarly oxidize Cu+ to prevent toxicity (Huffman et al., 2002; Djoko et al., 2008). PcoA can functionally substitute for CueO in E. coli, indicating that these proteins have redundant function.
E. coli CueO is among the best-characterized bacterial multicopper oxidases (MCOs). CueO is structurally similar to the large, cross-Kingdom family of MCOs [including ascorbate oxidase and the ferroxidases Fet3 and ceruloplasmin (Outten et al., 2000)] that oxidize substrates using oxidizing equivalents in molecular oxygen. This oxygen requirement renders oxidases inactive under anoxic conditions. CueO's active site consists of a trinuclear copper center MCO active site in which a fourth copper atom mediates electron transfer from the substrate (Roberts et al., 2002; Grass et al., 2004). The enzyme couples Cu+ oxidation with four-electron oxygen oxidation to water through the hydroxide-bridged fourth copper atom. Reactive oxygen intermediates generated during the reaction remain coordinated and are not released from the protein. It is curious that despite low cytoplasmic copper levels, CueO and PcoA exhibit a twin-arginine motif in their leader sequences, suggesting that they are translocated from the cytoplasm by the twin arginine translocation (Tat) pathway with copper-bound active sites (Huffman et al., 2002). Holo-protein translocation from the cytoplasm means that some amount of chaperone-bound copper must be delivered to these apo-proteins intracellularly. This indicates that intracellular copper may serve a biosynthetic role in this specific process. If MCOs ultimately evolved to prevent copper entry to the cytosol, it is possible that metallation by cytosolic copper is a form of feedback regulation in which higher cytosolic copper levels lead to higher MCO secretion. Further studies are necessary to discern this, and other, possibilities.
In addition to oxidizing periplasmic Cu+, E. coli CueO can also oxidize 2,3 dihydrobenzoic acid (DHB) (Grass et al., 2004). 2,3-DHB is the biosynthetic precursor to enterobactin, a catecholate siderophore, secreted during iron limitation. As enterobactin can reduce Cu2+ to Cu+, it has been hypothesized that CueO's 2,3-DHB oxidation activity is a strategy to prevent toxic Cu+ accumulation. While it may seem paradoxical to both synthesize and destroy a siderophore, an intracellular copper requirement for CueO secretion may ensure that it's siderophore destructive activity is only relevant in the presence of high copper levels. Together, these findings suggest that MCO's such as CueO help protect bacteria from copper stress by controlling copper ion oxidation states in oxic environments.
Superoxide dismutation. Recent work shows that yersiniabactin expression greatly facilitates pathogen survival within phagocytic cells in a copper- and NADPH oxidase system-dependent manner (Chaturvedi et al., 2013). In the presence of copper- and NADPH oxidase-derived superoxide, yersiniabactin production protects urinary pathogenic E. coli within cultured macrophage-like cell phagosomes. Superoxide's contribution to this phenotype suggests that yersiniabactin's cytoprotective effects may not be attributable to copper sequestration alone. Subsequent biochemical characterizations reveal that the copper-yersiniabactin complexes catalyze superoxide dismutation according to [Equations (6) and (7)]:
| (6) |
| (7) |
Copper-yersiniabactin confined within the phagolysosome may thus greatly diminish concentrations of superoxide (a reductant), while maintaining or increasing production of hydrogen peroxide (an oxidant). This may have the effect of minimizing reduced Cu+ concentrations while increasing oxidized—and less toxic—Cu2+ ion concentrations. Periplasmic Cu,Zn-SOD may similarly protect against copper stress, although there are distinctive pathogenic advantages to deploying a non-protein catalyst such as copper-yersiniabactin in the phagosomal microenvironment (Chaturvedi et al., 2013). Yersiniabactin may synergize with CueO and other mixed copper oxidases by binding Cu2+ product ions generated by these enzymes to form catalytic copper-yersiniabactin. While interactions such as these will require further experimental validation, they fit with an overall paradigm in which pathogens appear able to convert host-supplied copper into catalysts (mixed copper oxidases, copper-yersiniabactin, Cu,Zn-SOD) that help resist copper toxicity. SOD activity may promote bacterial survival in several pathologically important host niches and its connection with copper suggests new insights into host defense mechanisms that are critical to infection pathogenesis.
Prospects
Much remains to be understood about the mechanisms by which mammalian hosts deploy copper to resist infection, and how pathogenic bacteria respond to these strategies. ATP7A's emerging role in direct antibacterial immunity warrants its detailed study in mammalian cells that encounter bacterial pathogens. Cell type, pathogen, and regulatory activity may result in unforeseen interactions between copper and other innate immune effector molecules. Possible cooperation with mammalian copper absorption and trafficking may suggest routes by which copper-based immunity could be therapeutically supported. Both basic and translational research efforts will be necessary to understand these details.
The mechanisms by which pathogenic bacteria resist copper during mammalian infections merits further investigation. Studies conducted in bacterial cultures with environmental and pathogenic isolates provide an excellent starting point for infection models that may provide additional insights. The recent finding that yersiniabactin, a virulence-associated siderophore in E. coli binds copper during humans infections (Chaturvedi et al., 2012) and promotes microbial survival in phagocytic cells suggests that host microenvironments may reveal new copper resistance strategies (Chaturvedi et al., 2013). Yersiniabactin exemplifies the rich array of microbial secondary compounds that may include other copper-detoxifying microbial products. Metabolomic approaches, which are sensitive to the end products of multi-gene biosynthetic units, are well suited to discover additional copper-binding secondary compounds.
Copper's inherent toxicity has renewed interest in its use as an antimicrobial. Three hundred different copper and copper alloy surfaces are registered with the U.S. Environmental Protection Agency as antimicrobials and trials are underway to determine whether copper treated surfaces can significantly reduce nosocomial infections (http://www.epa.gov/pesticides/factsheets/copper-alloy-products.htm) (Grass et al., 2011). While these approaches may be useful in limiting nosocomial infections, it is worth noting that environmental copper-resistance loci have been isolated from Gram-negative bacteria that colonize agricultural areas repeatedly treated with copper salts. Given the linkage between copper resistance and virulence, it would be worth knowing whether sublethal copper exposures might effectively select for increased virulence in bacteria. Improved insight into bacterial copper resistance mechanisms in vivo and in environmental settings will be necessary to optimize antimicrobial uses of copper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Achard M. E., Stafford S. L., Bokil N. J., Chartres J., Bernhardt P. V., Schembri M. A., et al. (2012). Copper redistribution in murine macrophages in response to Salmonella infection. Biochem. J. 444, 51–57 10.1042/BJ20112180 [DOI] [PubMed] [Google Scholar]
- Achard M. E., Tree J. J., Holden J. A., Simpfendorfer K. R., Wijburg O. L., Strugnell R. A., et al. (2010). The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect. Immun. 78, 2312–2319 10.1128/IAI.01208-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adaikkalam V., Swarup S. (2005). Characterization of copABCD operon from a copper-sensitive Pseudomonas putida strain. Can. J. Microbiol. 51, 209–216 10.1139/w04-135 [DOI] [PubMed] [Google Scholar]
- Andoy N. M., Sarkar S. K., Wang Q., Panda D., Benitez J. J., Kalininskiy A., et al. (2009). Single-molecule study of metalloregulator CueR-DNA interactions using engineered Holliday junctions. Biophys. J. 97, 844–852 10.1016/j.bpj.2009.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C., Banci L., Bertini I., Rosato A. (2008). Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J. Proteome Res. 7, 209–216 10.1021/pr070480u [DOI] [PubMed] [Google Scholar]
- Arceneaux J. E., Boutwell M. E., Byers B. R. (1984). Enhancement of copper toxicity by siderophores in Bacillus megaterium. Antimicrob. Agents Chemother. 25, 650–652 10.1128/AAC.25.5.650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello J. M., Eren E., Gonzalez-Guerrero M. (2007). The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248 10.1007/s10534-006-9055-6 [DOI] [PubMed] [Google Scholar]
- Arnesano F., Banci L., Bertini I., Felli I. C., Luchinat C., Thompsett A. R. (2003a). A strategy for the NMR characterization of type II copper(II) proteins: the case of the copper trafficking protein CopC from Pseudomonas syringae. J. Am. Chem. Soc. 125, 7200–7208 10.1021/ja034112c [DOI] [PubMed] [Google Scholar]
- Arnesano F., Banci L., Bertini I., Mangani S., Thompsett A. R. (2003b). A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Natl. Acad. Sci. U.S.A. 100, 3814–3819 10.1073/pnas.0636904100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula P., Masarik M., Adam V., Eckschlager T., Stiborova M., Trnkova L., et al. (2012). Mammalian metallothioneins: properties and functions. Metallomics 4, 739–750 10.1039/c2mt20081c [DOI] [PubMed] [Google Scholar]
- Bagai I., Rensing C., Blackburn N. J., McEvoy M. M. (2008). Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47, 11408–11414 10.1021/bi801638m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Cooksey D. A. (1987). Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 169, 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I., Gray H., Steifel E., Valentine J. S. (2007). Biological Inorganic Chemistry: Structure and Reactivity, Sausalito, CA: University Science Books [Google Scholar]
- Beswick P. H., Hall G. H., Hook A. J., Little K., McBrien D. C., Lott K. A. (1976). Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem. Biol. Interact. 14, 347–356 10.1016/0009-2797(76)90113-7 [DOI] [PubMed] [Google Scholar]
- Beveridge S. J., Garrett I. R., Whitehouse M. W., Vernon-Roberts B., Brooks P. M. (1985). Biodistribution of 64Cu in inflamed rats following administration of two anti-inflammatory copper complexes. Agents Actions 17, 104–111 10.1007/BF01966692 [DOI] [PubMed] [Google Scholar]
- Borkow G. (2005). Copper as a biocidal tool. Curr. Med. Chem. 12, 2163–2175 10.2174/0929867054637617 [DOI] [PubMed] [Google Scholar]
- Boyne R., Arthur J. R. (1981). Effects of selenium and copper deficiency on neutrophil function in cattle. J. Comp. Pathol. 91, 271–276 10.1016/0021-9975(81)90032-3 [DOI] [PubMed] [Google Scholar]
- Brandel J., Humbert N., Elhabiri M., Schalk I. J., Mislin G. L., Albrecht-Gary A. M. (2012). Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans. 41, 2820–2834 10.1039/c1dt11804h [DOI] [PubMed] [Google Scholar]
- Brown N. L., Barrett S. R., Camakaris J., Lee B. T., Rouch D. A. (1995). Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17, 1153–1166 10.1111/j.1365-2958.1995.mmi_17061153.x [DOI] [PubMed] [Google Scholar]
- Brown N. L., Rouch D. A., Lee B. T. (1992). Copper resistance determinants in bacteria. Plasmid 27, 41–51 10.1016/0147-619X(92)90005-U [DOI] [PubMed] [Google Scholar]
- Brown N. L., Stoyanov J. V., Kidd S. P., Hobman J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163 10.1016/S0168-6445(03)00051-2 [DOI] [PubMed] [Google Scholar]
- Bull P. C., Thomas G. R., Rommens J. M., Forbes J. R., Cox D. W. (1993). The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat. Genet. 5, 327–337 10.1038/ng1293-327 [DOI] [PubMed] [Google Scholar]
- Burq V. (1867). Du cuivre contre le cholera: de l'immunite acquise par les ouvriers en cuivre par rapport au Cholera. Paris: G. Bailiere [Google Scholar]
- Cervantes C., Gutierrez-Corona F. (1994). Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol. Rev. 14, 121–137 10.1111/j.1574-6976.1994.tb00083.x [DOI] [PubMed] [Google Scholar]
- Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., et al. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 10.1126/science.1085950 [DOI] [PubMed] [Google Scholar]
- Chaturvedi K. S., Hung C. S., Crowley J. R., Stapleton A. E., Henderson J. P. (2012). The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736 10.1038/nchembio.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi K. S., Hung C. S., Giblin D. E., Urushidani S., Austin A. M., Dinauer M. C., et al. (2013). Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol. [Epub ahead of print]. 10.1021/cb400658k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelly J., Tumer Z., Tonnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., et al. (1993). Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet. 3, 14–19 10.1038/ng0193-14 [DOI] [PubMed] [Google Scholar]
- Chen K., Yuldasheva S., Penner-Hahn J. E., O'Halloran T. V. (2003). An atypical linear Cu(I)-S2 center constitutes the high-affinity metal-sensing site in the CueR metalloregulatory protein. J. Am. Chem. Soc. 125, 12088–12089 10.1021/ja036070y [DOI] [PubMed] [Google Scholar]
- Chiarla C., Giovannini I., Siegel J. H. (2008). Patterns of correlation of plasma ceruloplasmin in sepsis. J. Surg. Res. 144, 107–110 10.1016/j.jss.2007.03.024 [DOI] [PubMed] [Google Scholar]
- Chillappagari S., Miethke M., Trip H., Kuipers O. P., Marahiel M. A. (2009). Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 191, 2362–2370 10.1128/JB.01616-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. E., Stuart J., Sanders-Loehr J. (1987). Induction of siderophore activity in Anabaena spp. and its moderation of copper toxicity. Appl. Environ. Microbiol. 53, 917–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton R. F., Matthews D. M., Poisner R. (1965). Observations on the mechanism of absorption of copper by the small intestine. J. Physiol. 178, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton R. R., Pierre J. L. (2001). Old iron, young copper: from Mars to Venus. Biometals 14, 99–112 10.1023/A:1016710810701 [DOI] [PubMed] [Google Scholar]
- Crocker A., Lee C., Aboko-Cole G., Durham C. (1992). Interaction of nutrition and infection: effect of copper deficiency on resistance to Trypanosoma lewisi. J. Natl. Med. Assoc. 84, 697–706 [PMC free article] [PubMed] [Google Scholar]
- Danks D. M., Campbell P. E., Walker-Smith J., Stevens B. J., Gillespie J. M., Blomfield J., et al. (1972). Menkes' kinky-hair syndrome. Lancet 1, 1100–1102 10.1016/S0140-6736(72)91433-X [DOI] [PubMed] [Google Scholar]
- Delmar J. A., Su C. C., Yu E. W. (2013). Structural mechanisms of heavy-metal extrusion by the Cus efflux system. Biometals 26, 593–607 10.1007/s10534-013-9628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko K. Y., Franiek J. A., Edwards J. L., Falsetta M. L., Kidd S. P., Potter A. J., et al. (2012). Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect. Immun. 80, 1065–1071 10.1128/IAI.06163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko K. Y., Xiao Z., Wedd A. G. (2008). Copper resistance in E. coli: the multicopper oxidase PcoA catalyzes oxidation of copper(I) in Cu(I)Cu(II)-PcoC. Chembiochem 9, 1579–1582 10.1002/cbic.200800100 [DOI] [PubMed] [Google Scholar]
- Dollwet H., Sorenson J. R. J. (1985). Historic uses of copper compounds in medicine. Trace Elem. Med. 2, 80–87 [Google Scholar]
- Dupont C. L., Grass G., Rensing C. (2011). Copper toxicity and the origin of bacterial resistance–new insights and applications. Metallomics 3, 1109–1118 10.1039/c1mt00107h [DOI] [PubMed] [Google Scholar]
- Espariz M., Checa S. K., Audero M. E., Pontel L. B., Soncini F. C. (2007). Dissecting the Salmonella response to copper. Microbiology 153, 2989–2997 10.1099/mic.0.2007/006536-0 [DOI] [PubMed] [Google Scholar]
- Fan B., Rosen B. P. (2002). Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J. Biol. Chem. 277, 46987–46992 10.1074/jbc.M208490200 [DOI] [PubMed] [Google Scholar]
- Festa R. A., Jones M. B., Butler-Wu S., Sinsimer D., Gerads R., Bishai W. R., et al. (2011). A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 10.1111/j.1365-2958.2010.07431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney L. A., O'Halloran T. V. (2003). Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300, 931–936 10.1126/science.1085049 [DOI] [PubMed] [Google Scholar]
- Frangipani E., Slaveykova V. I., Reimmann C., Haas D. (2008). Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J. Bacteriol. 190, 6706–6717 10.1128/JB.00450-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S., Grass G., Rensing C., Nies D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185, 3804–3812 10.1128/JB.185.13.3804-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frausto da Silva J. J. R., Williams R. (1993). The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford: Oxford University Press [Google Scholar]
- Freinbichler W., Colivicchi M. A., Stefanini C., Bianchi L., Ballini C., Misini B., et al. (2011). Highly reactive oxygen species: detection, formation, and possible functions. Cell. Mol. Life Sci. 68, 2067–2079 10.1007/s00018-011-0682-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. (1974). Superoxide dismutases. Adv. Enzymol. Relat. Areas Mol. Biol. 41, 35–97 [DOI] [PubMed] [Google Scholar]
- Gold B., Deng H., Bryk R., Vargas D., Eliezer D., Roberts J., et al. (2008). Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 4, 609–616 10.1038/nchembio.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanz E., Kadlec K., Fessler A. T., Zarazaga M., Torres C., Schwarz S. (2013). Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob. Agents Chemother. 57, 3275–3282 10.1128/AAC.00171-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M., Raimunda D., Cheng X., Arguello J. M. (2010). Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78, 1246–1258 10.1111/j.1365-2958.2010.07402.x [DOI] [PubMed] [Google Scholar]
- Gordon S. (2008). Elie Metchnikoff: father of natural immunity. Eur. J. Immunol. 38, 3257–3264 10.1002/eji.200838855 [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C. (2001). CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286, 902–908 10.1006/bbrc.2001.5474 [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C., Solioz M. (2011). Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77, 1541–1547 10.1128/AEM.02766-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G., Thakali K., Klebba P. E., Thieme D., Muller A., Wildner G. F., et al. (2004). Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186, 5826–5833 10.1128/JB.186.17.5826-5833.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N., Kehl-Fie T. E., Ma Z., Adams K. W., Cowart D. M., Scott R. A., et al. (2011). Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 10.1074/jbc.M111.220012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumulec J., Masarik M., Krizkova S., Hlavna M., Babula P., Hrabec R., et al. (2012). Evaluation of alpha-methylacyl-CoA racemase, metallothionein and prostate specific antigen as prostate cancer prognostic markers. Neoplasma 59, 191–201 10.4149/neo_2012_025 [DOI] [PubMed] [Google Scholar]
- Gunn T. R., MacFarlane S., Phillips L. I. (1984). Difficulties in the neonatal diagnosis of Menkes' kinky hair syndrome–trichopoliodystrophy. Clin. Pediatr. 23, 514–516 10.1177/000992288402300915 [DOI] [PubMed] [Google Scholar]
- Guo Y., Smith K., Lee J., Thiele D. J., Petris M. J. (2004). Identification of methionine-rich clusters that regulate copper-stimulated endocytosis of the human Ctr1 copper transporter. J. Biol. Chem. 279, 17428–17433 10.1074/jbc.M401493200 [DOI] [PubMed] [Google Scholar]
- Gupta A., Matsui K., Lo J. F., Silver S. (1999). Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5, 183–188 10.1038/5545 [DOI] [PubMed] [Google Scholar]
- Hamza I., Faisst A., Prohaska J., Chen J., Gruss P., Gitlin J. D. (2001). The metallochaperone Atox1 plays a critical role in perinatal copper homeostasis. Proc. Natl. Acad. Sci. U.S.A. 98, 6848–6852 10.1073/pnas.111058498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y., Clasen S., Hasan N. M., Barry A. N., Lutsenko S. (2012). Functional partnership of the copper export machinery and glutathione balance in human cells. J. Biol. Chem. 287, 26678–26687 10.1074/jbc.M112.381178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig K., Bleuel C., Krauss G. J., Nies D. H. (2008). Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190, 5431–5438 10.1128/JB.00271-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C. G., Laurell C. B. (1948). Investigations in serum copper. II. Isolation of the copper containing protein and a description of some of its properties. Acta Chem. Scand. 2, 550–556 10.3891/acta.chem.scand.02-0550 [DOI] [Google Scholar]
- Huffman D. L., Huyett J., Outten F. W., Doan P. E., Finney L. A., Hoffman B. M., et al. (2002). Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 41, 10046–10055 10.1021/bi0259960 [DOI] [PubMed] [Google Scholar]
- Ilback N. G., Benyamin G., Lindh U., Friman G. (2003). Sequential changes in Fe, Cu, and Zn in target organs during early Coxsackievirus B3 infection in mice. Biol. Trace Elem. Res. 91, 111–124 10.1385/BTER:91:2:111 [DOI] [PubMed] [Google Scholar]
- Jones D. G., Suttle N. F. (1983). The effect of copper deficiency on the resistance of mice to infection with Pasteurella haemolytica. J. Comp. Pathol. 93, 143–149 10.1016/0021-9975(83)90052-X [DOI] [PubMed] [Google Scholar]
- Jones P. W., Taylor D., Williams D. R., Finney M., Iorwerth A., Webster D., et al. (2001). Using wound fluid analyses to identify trace element requirements for efficient healing. J. Wound Care 10, 205–208 [DOI] [PubMed] [Google Scholar]
- Jozefczak M., Remans T., Vangronsveld J., Cuypers A. (2012). Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 13, 3145–3175 10.3390/ijms13033145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaim W., Rall J. (1996). Copper–a ‘modern’ bioelement. Angew. Chem. Int. Ed. Engl. 35, 43–60 10.1002/anie.199600431 [DOI] [Google Scholar]
- Kaler S. G. (2011). ATP7A-related copper transport diseases-emerging concepts and future trends. Nat. Rev. Neurol. 7, 15–29 10.1038/nrneurol.2010.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin K. D. (1993). Metalloenzymes, structural motifs, and inorganic models. Science 261, 701–708 10.1126/science.7688141 [DOI] [PubMed] [Google Scholar]
- Kenney G. E., Rosenzweig A. C. (2012). Chemistry and biology of the copper chelator methanobactin. ACS Chem. Biol. 7, 260–268 10.1021/cb2003913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp L. W., Lin S. J., Yuan D. S., Klausner R. D., Culotta V. C., Gitlin J. D. (1997). Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J. Biol. Chem. 272, 9221–9226 10.1074/jbc.272.14.9221 [DOI] [PubMed] [Google Scholar]
- Koller L. D., Mulhern S. A., Frankel N. C., Steven M. G., Williams J. R. (1987). Immune dysfunction in rats fed a diet deficient in copper. Am. J. Clin. Nutr. 45, 997–1006 [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W. (2004). Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 10.1038/nrm1354 [DOI] [PubMed] [Google Scholar]
- La Fontaine S., Ackland M. L., Mercer J. F. (2010). Mammalian copper-transporting P-type ATPases, ATP7A and ATP7B: emerging roles. Int. J. Biochem. Cell Biol. 42, 206–209 10.1016/j.biocel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Grass G., Rensing C., Barrett S. R., Yates C. J., Stoyanov J. V., et al. (2002). The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295, 616–620 10.1016/S0006-291X(02)00726-X [DOI] [PubMed] [Google Scholar]
- Leszczyszyn O. I., Zeitoun-Ghandour S., Sturzenbaum S. R., Blindauer C. A. (2011). Tools for metal ion sorting: in vitro evidence for partitioning of zinc and cadmium in C. elegans metallothionein isoforms. Chem. Commun. 47, 448–450 10.1039/c0cc02188a [DOI] [PubMed] [Google Scholar]
- Linder M. C., Hazegh-Azam M. (1996). Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63, 797S–811S [DOI] [PubMed] [Google Scholar]
- Liochev S. I. (1999). The mechanism of “Fenton-like” reactions and their importance for biological systems. A biologist's view. Met. Ions Biol. Syst. 36, 1–39 [PubMed] [Google Scholar]
- Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., et al. (2007). CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 10.1038/nchembio844 [DOI] [PubMed] [Google Scholar]
- Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., et al. (2010). Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 10.1038/nature09395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L., Imlay J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L., Rensing C., Imlay J. A. (2007). Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189, 1616–1626 10.1128/JB.01357-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani D., Barre O., Gerber S. D., Solioz M. (2008). Characterization of the CopR regulon of Lactococcus lactis IL1403. J. Bacteriol. 190, 536–545 10.1128/JB.01481-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matousek De Abel De La Cruz A. J., Burguera J. L., Burguera M., Anez N. (1993). Changes in the total content of iron, copper, and zinc in serum, heart, liver, spleen, and skeletal muscle tissues of rats infected with Trypanosoma cruzi. Biol. Trace Elem. Res. 37, 51–70 10.1007/BF02789401 [DOI] [PubMed] [Google Scholar]
- Mealman T. D., Bagai I., Singh P., Goodlett D. R., Rensing C., Zhou H., et al. (2011). Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry 50, 2559–2566 10.1021/bi102012j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealman T. D., Blackburn N. J., McEvoy M. M. (2012). Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr. Top. Membr. 69, 163–196 10.1016/B978-0-12-394390-3.00007-0 [DOI] [PubMed] [Google Scholar]
- Menkes J. H., Alter M., Steigleder G. K., Weakley D. R., Sung J. H. (1962). A sex-linked recessive disorder with retardation of growth, peculiar hair, and focal cerebral and cerebellar degeneration. Pediatrics 29, 764–779 [PubMed] [Google Scholar]
- Mercer J. F., Livingston J., Hall B., Paynter J. A., Begy C., Chandrasekharappa S., et al. (1993). Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat. Genet. 3, 20–25 10.1038/ng0193-20 [DOI] [PubMed] [Google Scholar]
- Mermod M., Magnani D., Solioz M., Stoyanov J. V. (2012). The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals 25, 33–43 10.1007/s10534-011-9510-x [DOI] [PubMed] [Google Scholar]
- Munson G. P., Lam D. L., Outten F. W., O'Halloran T. V. (2000). Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182, 5864–5871 10.1128/JB.182.20.5864-5871.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesha R. K., Natesha R., Victory D., Barnwell S. P., Hoover E. L. (1992). A prognostic role for ceruloplasmin in the diagnosis of indolent and recurrent inflammation. J. Natl. Med. Assoc. 84, 781–784 [PMC free article] [PubMed] [Google Scholar]
- Nevitt T., Ohrvik H., Thiele D. J. (2012). Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 1823, 1580–1593 10.1016/j.bbamcr.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberne P. M., Hunt C. E., Young V. R. (1968). The role of diet and the reticuloendothelial system in the response of rats to Salmonella typhilmurium infection. Br. J. Exp. Pathol. 49, 448–457 [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Silver S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199 10.1007/BF01569902 [DOI] [PubMed] [Google Scholar]
- Odermatt A., Krapf R., Solioz M. (1994). Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem. Biophys. Res. Commun. 202, 44–48 10.1006/bbrc.1994.1891 [DOI] [PubMed] [Google Scholar]
- Odermatt A., Solioz M. (1995). Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 270, 4349–4354 10.1074/jbc.270.9.4349 [DOI] [PubMed] [Google Scholar]
- Osman D., Cavet J. S. (2008). Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65, 217–247 10.1016/S0065-2164(08)00608-4 [DOI] [PubMed] [Google Scholar]
- Osman D., Cavet J. S. (2011). Metal sensing in Salmonella: implications for pathogenesis. Adv. Microb. Physiol. 58, 175–232 10.1016/B978-0-12-381043-4.00005-2 [DOI] [PubMed] [Google Scholar]
- Osman D., Waldron K. J., Denton H., Taylor C. M., Grant A. J., Mastroeni P., et al. (2010). Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285, 25259–25268 10.1074/jbc.M110.145953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterberg R., Ligaarden R., Persson D. (1979). Copper(I) complexes of penicillamine and glutathione. J. Inorg. Biochem. 10, 341–355 10.1016/S0162-0134(00)80200-7 [DOI] [PubMed] [Google Scholar]
- Outten C. E., O'Halloran T. V. (2001). Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 10.1126/science.1060331 [DOI] [PubMed] [Google Scholar]
- Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677 10.1074/jbc.M104122200 [DOI] [PubMed] [Google Scholar]
- Outten F. W., Outten C. E., Hale J., O'Halloran T. V. (2000). Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275, 31024–31029 10.1074/jbc.M006508200 [DOI] [PubMed] [Google Scholar]
- Paulsen I. T., Brown M. H., Skurray R. A. (1996). Proton-dependent multidrug efflux systems. Microbiol. Rev. 60, 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peariso K., Huffman D. L., Penner-Hahn J. E., O'Halloran T. V. (2003). The PcoC copper resistance protein coordinates Cu(I) via novel S-methionine interactions. J. Am. Chem. Soc. 125, 342–343 10.1021/ja028935y [DOI] [PubMed] [Google Scholar]
- Pena M. M., Lee J., Thiele D. J. (1999). A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129, 1251–1260 [DOI] [PubMed] [Google Scholar]
- Petersen C., Moller L. B. (2000). Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene 261, 289–298 10.1016/S0378-1119(00)00509-6 [DOI] [PubMed] [Google Scholar]
- Petris M. J., Smith K., Lee J., Thiele D. J. (2003). Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J. Biol. Chem. 278, 9639–3646 10.1074/jbc.M209455200 [DOI] [PubMed] [Google Scholar]
- Pontel L. B., Audero M. E., Espariz M., Checa S. K., Soncini F. C. (2007). GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol. Microbiol. 66, 814–825 10.1111/j.1365-2958.2007.05963.x [DOI] [PubMed] [Google Scholar]
- Pontel L. B., Soncini F. C. (2009). Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 73, 212–225 10.1111/j.1365-2958.2009.06763.x [DOI] [PubMed] [Google Scholar]
- Portmann R., Poulsen K. R., Wimmer R., Solioz M. (2006). CopY-like copper inducible repressors are putative ‘winged helix’ proteins. Biometals 19, 61–70 10.1007/s10534-005-5381-3 [DOI] [PubMed] [Google Scholar]
- Prohaska J. R., Lukasewycz O. A. (1990). Effects of copper deficiency on the immune system. Adv. Exp. Med. Biol. 262, 123–143 10.1007/978-1-4613-0553-8_11 [DOI] [PubMed] [Google Scholar]
- Qin Z., Konaniah E. S., Neltner B., Nemenoff R. A., Hui D. Y., Weintraub N. L. (2010). Participation of ATP7A in macrophage mediated oxidation of LDL. J. Lipid Res. 51, 1471–1477 10.1194/jlr.M003426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher C., Masepohl B. (2012). Copper-responsive gene regulation in bacteria. Microbiology 158, 2451–2464 10.1099/mic.0.058487-0 [DOI] [PubMed] [Google Scholar]
- Rauser W. E. (1999). Structure and function of metal chelators produced by plants: the case for organic acids, amino acids, phytin, and metallothioneins. Cell Biochem. Biophys. 31, 19–48 10.1007/BF02738153 [DOI] [PubMed] [Google Scholar]
- Rensing C., Fan B., Sharma R., Mitra B., Rosen B. P. (2000). CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 652–656 10.1073/pnas.97.2.652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Jara A., Latorre M., Lopez G., Bourgogne A., Murray B. E., Cambiazo V., et al. (2010). Genome-wide transcriptome analysis of the adaptive response of Enterococcus faecalis to copper exposure. Biometals 23, 1105–1112 10.1007/s10534-010-9356-7 [DOI] [PubMed] [Google Scholar]
- Roberts S. A., Weichsel A., Grass G., Thakali K., Hazzard J. T., Tollin G., et al. (2002). Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 2766–2771 10.1073/pnas.052710499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland J. L., Niederweis M. (2012). Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis 92, 202–210 10.1016/j.tube.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland J. L., Niederweis M. (2013). A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J. Bacteriol. 195, 3724–3733 10.1128/JB.00546-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L., Linkevicius M., Lytsy B., Melhus A., Andersson D. I. (2012). Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67, 74–83 10.1093/jac/dkr405 [DOI] [PubMed] [Google Scholar]
- Shafeeq S., Yesilkaya H., Kloosterman T. G., Narayanan G., Wandel M., Andrew P. W., et al. (2011). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270 10.1111/j.1365-2958.2011.07758.x [DOI] [PubMed] [Google Scholar]
- Shi J., Lindsay W. P., Huckle J. W., Morby A. P., Robinson N. J. (1992). Cyanobacterial metallothionein gene expressed in Escherichia coli. Metal-binding properties of the expressed protein. FEBS Lett. 303, 159–163 10.1016/0014-5793(92)80509-F [DOI] [PubMed] [Google Scholar]
- Shoeb E., Badar U., Akhter J., Shams H., Sultana M., Ansari M. A. (2012). Horizontal gene transfer of stress resistance genes through plasmid transport. World J. Microbiol. Biotechnol. 28, 1021–1025 10.1007/s11274-011-0900-6 [DOI] [PubMed] [Google Scholar]
- Singh S. K., Grass G., Rensing C., Montfort W. R. (2004). Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 186, 7815–7817 10.1128/JB.186.22.7815-7817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. D., Botero S., Levander O. A. (2008). Copper deficiency increases the virulence of amyocarditic and myocarditic strains of coxsackievirus B3 in mice. J. Nutr. 138, 849–855 [DOI] [PubMed] [Google Scholar]
- Stadtman E. R. (2006). Protein oxidation and aging. Free Radic. Res. 40, 1250–1258 10.1080/10715760600918142 [DOI] [PubMed] [Google Scholar]
- Stoyanov J. V., Hobman J. L., Brown N. L. (2001). CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39, 502–511 10.1046/j.1365-2958.2001.02264.x [DOI] [PubMed] [Google Scholar]
- Stoyanov J. V., Magnani D., Solioz M. (2003). Measurement of cytoplasmic copper, silver, and gold with a lux biosensor shows copper and silver, but not gold, efflux by the CopA ATPase of Escherichia coli. FEBS Lett. 546, 391–394 10.1016/S0014-5793(03)00640-9 [DOI] [PubMed] [Google Scholar]
- Strausak D., Solioz M. (1997). CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J. Biol. Chem. 272, 8932–8936 10.1074/jbc.272.14.8932 [DOI] [PubMed] [Google Scholar]
- Sullivan J. L., Ochs H. D. (1978). Copper deficiency and the immune system. Lancet 2, 686 10.1016/S0140-6736(78)92806-4 [DOI] [PubMed] [Google Scholar]
- Tanzi R. E., Petrukhin K., Chernov I., Pellequer J. L., Wasco W., Ross B., et al. (1993). The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat. Genet. 5, 344–350 10.1038/ng1293-344 [DOI] [PubMed] [Google Scholar]
- Teitzel G. M., Geddie A., De Long S. K., Kirisits M. J., Whiteley M., Parsek M. R. (2006). Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188, 7242–7256 10.1128/JB.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetaz T. J., Luke R. K. (1983). Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 154, 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaden J. T., Lory S., Gardner T. S. (2010). Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa. J. Bacteriol. 192, 2557–2568 10.1128/JB.01528-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumoorthy N., Shyam Sunder A., Manisenthil Kumar K., Senthil Kumar M., Ganesh G., Chatterjee M. (2011). A review of metallothionein isoforms and their role in pathophysiology. World J. Surg. Oncol. 9, 54 10.1186/1477-7819-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigui H., Dudyk P., Sum J., Shuman H. A., Faucher S. P. (2013). Analysis of the transcriptome of Legionella pneumophila hfq mutant reveals a new mobile genetic element. Microbiology 159(Pt 8), 1649–1660 10.1099/mic.0.067983-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufft L. S., Nockels C. F., Fettman M. J. (1988). Effects of Escherichia coli on iron, copper, and zinc metabolism in chicks. Avian Dis. 32, 779–786 10.2307/1590998 [DOI] [PubMed] [Google Scholar]
- Vancampen D. R., Mitchell E. A. (1965). Absorption of Cu-64, Zn-65, Mo-99, and Fe-59 from ligated segments of the rat gastrointestinal tract. J. Nutr. 86, 120–124 [DOI] [PubMed] [Google Scholar]
- Vats N., Lee S. F. (2001). Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiology 147, 653–662 [DOI] [PubMed] [Google Scholar]
- Voloudakis A. E., Bender C. L., Cooksey D. A. (1993). Similarity between copper resistance genes from Xanthomonas campestris and Pseudomonas syringae. Appl. Environ. Microbiol. 59, 1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rozycki T., Nies D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96, 115–139 10.1007/s10482-008-9284-5 [DOI] [PubMed] [Google Scholar]
- Voruganti V. S., Klein G. L., Lu H. X., Thomas S., Freeland-Graves J. H., Herndon D. N. (2005). Impaired zinc and copper status in children with burn injuries: need to reassess nutritional requirements. Burns 31, 711–716 10.1016/j.burns.2005.04.026 [DOI] [PubMed] [Google Scholar]
- Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. (1993). Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat. Genet. 3, 7–13 10.1038/ng0193-7 [DOI] [PubMed] [Google Scholar]
- Vulpe C. D., Kuo Y. M., Murphy T. L., Cowley L., Askwith C., Libina N., et al. (1999). Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21, 195–199 10.1038/5979 [DOI] [PubMed] [Google Scholar]
- Wagner D., Maser J., Lai B., Cai Z., Barry C. E., 3rd., Honer Zu Bentrup K., et al. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174, 1491–1500 [DOI] [PubMed] [Google Scholar]
- Waidner B., Melchers K., Stahler F. N., Kist M., Bereswill S. (2005). The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J. Bacteriol. 187, 4683–4688 10.1128/JB.187.13.4683-4688.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hodgkinson V., Zhu S., Weisman G. A., Petris M. J. (2011). Advances in the understanding of mammalian copper transporters. Adv. Nutr. 2, 129–137 10.3945/an.110.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. K., Hoye E. A., Talaat A. M. (2008). The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 10.1128/JB.01847-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C., Lee J., Kambe T., Fritsche K., Petris M. J. (2009). A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956 10.1074/jbc.M109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. R., Morgan A. G., Rouch D. A., Brown N. L., Lee B. T. (1993). Copper-resistant enteric bacteria from United Kingdom and Australian piggeries. Appl. Environ. Microbiol. 59, 2531–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschendorf F., Ackart D., Shrestha T. B., Hascall-Dove L., Nolan S., Lamichhane G., et al. (2011). Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 1621–1626 10.1073/pnas.1009261108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F. F., Imlay J. A. (2012). Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 78, 3614–3621 10.1128/AEM.07368-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Davis A. V., Balakrishnan G., Stasser J. P., Staehlin B. M., Focia P., et al. (2008). Cu(I) recognition via cation-pi and methionine interactions in CusF. Nat. Chem. Biol. 4, 107–109 10.1038/nchembio.2007.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Ishihama A. (2005). Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56, 215–227 10.1111/j.1365-2958.2005.04532.x [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Ishihama A. (2006). Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70, 1688–1695 10.1271/bbb.60024 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Furuta S., Niki E. (1993). Effects of metal chelating agents on the oxidation of lipids induced by copper and iron. Biochim. Biophys. Acta 1210, 81–88 10.1016/0005-2760(93)90052-B [DOI] [PubMed] [Google Scholar]
- Zhou B., Gitschier J. (1997). hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 10.1073/pnas.94.14.7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Udagedara S. R., Sze C. M., Ryan T. M., Howlett G. J., Xiao Z., et al. (2012). PcoE–a metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J. Inorg. Biochem. 115, 186–197 10.1016/j.jinorgbio.2012.04.009 [DOI] [PubMed] [Google Scholar]