Abstract
Saliva of bloodsucking animals contains dozens to hundreds of proteins that counteract their hosts’ hemostasis, inflammation, and immunity. It was previously observed that salivary proteins involved in hematophagy are much more divergent in their primary sequence than those of housekeeping function, when comparisons were made between closely related organisms. While this pattern of evolution could result from relaxed selection or drift, it could alternatively be the result of positive selection driven by the intense pressure of the host immune system. We investigated the polymorphism of five different genes associated with blood feeding in the mosquito Anopheles gambiae and obtained evidence in four genes for sites with signatures of positive selection. These results add salivary gland genes from bloodsucking arthropods to the small list of genes driven by positive selection.
Keywords: Evolution, hematophagy, salivary glands
Introduction
The neutral theory of evolution “holds that at the molecular level most evolutionary change and most of the variability within a species are caused not by selection but by random drift of mutant genes that are selectively equivalent.” Accordingly, “for each protein the rate of evolution in terms of amino acid substitutions per year is approximately constant and about the same in various lineages” (Kimura 1979). Against this pervasive random drift, positive and negative selection occurs. While negative selection removes deleterious mutations, positive (Darwinian) selection selects beneficial ones. The neutral theory also states that “molecules or parts of a molecule subjected to a relatively small degree of functional constraint evolve at a higher rate than those subjected to stronger constraints” because the latter is under a higher rate of negative selection. Positive selection is predicted to be a rare phenomenon. Indeed, most of the selection signatures deviating from neutral expectations are found for negative selection. Exceptionally, cases for positive selection are found in genes associated with immunity, mate and self-recognition, or with genes associated with virulence in pathogen recognition (Aguileta et al. 2009; Clark and Swanson 2005; Meslin et al. 2012; Swanson 2003; Tiffin and Moeller 2006; Yang and Swanson 2002). We here present evidence for a new class of positively selected genes, those coding for salivary proteins involved in blood sucking by arthropods, specifically the case for salivary proteins of the malaria vector, Anopheles gambiae.
When searching for blood, mosquitoes penetrate their host’s skin with their proboscis while salivating into the wound. Saliva has antihemostatic components including antiplatelet, vasodilatory, and anticlotting components (Ribeiro et al. 2010). Non-salivating mosquitoes take a much longer period of time to feed, indicating saliva to be important for blood acquisition (Ribeiro et al. 1984), which is needed for egg development. On the other hand, host allergy to mosquito bites may be disruptive to the meal attempt or even fatal to the mosquito due to host behavioral defensive responses (Gillett 1967). It is thus possible that host immune responses may create a fast-evolving scenario for salivary proteins that are involved in blood feeding. Indeed, interspecific comparison of mosquito polypeptides deduced from transcriptome analyses showed that salivary (S) proteins are much more diverse than housekeeping ones (H). In the anophelines An. gambiae–An. stephensi, S proteins show an average identity of 62.4 ± 15.4% versus the 93.1 ± 5.9% found for H proteins (Valenzuela et al. 2003). Similar results were obtained comparing the culicines Ae. aegypti–Ae. albopictus, where 94.0 ± 1.3% (H proteins) and 71.5 ± 1.1% identities (S proteins) were found (Arca et al. 2007).
We hypothesized that the higher divergence of orthologous S proteins in mosquitoes may be the result of the evolutionary pressure of the host immune system on proteins that are essential for blood feeding and, therefore, strongly affect mosquito reproduction/fitness (Ribeiro et al. 2010). In such a scenario, host antibody response to S proteins may favour selection of protein variants with conserved biologic functions but with different antigenic properties. This seems to be the case for the vasodilator maxadilan in different populations of the sand fly Lutzomyia longipalpis (Milleron et al. 2004). It remains to be tested, though, whether the divergence observed among salivary proteins of blood feeding insects is due to relaxed selection and drift, or accelerated by positive selection. (Borghans et al. 2004).
Results and Discussion
No information is presently available on the degree of polymorphism of salivary genes in natural mosquito populations. Therefore, as a first step to test the working hypothesis, we started an analysis of salivary gene polymorphism in a natural An. gambiae population (S form) collected in October 2008 in the village of Soumousso (Bobo-Dioulasso area, Burkina Faso). Five salivary genes—whose expression is either specific or highly enriched in female salivary glands (SGs) and whose functions are known or predicted to be important for blood feeding—were selected. The apyrase gene (Apy, AGAP011971) encodes an ATP-diphosphohydrolase that catalyses the hydrolysis of ATP and ADP to AMP and inorganic phosphate. The physiological role of mosquito salivary apyrase is to facilitate blood feeding by inhibiting ADP-induced platelet recruitment and aggregation. Because the An. gambiae apyrase gene is too long for convenient PCR amplification and sequencing as a single fragment, it was amplified by PCR as three partially overlapping fragments named ApyF1, ApyF2, and ApyF3. The D7-related 2 gene (D7r2, AGAP008282) is a member of the D7 family of proteins, which is widespread among bloodfeeding Nematocera (Ribeiro et al. 2010). In the mosquito An. gambiae, the D7 is a multigene family and the D7r2 protein has been shown to bind with high-affinity biogenic amines (as serotonin and norepinephrine) and proposed to act by antagonizing the vasoconstrictor, platelet-aggregating, and pain-inducing properties of host amines (Calvo et al. 2006). The gSG6 gene (AGAP000150) encodes a small protein whose specific function is still unknown, but it is also involved in hematophagy, as gene silencing by RNAi affects bloodfeeding ability by increasing mosquito probing time (Lombardo et al. 2009). gSG7 (AGAP008216) is highly related to the An. stephensi anophensin, which acts on the kallikrein-kinin system and inhibits bradykinin release (Isawa et al. 2007). In An. gambiae, two members of the gSG7 family—gSG7 and gSG7-2—share 43% identity at the amino acid level and show a tandem arrangement. gVAG (AGAP006421) is a member of the widely spread insect Antigen 5 family with similarity to venom allergens from ants and wasps. The function of the gVAG-encoded protein is unknown, but a member of this family is commonly found in the saliva of bloodfeeding insects (Ribeiro and Arca 2009), and a salivary triatomine member of the family was shown to display anti-oxidant properties (Assumpcao et al. 2013). Because we are using PCR-based mini libraries to obtain sequence information of the above genes, and because PCR may create sequence errors and artifactually increase the degree of polymorphism of these genes, the rpS7 gene (AGAP010592), encoding the ribosomal protein S7 was also chosen as a conserved housekeeping internal control gene. Throughout the text, we will compare our results obtained for the salivary genes to 109 An. gambiae previously reported genes (Cohuet et al. 2008) and to other published data as indicated in the relevant sections. The chromosomal location of the above mentioned genes and some relevant features of the PCR amplified fragments are summarized in Table 1. The salivary genes analyzed here do not fall in the genomic islands of speciation as initially identified by microarray studies (Turner et al. 2005) and then extended by genome-wide scans (Lawniczak et al. 2010).
Table 1.
Features of the amplified PCR fragments
| Gene | Location | Size | Coding | Non-coding | 5′-flank | 5′-UTR | Intron | 3′-UTR | 3′-flank |
|---|---|---|---|---|---|---|---|---|---|
| ApyF1 | 3L:45A | 866 | 579 (3) | 287 | 131 | 17 | 139 (2) | — | — |
| ApyF2 | 3L:45A | 867 | 698 (3) | 169 | — | — | 169 (2) | — | — |
| ApyF3 | 3L:45A | 800 | 610 (2) | 190 | — | — | 76 (1) | 114 | — |
| D7r2 | 3R:30C | 1058 | 507 (2) | 551 | 157 | 112 | 149 (2) | 70 | 63 |
| gSG6 | X:4B | 836 | 348 (1) | 488 | 52 | 78 | — | 138 | 220 |
| gSG7 | 3R:30A | 875 | 438 (3) | 437 | 85 | 37 | 172 | 82 | 61 |
| gVAG | 2L:24D | 1204 | 783 (3) | 421 | 59 | 22 | 158 (2) | 126 | 56 |
| rpS7 | 3L:39B | 854 | 579 (3) | 275 | — | 2 | 231 (2) | 42 | — |
For each gene analyzed, the chromosomal location and the size in bp of the amplified fragments, coding and non-coding regions, introns, flankings, and UTRs are shown. Numbers in parentheses refer to number of exons and introns, respectively.
NUCLEOTIDE DIVERSITY
Nucleotide diversity—a measure of the degree of polymorphism within a population and determined as the average number of nucleotide differences per site between any two randomly selected DNA sequences from a sample population (Nei and Li 1979)—was computed for salivary genes and rpS7 by DnaSP (Librado and Rozas 2009), and the results of this analysis are summarized in Table 2.
Table 2.
Nucleotide polymorphism in coding and non-coding regions of salivary genes and rpS7 a
| m | seq | Coding
|
Non-Coding
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sites | S | h (Hd) | π | π/πrpS7 b | syn | dS c | nsyn | dN c | dN/dS | P | sites | S | h (Hd) | π | π/πrpS7 b | πnc/πc | |||
| ApyF1 | 81 | 172 | 504 | 51 | 65 (0.97) | 0.00925 | 3.78 | 35 | 0.02102 | 16 | 0.00279 | 0.13 | ** | 251 | 38 | 60 (0.98) | 0.01696 | 5.19 | 1.83 |
|
| |||||||||||||||||||
| ApyF2 | 63 | 181 | 654 | 54 | 98 (0.99) | 0.00943 | 3.85 | 36 | 0.02331 | 18 | 0.00350 | 0.15 | *** | 169 | 40 | 48 (0.92) | 0.04028 | 12.32 | 4.27 |
|
| |||||||||||||||||||
| ApyF3 | 84 | 150 | 585 | 64 | 74 (0.98) | 0.00783 | 3.20 | 39 | 0.01729 | 25 | 0.00350 | 0.20 | ** | 166 | 54 | 35 (0.84) | 0.03376 | 10.32 | 4.31 |
|
| |||||||||||||||||||
| D7r2 | 74 | 140 | 441 | 53 | 57 (0.97) | 0.01299 | 5.30 | 34 | 0.03773 | 19 | 0.00317 | 0.08 | *** | 262 | 64 | 71 (0.98) | 0.04377 | 13.39 | 3.37 |
|
| |||||||||||||||||||
| gSG6 | 70 | 148 | 261 | 14 | 17 (0.54) | 0.00340 | 1.39 | 9 | 0.00829 | 5 | 0.00148 | 0.18 | ns | 443 | 91 | 109 (0.99) | 0.02402 | 7.35 | 7.06 |
|
| |||||||||||||||||||
| gSG7 | 81 | 163 | 360 | 54 | 65 (0.97) | 0.01280 | 5.22 | 26 | 0.02169 | 28 | 0.00998 | 0.46 | ns | 398 | 71 | 70 (0.98) | 0.01601 | 4.90 | 1.25 |
|
| |||||||||||||||||||
| gVAG | 75 | 149 | 717 | 86 | 89 (0.99) | 0.01744 | 7.12 | 62 | 0.05715 | 24 | 0.00309 | 0.05 | *** | 377 | 105 | 96 (0.99) | 0.03471 | 10.61 | 1.99 |
|
| |||||||||||||||||||
| rpS7 | 77 | 168 | 558 | 19 | 25 (0.72) | 0.00245 | 1.00 | 8 | 0.00875 | 11 | 0.00072 | 0.08 | ns | 231 | 15 | 14 (0.43) | 0.00327 | 1.00 | 1.33 |
|
| |||||||||||||||||||
| Average salivary | — | — | — | — | — | 0.01109 | — | — | 0.02908 | — | 0.00419 | 0.186 | — | — | — | — | 0.02976 | — | — |
|
| |||||||||||||||||||
| Total | — | — | 4080 | 395 | — | — | — | 249 | — | 146 | — | — | — | 2297 | 478 | — | — | — | — |
Number of mosquitoes (m), sequences (seq), sites, polymorphic sites (S), haplotypes (h) with haplotype diversity (Hd), nucleotide diversity (π), nucleotide diversity in salivary genes versus rpS7 (π/πrpS7), number of synonymous (syn) and nonsynonymous (nsyn) polymorphic sites, rates of synonymous (dS) and nonsynonimous (dN) substitutions, ratio dN/dS.
P-value of the two-tailed test of neutral evolution and nucleotide diversity in non-coding versus coding regions (πnc/πc) are reported.
Nucleotide diversity was computed by DnaSP 5.10.01 using the gaps/missing data option (i.e., gaps/missing data were excluded only in pairwise comparisons). Synonymous/nonsynonymous substitutions and Z-test of selection were computed by MEGA5 with the pairwise deletion option and variance estimated by the bootstrap method (ns, not significant, P > 0.05; *, P< 0.05; **, P < 0.01; ***, P < 0.001).
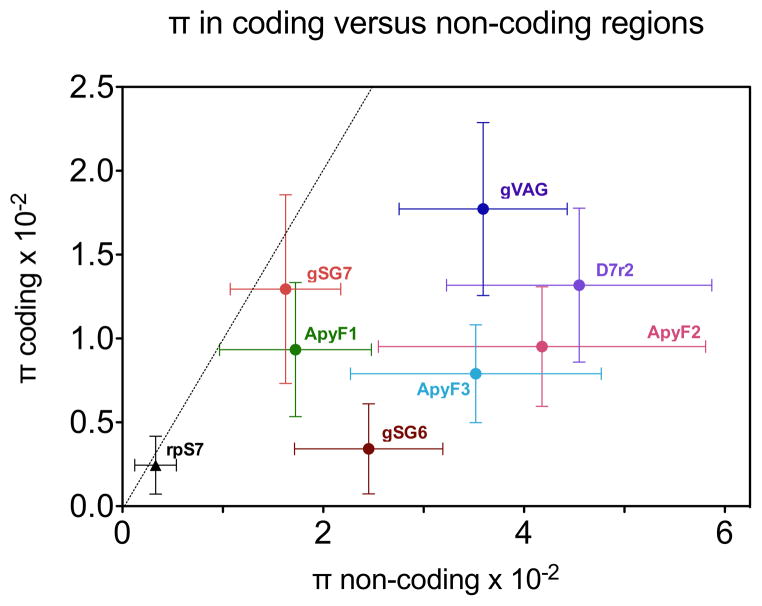
In all cases, nucleotide diversity of salivary genes was higher than rpS7, both in coding (πc) and in non-coding (πnc) regions (see columns π/πS7 in Table 2). gSG6 showed the lowest nucleotide diversity in the coding region, comparable to that of rpS7 (0.0034/0.00245 = 1.39), however, for the other salivary genes, πc was 3.2-to 7.1-fold (ApyF3 and gVAG, respectively) higher as compared with rpS7. A similar situation was found for non-coding regions where salivary genes showed a πnc that was 4.9- to 13.4-fold (gSG7 and D7r2, respectively) higher than rpS7. These differences were in most cases significant (Table 2) and can be visualized in Figure 1, where nucleotide diversity in coding versus non-coding regions with 95% C.I. is reported.
Fig. 1. Nucleotide diversity (π) and 95% CI in coding and non-coding regions.
Nucleotide diversity π and SE were calculated by Mega, using the pairwise deletion option. Diagonal line marks equal diversity of coding and non-coding regions.
As expected according to purifying (negative) selection, diversity in non-coding regions was higher than in coding regions in all studied cases (see column πnc/πc in Table 2). It is noteworthy that among salivary genes gSG6 showed the highest ratio πnc/πc (7.06), suggesting that this gene is under strong purifying selection, which limits diversity in the coding region. This is also indicated by the low gSG6 haplotype diversity (0.54). Notice that this is the only gene studied located in the X chromosome, which is known for overall lower genetic diversity in comparison to autosomes (Cohuet et al. 2008; Wilding et al. 2009). On the other side the lowest πnc/πc ratio was found for gSG7 that exhibits comparable diversity in non-coding versus coding region (0.0160/0.0128 = 1.25); this observation suggests that the encoded protein may evolve at a very fast rate, perhaps also as a consequence of the existence of two different family members, gSG7 and gSG7-2 (Arca et al. 2005).
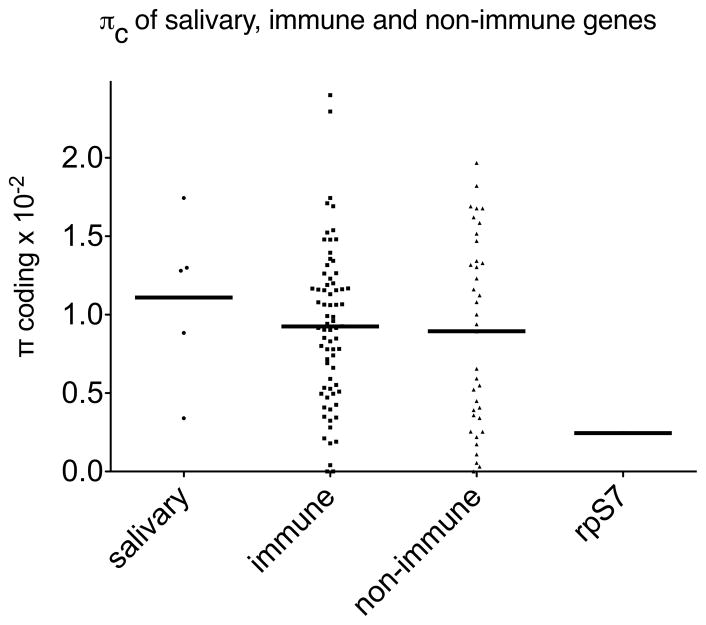
The average nucleotide diversity of salivary genes in coding and non-coding regions was 0.01109 (πc) and 0.02976 (πnc), respectively; that is 4.5 and 9.1 times higher than rpS7. When we compared the average nucleotide diversity in coding regions of salivary genes to corresponding values found for 72 immune and 37 non-immune genes in an An. gambiae S population from Cameroon (Cohuet et al. 2008) we also found higher diversity in the salivary genes, although these differences were not statistically significant (Fig. 2). For the coding regions, this corresponded approximately to 11.1 single-nucleotide polymorphisms (SNPs) per kb for salivary genes, 2.4 SNPs per kb for rpS7, and to 9.2 and 8.9 SNPs per kb for immune and non-immune genes, respectively. A similar value of 7.9 SNPs per kb was also found in another study focused on An. gambiae genes with potential roles in pathogen-vector interactions (Morlais et al. 2004), whereas 7 SNPs per kb were found for a set of 50 Anopheles funestus genes (Wondji et al. 2007). It should be noted that the lower average nucleotide diversity reported by (Morlais et al. 2004) may be connected to the use in this study of An. gambiae laboratory strains rather than natural field populations.
Fig. 2. Average nucleotide diversity in coding regions of An. gambiae salivary, immune and non-immune genes.
Scatter plot comparing mean values of nucleotide diversity of salivary genes and rpS7 analyzed in this study to 72 immune and 37 non-immune genes from an An. gambiae S population from Cameroon (additional files 3 and 4 from Cohuet et al. 2008 (Cohuet et al. 2008)).
The low nucleotide diversity value found for the rpS7 gene (0.0024) is not surprising, as ribosomal proteins are expected to be under strong selective constraints. In Aedes aegypti, rpS11 and rpL31 showed no variation in both coding and non-coding regions (π = 0.0000), whereas rpL17A had πc = 0.0024 and πnc = 0.0096 (Morlais et al. 2003). For other An. gambiae housekeeping genes—such as those encoding β-tubulin (π = 0.0019), integrin (π = 0.0022), and laminin (π = 0.0034)—nucleotide diversity was comparable to the value found here for rpS7 (Morlais et al. 2004). These results for the rpS7 gene indicate that our PCR-based methodology is not artifactually increasing polymorphism.
Overall, a total of 873 SNPs were identified in our study, 395 in coding and 478 in non-coding regions. As expected from the nucleotide diversity results, haplotypes for exons were less numerous for rpS7 and gSG6 (19 and 14, respectively) and more abundant for the remaining SG genes (range: 53–86) (Table 2).
SYNONYMOUS AND NON-SYNONYMOUS SUBSTITUTIONS
For all genes analyzed, the rate of synonymous substitutions (dS) was higher compared with the rate of corresponding non-synonymous substitutions (dN) (i.e., dS > dN, Table 2). On average, salivary genes showed a dS ~3.3-fold higher as compared to rpS7, with gSG6 getting the lowest value—comparable to rpS7—and gVAG with a value ~6.5-fold higher than rpS7. The rate of dN was on average ~5.8-fold higher in salivary genes than in rpS7; gSG6 again showed the lowest value (~2× rpS7) and gSG7 the highest (~13.8× rpS7).
According to the classical view of neutral evolution, the average ratio dN/dS for all codons in a gene should be 1 for neutrally evolving genes, < 1 in negatively selected genes, and > 1 in positively selected genes (Kimura 1977). In all cases, the ratio was < 1. P values obtained from the Z statistics were significantly different from zero for all apyrase coding sequences, as well as for the D7r2 and gVAG exons (Table 2). rpS7, gSG6, and gSG7 exons had dN/dS not significantly different from zero.
We also applied Tajima’s D test to our data. All SG gene models yielded negative values (results not shown) indicative of non-neutral evolution and an excess of low-frequency alleles, but the values were not statistically significant; however, the same was true for rpS7, suggesting this could reflect a population-size expansion, which is the expected situation with our sample that was collected at the end of the rainy season following large (at least 100 ×) population expansion.
Overall, these data confirm the low degree of polymorphism of gSG6, a gene located in the X chromosome, further supporting the idea that this gene is under strong purifying selection. It should also be noted that the low dN/dS ratios found for D7r2 (0.08) and for gVAG (0.05)—two salivary genes with high nucleotide diversity values in both coding and non-coding regions—suggest the existence of strong evolutionary constraints negatively selecting replacement substitutions.
On the other hand, in comparison with the other salivary genes, gSG7 shows the highest dN (3.1–6.7×) and dN/dS ratio (2.5–9.2×) as well as the lowest nucleotide diversity in non-coding regions, suggesting that the gSG7 protein may evolve at a very fast evolutionary rate.
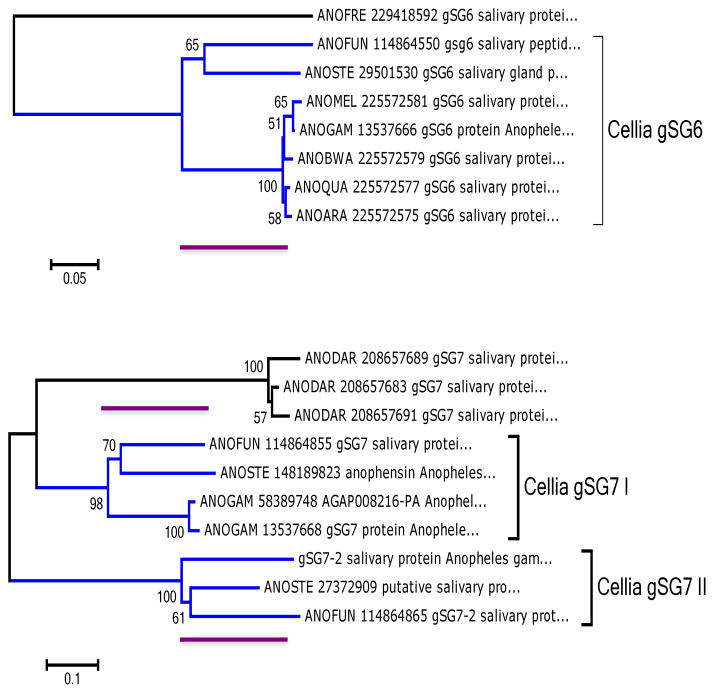
Further support for the evidence of gSG6 being under purifying selection and gSG7 evolving at a very fast rate comes also from the analysis of bootstrapped phylograms for the gSG6 and gSG7 family of proteins (Fig. 3). If gSG6 is under purifying selection, it should not diverge too much within the subgenus Cellia, where we have information for An. funestus and An. stephensi in addition to several species in the An. gambiae complex. In comparison, we have the duo gSG7 and gSG7-2, also having the orthologs for An. funestus and An. stephensi. The way the graphs are constructed, the divergence distances (see three unmarked red bars, all of the same size) appear the same for the Cellia clades, but notice that the amino acid diversity bar is 0.05 for gSG6 and 0.1 for gSG7, indicating twice the speed of divergence for gSG7 when compared with gSG6.
Fig. 3. Phylograms of the gSG6 and gSG7 family of proteins.
The bootstraped phylograms were obtained by MEGA5. Numbers on branches indicate bootstrap value for 100 trials. Black bars indicate 5% or 10% distance in aa sequence for gSG6 or gSG7, respectively. The three unmarked red bars point at the divergence distances within the Cellia clades. Sequences are indicated by their NCBI accession number with the only exception of the An. gambiae gSG7-2. Sequences from An. gambiae (ANOGAM), An. melas (ANOMEL), An. bwambae (ANOBWA), An. quadriannulatus A (ANOQUA), An. arabiensis (ANOARA), An. funestus (ANOFUN) and An. stephensi (ANOSTE) were analyzed. The gSG6 sequence from An. freeborni (ANOFRE) and the gSG7 sequences from An. darlingi (ANODAR) were used as outgroups.
SELECTION SIGNATURES ON CODING SEQUENCES
According to the classical theory of neutral evolution, ratios of dN/dS < 1 (Table 2) indicate overall negative selection acting on the salivary genes; however, these results reflect the average values for all codons on the genes. To test for positive selection in individual codons, the web version of the HyPhy program (Pond et al. 2005) was run with the fixed effects likelihood (FEL) (Kosakovsky Pond and Frost 2005) model of nucleotide substitution, which tests for pervasive diversifying selection. For this test, we also included orthologs of additional anopheline species when they were known. Remarkably, nearly all SG genes (except for gSG6 and D7r2) showed one or more codons with significant signatures of positive selection (Table 3 and supplemental Fig. S1); but no positive selection signature, as expected, was detected for RpS7. We also ran the FEL model in a set of immune gene sequences from a previous work that detected a single codon under positive selection from an An. gambiae immune gene (Lehmann et al. 2009) and obtained the same codon result reported for the antimicrobial gambicin, indicating that the tests used are in concordance with other tests for codon selection bias. The FEL model thus indicates that the SG genes analyzed are under a greater evolutionary pressure than previously reported immune genes. We have also tested for positive selection using the mixed effects model of evolution (MEME) (Murrell et al. 2012), which takes into consideration episodic and pervasive positive selection at the level of single sites. This model identified additional codons under positive selection in all SG genes but not on RpS7 (Table 4).
Table 3.
Codon-based analysis for positive selection on anopheline salivary gland genes using the FEL model (Kosakovsky Pond and Frost 2005)
| Gene | Additional Species a | Codon | dS | dN | dN/dS | Normalized dN-dS | P-value |
|---|---|---|---|---|---|---|---|
| Apyrase F2 | Af | 82 | 0.00E + 00 | 5.1864 | Infinite | 8.1419 | 0.0427 |
| Apyrase F2 | Af | 142 | 0.00E + 00 | 5.4695 | Infinite | 8.5863 | 0.0484 |
| Apyrase F2 | Af | 183 | 0.00E + 00 | 2.8801 | Infinite | 4.5214 | 0.0356 |
| Apyrase F2 | Af | 215 | 0.00E + 00 | 4.281 | Infinite | 6.7205 | 0.0156 |
| Apyrase F3 | Af | 1 | 0.00E + 00 | 2.9096 | Infinite | 5.0588 | 0.0315 |
| gVAG | Af, As | 1 | 0.00E + 00 | 5.4862 | Infinite | 1.4124 | 0.0085 |
| gVAG | Af, As | 29 | 0.00E + 00 | 3.4076 | Infinite | 0.8772 | 0.0319 |
| gSG7 | Af, As | 116 | 0.00E + 00 | 4.3685 | Infinite | 2.2244 | 0.0190 |
Additional orthologs from: Af, Anopheles funestus; As, Anopheles stephensi.
Table 4.
Codon-based analysis for positive selection on anopheline salivary gland genes using the MEME model (Murrell et al. 2012) a
| Gene | Codon | α | β− | Pr[β=β−] | β+ | Pr[β=β+] | p-value | Additional Speciesb |
|---|---|---|---|---|---|---|---|---|
| Apyrase F1 | 12 | 3.0384 | 3.0384 | 0.8043 | 90.4688 | 0.1957 | 0.0457 | Af |
| Apyrase F1 | 135 | 0.0000 | 0 | 0.8681 | 16.2959 | 0.1319 | 0.0378 | Af |
| Apyrase F1 | 159 | 0.0000 | 0 | 0.7650 | 14.1306 | 0.2350 | 0.0123 | Af |
| Apyrase F2 | 82 | 0.9646 | 0 | 0.2890 | 10.2055 | 0.7110 | 0.0167 | Af |
| Apyrase F2 | 103 | 0.0000 | 0 | 0.4442 | 3.2204 | 0.5558 | 0.0388 | Af |
| Apyrase F2 | 126 | 0.9529 | 0 | 0.2738 | 9.1552 | 0.7262 | 0.0270 | Af |
| Apyrase F2 | 142 | 0.8862 | 0 | 0.3315 | 7.0673 | 0.6685 | 0.0474 | Af |
| Apyrase F2 | 150 | 0.9113 | 0 | 0.2397 | 5.9028 | 0.7603 | 0.0396 | Af |
| Apyrase F2 | 178 | 0.0000 | 0 | 0.4297 | 4.1467 | 0.5703 | 0.0305 | Af |
| Apyrase F3 | 1 | 0.0000 | 0 | 0.0394 | 4.7934 | 0.9606 | 0.0402 | Af |
| Apyrase F3 | 131 | 0.9034 | 0 | 0.8818 | 17.0053 | 0.1183 | 0.0446 | Af |
| gVAG | 1 | 0.6061 | 0 | 0.4145 | 12.9717 | 0.5855 | 0.0015 | Af As |
| gVAG | 10 | 0.7756 | 0 | 0.8737 | 13.4465 | 0.1263 | 0.0442 | Af As |
| gVAG | 157 | 0.0000 | 0 | 0.9763 | 68.0998 | 0.0237 | 0.0135 | Af As |
| gVAG | 181 | 0.0000 | 0 | 0.9653 | 31.2272 | 0.0347 | 0.0290 | Af As |
| gVAG | 222 | 0.3598 | 0 | 0.9485 | 31.8775 | 0.0515 | 0.0243 | Af As |
| D7 | 4 | 0.0000 | 0 | 0.3826 | 2.1504 | 0.6174 | 0.0235 | Af As Adi Ada |
| D7 | 24 | 1.0467 | 0 | 0.8158 | 21.3375 | 0.1842 | 0.0095 | Af As Adi Ada |
| gSG6 | 31 | 0.0000 | 0 | 0.4219 | 2.4419 | 0.5781 | 0.0353 | Af As Aq Aa Am Ab Afr |
| gSG7 | 66 | 0.0000 | 0 | 0.3296 | 2.0876 | 0.6704 | 0.0452 | Af As |
| gSG7 | 112 | 0.5861 | 0 | 0.7757 | 11.2925 | 0.2243 | 0.0376 | Af As |
| gSG7 | 116 | 0.2374 | 0 | 0.4136 | 17.5509 | 0.5864 | 0.0012 | Af As |
Synonymous (α) and non-synonymous (β) substitution rates where the proportion of branches with β > α is significantly greater than 0.
Additional orthologs from: Aa, Anopheles arabiensis; Ab, Anopheles buambae; Ada, Anopheles darlingi; Adi, Anopheles dirus; Af, Anopheles funestus; Afr, Anopheles freeborni; Am, Anopheles melas; Aq, Anopheles quadriannulatus; As, Anopheles stephensi.
Conclusions
We have found that salivary genes important for blood acquisition in An. gambiae mosquitoes display high rates of polymorphisms and the presence of codons with signals for both positive and negative selection. Characterization of this high content of genetic variation is based on individuals sampled from the current generation of a wild population and—except for the codon-based analysis of selection signatures—does not involve comparison with orthologous genes of other mosquito species. Our data are also restricted in space and time. Speculation about the mechanisms leading to the genesis of the observed genetic diversity and, possibly, the stability of polymorphism equilibrium, is therefore conditional on the sample characteristics. In particular, the time scale of the mechanism of selection (current generation vs. recent past vs. distant past), its duration (short lived vs. long lived), and the possible contribution of mechanisms that maintain the presence of polymorphisms through selection varying in space and time will be difficult to uncover (Hedrick 2012).
Several evolutionary scenarios could account for the evolution of mosquito SG genes whose products are interacting with the host’s immune system. An “arms race” scenario (Dawkins and Krebs 1979) would create a series of selective sweeps in both host and mosquito genomes (Lehmann et al. 2009), resulting in a significantly higher ratio of non-synonymous to synonymous substitutions for antigenically important codons (Hughes and Nei 1988; Pritchard 2010). This scenario is supported by our data, as nearly all SG genes (except for gSG6 and D7r2) showed one or more codons with significant signatures of positive selection (Table 3 and supplemental Fig. S1). The short generation time of mosquitoes as compared to humans adds plausibility to this scenario as implied by stability analysis (Tellier and Brown 2007).
The possibility of genetic polymorphism maintained by arms race dynamics makes one wonder about the genomic sites in humans also involved in this interaction. Recent genomic surveys in humans have suggested that many loci have been under positive selection, and these sites become natural candidates as targets of future investigations (Akey 2009; Bustamante et al. 2005; Leffler et al. 2013); however, the location of these sweeps in the human genome is not apparent. Perhaps they would be located in genes targeting mast cell functions, considering their pivotal role in allergic reactions to arthropods bites, but they could involve all those associated with the adaptive immune response, which are also affected by myriads of pathogens. Previous approaches to uncover the impact of pathogens on human genomes (Altmann et al. 2012) might prove untenable in this context, because locating control human populations with extensive less contact with bloodsucking arthropods may be difficult.
Alternatively, a scenario of “trench warfare” (Tellier and Brown 2007) leading to diversifying selection—such as proposed for MHC or HLA genes in humans or in pathogen recognition-coding genes in plants (Hughes and Nei 1988; Tiffin and Moeller 2006)—presents dN/dS of the range reported in Table 2 for salivary genes when compared with the exons coding for mammalian antigen-recognition sites of the MHC (0.5–0.1) (Hughes and Nei 1988). This HLA evolutionary scenario, however, favors overdominance or heterozygotic advantage and not necessarily rare allele advantage, which would be the preferred scenario for a gene that is rewarded by selection at low frequency. This entanglement of overdominance and negative balancing selection, among other evolutionary forces, has been addressed by several authors with regard to the hypervariable region of MHC genes (Mona et al. 2008), where positive selection of codons is widespread. Accordingly, the scenario of negative frequency-dependent selection, also called to explain diversity of immune genes in An. gambiae (Lehmann et al. 2009), seems also to apply to SG genes associated with hematophagy but with a larger impression on salivary than immune genes. We are cautious about the evolutionary interpretation of these scenarios, which is necessarily speculative; other models may also be possible.
Mosquito salivary genes associated with blood feeding are thus shown here to be under strong positive selection, explaining their fast rate of divergence. This class of genes, which may eventually be extended to all salivary genes of hematophagous animals, may thus be added to the somewhat rare group of positively selected gene classes, similarly to those pathogen virulence-linked genes under positive selection (Aguileta et al. 2009; Tiffin and Moeller 2006); “”
Experimental Procedures
MOSQUITO SAMPLES AND SPECIES/FORM IDENTIFICATION
Mosquitoes used in this study were collected in October 2008 in the Bobo-Dioulasso area (Burkina Faso, West Africa) in the village of Soumousso (11°01′ N–4°03′ W). Indoor daytime-resting mosquitoes were collected in human dwellings by pyrethroid spray catches. An. gambiae s.l. were morphologically identified and individually stored under silica gel. DNA was extracted from individual mosquitoes by DNAzol® reagent (Invitrogen) and specimens identified to species and molecular forms following the PCR-RFLP protocol as described (Fanello et al. 2002).
PCR AMPLIFICATION AND MINI-LIBRARIES CONSTRUCTION
Ninety-two An. gambiae S (59 females, 33 males) were used for PCR amplification of SG and ribosomal protein S7 genes. Gene-specific oligonucleotide primers used for PCR amplifications of the different genes or gene fragments are listed in supplemental Table S1. PCR conditions were the following: 5 min 94°C, followed by 35 amplification cycles (1 min 94°C, 1 min annealing at the temperature specified in supplemental Table S1, 1 min 68°C), and a final elongation step of 7 min at 68°C. Platinum® Taq DNA Polymerase High Fidelity (Invitrogen) was used to minimize PCR error rates. PCR fragments obtained from each individual mosquito for each gene were quantified by densitometry using Quantity One® 1-D Analysis Software (Bio-Rad Laboratories). For each gene, a pool was constructed by mixing equimolar amounts of the PCR products amplified from the different individual mosquitoes. Because amplification and/or quantification was not successful in all cases, the pools were assembled from a minimum of 63 (ApyF2) up to a maximum of 84 (ApyF3) individual mosquitoes as detailed in Table 2. PCR fragments from each pool were gel purified by the GenElute Gel Extraction Kit (Sigma) and cloned into the plasmid vector pCR2.1 (Invitrogen) to construct, for each selected gene, a mini-library.
SEQUENCING, SEQUENCE HANDLING, AND SEQUENCE ANALYSES
The mini-libraries were plated and 192 recombinant (white) clones/mini-library were randomly picked and transferred to 96-well plates containing 10 μl of H2O per well. The bacterial suspensions were used for PCR amplification by M13 reverse and T7 primers and double-strand sequenced. Approximately 200–250 ng of each PCR product was transferred to a 96-well PCR microtiter plate (Applied Biosystems) and frozen at −20°C. Samples were shipped on dry ice to the Rocky Mountain Laboratories Genomics Unit with primers and template combined together in an ABI 96-well optical reaction plate (P/N 4306737) following the manufacturer’s recommended concentrations. Sequencing reactions were set up as recommended by Applied Biosystems BigDye® Terminator v3.1 cycle sequencing kit. Sequences were aligned by the cap3 assembler (Huang and Madan 1999) and then edited by BioEdit version 7.1.3.0 (Hall 1999). They were first cleaned from vector and oligonucleotide primer sequences. Then each clone was carefully inspected by comparing sequences from the two different strands. To minimize introduction of potential sequencing errors, a conservative criterion was adopted: (i) when sequences on the two strands showed large discordance, they were considered as low quality and discarded; (ii) when differences on the two sequenced strands were limited to a single or a few positions, the nucleotide conforming to the consensus was selected. Coding sequences were reconstructed by joining the different exons. Regions encoding signal peptides and stop codons were excluded from the following analyses. Non-coding sequences were assembled joining flanking UTRs and introns. To minimize introduction of mutations due to PCR amplification or sequencing errors, only parsimony informative sites were considered; therefore, singletons were excluded from coding and non-coding regions by introducing in the corresponding position an N. We have also included in our analysis a gene coding for a ribosomal protein (rpS7 AGAP010592), which should be under strong negative selection pressure, as an internal control of our procedures. In the following sequence analyses, the pairwise deletion option was used so these positions were not considered. This approach may imply an underestimation of variability of salivary genes. Nucleotide diversity as reported in Table 2 was estimated by DnaSP version 5.10.01 (Librado and Rozas 2009) using the option “gaps/missing data,” which excludes gaps and missing data only in the pairwise comparisons. Standard errors and rates of synonymous and non-synonymous substitutions were estimated by MEGA version 5 (Tamura et al. 2011) using the bootstrap method and the pairwise deletion option. Data on nucleotide diversity in coding regions of 72 immune and 37 non-immune genes from an An. gambiae S population from Cameroon are from Cohuet and collaborators (Cohuet et al. 2008) and were retrieved from Supplemental Information (SI) files 3 and 4.
SELECTION SIGNATURES BASED ON PROBABILISTIC CODON SUBSTITUTION MODELS
Selection signatures were identified by fitting probabilistic codon substitution models to the coding sequences after excluding singleton polymorphic sites that were changed to an N, as indicated above. This was done to avoid inclusion of PCR or sequencing errors. Orthologs from other anopheline species were added to the An. gambiae sequences when known, and these are indicated in Table 3. Each dataset was tested for the presence of recombination using the program GARD (Pond et al. 2006). These tests resulted negative. Codon substitution models based on the rate of synonymous/non-synonymous sites (Yang 1997) were fitted using the methods implemented in the program HyPhy, procedures FEL (Kosakovsky Pond and Frost 2005) and MEME (Murrell et al. 2012) accessed through the Datamonkey webserver (http://datamonkey.org) (Delport et al. 2010; Kosakovsky Pond and Frost 2005).
Data Availability
All sequences used are available as supplemental files to this manuscript.
Supplementary Material
Evolutionary fingerprinting of salivary genes displaying approximate sampling distributions of synonymous and nonsynonymous rates. Rate plots are drawn on the log scale with the diagonal line corresponding to neutrality (equal rates). The ellipses reflect a Gaussian-approximated variance in each individual rate estimate, and colored pixels show the density of the posterior sample of the distribution for a given rate. Points above the line correspond to positive selection, and points below the line to negative selection. Plot generated using procedure EVF from datamonkey.org (Pond et al. 2010).
Supplemental Table S1. Gene-specific oligonucleotide primers used for PCR amplifications of the different genes or gene fragments.
Supplemental File S1. Data for all sequences used.
Acknowledgments
We thank C. Rizzo (Sapienza University) and R. Dabiré (IRSS/Centre Muraz, Bobo-Dioulasso, Burkina Faso) for help with mosquito collection and B. R. Marshall (NIAID) for manuscript editing. This work was supported by funds from the INFRAVEC project (228421) to B.A. and partly by the EVIMalaR NoE (242095). C.J.S. was funded by CNPq and FAPERJ. J.M.C.R. and V.M.P were supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA.
Because J.M.C.R. and V.M.P. are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- FEL
fixed effects likelihood
- H
housekeeping
- MEME
mixed effects model of evolution
- S
salivary
- SG
salivary gland
- SNP
single-nucleotide polymorphism
Footnotes
Author contributions: B.A. and J.M.C.R. experimental design, data analysis, and manuscript writing. C.J.S., data analysis and manuscript writing. V.M.P, G.S., F.L., and M.P., DNA extraction and sequencing, and manuscript writing.
The authors declare no conflict of interest.
References cited
- Aguileta G, Refregier G, Yockteng R, Fournier E, Giraud T. Rapidly evolving genes in pathogens: methods for detecting positive selection and examples among fungi, bacteria, viruses and protists. Infect Genet Evol. 2009;9:656–70. doi: 10.1016/j.meegid.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–22. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann DM, Balloux F, Boyton RJ. Diverse approaches to analysing the history of human and pathogen evolution: how to tell the story of the past 70 000 years. Philos Trans R Soc Lond B Biol Sci. 2012;367:765–9. doi: 10.1098/rstb.2011.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–27. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Assumpcao TC, Ma D, Schwarz A, Reiter K, Santana JM, Andersen JF, Ribeiro JM, Nardone G, Yu LL, Francischetti IM. Salivary Antigen-5/CAP family members are Cu2+-dependent antioxidant enzymes that scavenge O2− and inhibit collagen-induced platelet aggregation and neutrophil oxidative burst. J Biol Chem. 2013;288:14341–61. doi: 10.1074/jbc.M113.466995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghans JA, Beltman JB, De Boer RJ. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–9. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–7. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–42. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet A, Krishnakumar S, Simard F, Morlais I, Koutsos A, Fontenille D, Mindrinos M, Kafatos FC. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics. 2008;9:227. doi: 10.1186/1471-2164-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–7. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamaza F, Della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–4. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Gillett JD. Natural selection and feeding speed in a blood sucking insect. Proc R Soc Ser B. 1967;167:316–329. doi: 10.1098/rspb.1967.0029. [DOI] [PubMed] [Google Scholar]
- Hall Ta. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hedrick PW. What is the evidence for heterozygote advantage selection? Trends Ecol Evol. 2012;27:698–704. doi: 10.1016/j.tree.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–70. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Isawa H, Orito Y, Iwanaga S, Jingushi N, Morita A, Chinzei Y, Yuda M. Identification and characterization of a new kallikrein-kinin system inhibitor from the salivary glands of the malaria vector mosquito Anopheles stephensi. Insect Biochem Mol Biol. 2007;37:466–77. doi: 10.1016/j.ibmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–6. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Sci Am. 1979;241:98–100. 102, 108. doi: 10.1038/scientificamerican1179-98. passim. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–22. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, Redmond S, Fulton L, Appelbaum E, Godfrey J, Farmer C, Chinwalla A, Yang SP, Minx P, Nelson J, Kyung K, Walenz BP, Garcia-Hernandez E, Aguiar M, Viswanathan LD, Rogers YH, Strausberg RL, Saski CA, Lawson D, Collins FH, Kafatos FC, Christophides GK, Clifton SW, Kirkness EF, Besansky NJ. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–4. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Segurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, Donnelly P, McVean G, Przeworski M. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–82. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hume JC, Licht M, Burns CS, Wollenberg K, Simard F, Ribeiro JM. Molecular evolution of immune genes in the malaria mosquito Anopheles gambiae. PLoS ONE. 2009;4:e4549. doi: 10.1371/journal.pone.0004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Ronca R, Rizzo C, Mestres-Simon M, Lanfrancotti A, Curra C, Fiorentino G, Bourgouin C, Ribeiro JM, Petrarca V, Ponzi M, Coluzzi M, Arca B. The Anopheles gambiae salivary protein gSG6: an anopheline-specific protein with a blood-feeding role. Insect Biochem Mol Biol. 2009;39:457–66. doi: 10.1016/j.ibmb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslin C, Mugnier S, Callebaut I, Laurin M, Pascal G, Poupon A, Goudet G, Monget P. Evolution of genes involved in gamete interaction: evidence for positive selection, duplications and losses in vertebrates. PLoS One. 2012;7:e44548. doi: 10.1371/journal.pone.0044548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milleron RS, Ribeiro JM, Elnaime D, Soong L, Lanzaro GC. Negative effect of antibodies against maxadilan on the fitness of the sand fly vector of American visceral leishmaniasis. Am J Trop Med Hyg. 2004;70:278–85. [PubMed] [Google Scholar]
- Mona S, Crestanello B, Bankhead-Dronnet S, Pecchioli E, Ingrosso S, D’Amelio S, Rossi L, Meneguz PG, Bertorelle G. Disentangling the effects of recombination, selection, and demography on the genetic variation at a major histocompatibility complex class II gene in the alpine chamois. Mol Ecol. 2008;17:4053–67. doi: 10.1111/j.1365-294x.2008.03892.x. [DOI] [PubMed] [Google Scholar]
- Morlais I, Mori A, Schneider JR, Severson DW. A targeted approach to the identification of candidate genes determining susceptibility to Plasmodium gallinaceum in Aedes aegypti. Mol Genet Genomics. 2003;269:753–64. doi: 10.1007/s00438-003-0882-7. [DOI] [PubMed] [Google Scholar]
- Morlais I, Poncon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. Am J Trop Med Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–73. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Pond SL, Scheffler K, Gravenor MB, Poon AF, Frost SD. Evolutionary fingerprinting of genes. Mol Biol Evol. 2010;27:520–36. doi: 10.1093/molbev/msp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW. GARD: a genetic algorithm for recombination detection. Bioinf Appl Note. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Pritchard JK. How we are evolving. Sci Am. 2010;303:40–7. doi: 10.1038/scientificamerican1010-40. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Mans BJ, Arca B. An insight into the sialome of blood-feeding Nematocera. Insect Biochem Mol Biol. 2010;40:767–84. doi: 10.1016/j.ibmb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Arca B. From sialomes to the sialoverse: An insight into the salivary potion of blood feeding insects. Adv Insect Physiol. 2009;37:59–118. [Google Scholar]
- Ribeiro JMC, Rossignol PA, Spielman A. Role of mosquito saliva in blood vessel location. J Exp Biol. 1984;108:1–7. doi: 10.1242/jeb.108.1.1. [DOI] [PubMed] [Google Scholar]
- Swanson WJ. Adaptive evolution of genes and gene families. Curr Opin Genet Dev. 2003;13:617–22. doi: 10.1016/j.gde.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier A, Brown JK. Stability of genetic polymorphism in host-parasite interactions. Proc Biol Sci. 2007;274:809–17. doi: 10.1098/rspb.2006.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Moeller DA. Molecular evolution of plant immune system genes. Trends Genet. 2006;22:662–70. doi: 10.1016/j.tig.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Wilding CS, Weetman D, Steen K, Donnelly MJ. High, clustered, nucleotide diversity in the genome of Anopheles gambiae revealed through pooled-template sequencing: implications for high-throughput genotyping protocols. BMC Genomics. 2009;10:320. doi: 10.1186/1471-2164-10-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji CS, Hemingway J, Ranson H. Identification and analysis of single nucleotide polymorphisms (SNPs) in the mosquito Anopheles funestus, malaria vector. BMC Genomics. 2007;8:5. doi: 10.1186/1471-2164-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol. 2002;19:49–57. doi: 10.1093/oxfordjournals.molbev.a003981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionary fingerprinting of salivary genes displaying approximate sampling distributions of synonymous and nonsynonymous rates. Rate plots are drawn on the log scale with the diagonal line corresponding to neutrality (equal rates). The ellipses reflect a Gaussian-approximated variance in each individual rate estimate, and colored pixels show the density of the posterior sample of the distribution for a given rate. Points above the line correspond to positive selection, and points below the line to negative selection. Plot generated using procedure EVF from datamonkey.org (Pond et al. 2010).
Supplemental Table S1. Gene-specific oligonucleotide primers used for PCR amplifications of the different genes or gene fragments.
Supplemental File S1. Data for all sequences used.
Data Availability Statement
All sequences used are available as supplemental files to this manuscript.