Abstract
Background
The neuroepithelia of the choroid plexus (CP) in the brain and the ciliary body (CB) of the eye have common embryological origins and share similar micro-structure and functions. The CP epithelium (CPE) and the non-pigmented epithelium (NPE) of the CB produce the cerebrospinal fluid (CSF) and the aqueous humor (AH) respectively. Production and outflow of the CSF determine the intracranial pressure (ICP); production and outflow of the AH determine the intraocular pressure (IOP). Together, the IOP and ICP determine the translaminar pressure on the optic disc which may be involved in the pathophysiology of primary open angle glaucoma (POAG). The aim of this study was to compare the molecular machinery of the secretory neuroepithelia of the CP and CB (CPE versus NPE) and to determine their potential role in POAG.
Methods
We compared the transcriptomes and functional annotations of healthy human CPE and NPE. Microarray and bioinformatic studies were performed using an Agilent platform and the Ingenuity Knowledge Database (IPA).
Results
Based on gene expression profiles, we found many similar functions for the CPE and NPE including molecular transport, neurological disease processes, and immunological functions. With commonly-used selection criteria (fold-change > 2.5, p-value < 0.05), 14% of the genes were expressed significantly differently between CPE and NPE. When we used stricter selection criteria (fold-change > 5, p-value < 0.001), still 4.5% of the genes were expressed differently, which yielded specific functions for the CPE (ciliary movement and angiogenesis/hematopoiesis) and for the NPE (neurodevelopmental properties). Apart from a few exceptions (e.g. SLC12A2, SLC4A4, SLC4A10, KCNA5, and SCN4B), all ion transport protein coding genes involved in CSF and AH production had similar expression profiles in CPE and NPE. Three POAG disease genes were expressed significantly higher in the CPE than the NPE, namely CDH1, CDKN2B and SIX1.
Conclusions
The transcriptomes of the CPE and NPE were less similar than we previously anticipated. High expression of CSF/AH production genes and candidate POAG disease genes in the CPE and NPE suggest that both might be involved in POAG.
Background
Neuroepithelial cells, which themselves are derived from embryonic stem cells, can be viewed as neural progenitor cells [1,2]. They appear during embryonic development and form the neuroectoderm of the neural tube [3]. They give rise to multiple cell types, such as neurons, astrocytes and oligodendrocytes. In addition, in specialized areas of the central nervous system (CNS), these cells retain their epithelial nature and differentiate into fluid-secreting cells: the neuroepithelia of the ocular ciliary body and the choroid plexus epithelium of the brain [4-9]. These tight single-cell layers function together with the outer blood-retina barrier and the blood–brain barrier to maintain CNS homeostasis and are implicated in diseases like age-related macular degeneration, primary open-angle glaucoma (POAG) and Alzheimer’s disease.
In the literature, transcriptome analysis and functional predictions of human neuroepithelia, including the human retinal pigment epithelium [10] and ciliary body [11] in the eye and the choroid plexus epithelium (CPE) of the brain [12] have been described, but most studies are difficult to compare since they used different sampling methods, methodologies and bioinformatic techniques. Previously we determined the RNA expression profiles and predicted functional properties of five adult human neuroepithelia using a single microarray and bioinformatics platform: the central and peripheral retinal pigment epithelium [13-15], the pigmented and non-pigmented neural epithelia of the ocular ciliary body (CB) [16], and the choroid plexus neural epithelium (CPE) of the brain [17].
In the current study, we set out to compare the gene expression profiles and functional annotations of two, apparently closely related neural epithelia: the CPE, which produces the cerebrospinal fluid (CSF) of the brain and the CB epithelium (CBE), which produces aqueous humor (AH) of the eye.
The CPE and CBE resemble each other in many ways, as reviewed by Straziella and Ghersi-Egea [18] for the CPE and by Coca-Prados and Escribano [19] for the CBE. In summary, both epithelia are lobed neuroectodermal cell layers sealed by tight junctions, thereby creating diffusion barriers for most hydrophilic molecules between the blood and CNS. Both CPE and CBE produce colorless fluids, the CSF and AH, respectively. The CPE and CBE regulate production, composition and reabsorption of these fluids and are involved in intra-cranial and intra-ocular pressure dynamics, respectively [20,21]. Both the CPE and CBE form villi that overly a loose extracellular matrix and fenestrated choroidal capillaries.
There are also major differences between the CPE and CBE. Obviously, they are located in different parts of the CNS: the CPE is located in the lateral, third and fourth ventricles of the brain, whereas the CBE resides in the posterior chamber of the eye. A major difference is that the CBE consists of two adjacent neuroectodermal layers: the non-pigmented (NPE) and the pigmented epithelium (PE), whereas the CPE consists of a single epithelial layer. Using large scale gene expression analysis and functional annotation, we recently showed that the NPE and PE layers are highly comparable: only 1% of the transcriptome was different between the NPE and PE [16].
The CPE and CBE are involved in the pressure dynamics in the brain and eye, respectively. The pressures of the ocular and cranial fluids uniquely meet at one location: the optic disc, situated at the point where the optic nerve leaves the eye. The optic disc contains the lamina cribrosa that supports the retinal nerve fibers where they leave the eye. The difference between the posteriorly directed IOP and anteriorly directed ICP on the optic disk is the translaminar pressure gradient. This translaminar pressure ultimately determines the forces applied to the lamina cribrosa and these forces appear to play a key role in a common age-related eye disease POAG. POAG is a progressive optic neuropathy with abnormal cupping of the optic nerve head (excavation) and loss of retinal ganglion cells. The pathogenesis of POAG is largely unknown, but an increased IOP is a major risk factor. Recent findings suggest that it is actually the translaminar pressure, rather than the IOP, that forms the risk factor for POAG and its subtype normal tension glaucoma (NTG) [22-29].
Since the CPE and CBE have a similar origin, structure and function, and together are involved in translaminar pressure dynamics, we were interested in the gene expression-based functional comparison of the human CPE and CBE. Previously, we found that the gene expression of the two ciliary epithelia, PE and NPE, resembled each other closely [16]. For the present study we chose to compare the CPE with the NPE (and not the PE), since the NPE is non-pigmented and contains tight junctions, just like the CPE. The aims of this study were (1) to compare the gene expression profiles and functional annotation of the CPE and NPE in order to determine common properties and potential specific functions and (2) to determine CPE/NPE highly and specifically expressed genes coding for ion transport proteins involved in CSF/AH production and (3) for genes previously associated with POAG.
Methods
Ethics statement
This study was performed after institutional approval of the Netherlands Institute for Neuroscience, Amsterdam, Netherlands. The human CPE material was obtained from the Netherlands Brain Bank (Amsterdam, Netherlands). The human donor eyes were provided by the Corneabank Beverwijk, Netherlands. In accordance with the international declaration of Helsinki, the NBB and the Corneabank obtained permission from the donors for, respectively, brain and eye autopsy and the use of clinical information for research purposes. All human data were analyzed anonymously.
Gene expression data of the human choroid plexus and ciliary body epithelia
We used the gene expression data from our previously-published microarray studies of healthy human CPE (seven samples) (GSE49974; [17]) and NPE (seven samples) (GSE37957; [16]). The age of the donors varied between 39 and 73 years. Donors had neither history of any brain or eye disease nor any malignancies. The seven human CPE donors were all male, five human CBE donors were male and two female. Since several external or genetic factors may have influence on our human gene expression data, we could detect only consistent and significant similarities or differences between the selected samples. All RNA samples from each tissue were analyzed separately by microarray. The microarrays were performed against the same common reference sample, being human retinal pigment epithelium (RPE)/choroid. In this way, the CPE and NPE gene-expression data could be compared with each other. Human choroid plexus tissue was removed from the lateral ventricles of fresh post mortem brains. Whereas the gross anatomy of the CPE became somewhat distorted, the neuroepithelium could be identified easily on cryosections for laser dissection. Donor eyes were snap-frozen in their entirety, which maintained the overall structure of the ciliary body. Detailed description of cell sampling, RNA processing, microarray hybridization, and confirmation methodology of the microarray data were described elsewhere previously [16].
Data analysis
The microarray image files were analyzed and processed by Agilent Feature Extraction Software (Agilent Technologies, version 9.5.3.1) and mean intensities (log2) were given to the spots. These mean intensities were normalized between arrays of the CPE and NPE in R (version 2.14.0 for Windows, R Development Core Team, 2009; NormalizeBetweenArrays with the ‘aquantile’ method). Next, the genes were ranked by expression level and assigned percentile ranks [13]. The expression datasets were functionally annotated using the Ingenuity Knowledge Base (Ingenuity® Systems, version 11631407, http://www.ingenuity.com; assessed at August 27, 2013). The ingenuity core-analysis yields information about biological functions, functional molecular networks, and canonical pathways. Functional molecular networks represent the interactions of molecules leading to biological functions that are attributed to a certain dataset of genes. Canonical pathways are the simplest representation of an (established) molecular pathway. The Ingenuity database contains millions of molecular and functional data points from experimental studies, available from both literature and online experimental databases such as PUBMED, the GWAS Database, GEO, and OMIM.
For the first functional analysis, genes were selected with the highest expression values (expression above 90th percentile [P90]) in both the CPE and NPE. This means that these genes have an expression intensity that falls into the highest 10% intensity values of the CPE and NPE microarrays [13,16]. These genes are referred to as the “Highly expressed CPE and NPE genes”. Next, the gene expression data of the CPE and NPE was compared with a t-test in R and Benjamini-Hochberg correction for multiple testing (version 2.14.0 for Windows, R Development Core Team, 2009). Our initial selection criteria for differentially expressed genes were a fold-change (FC) >2.5 and a p-value <0.05. To identify larger and more specific differences, a relatively strict criteria of FC > 5 and a p-value < 0.001 was used, and this resulted in a set of genes referred to as the “Significantly differentially expressed CPE and NPE genes”. These significantly differentially expressed CPE and NPE genes were functionally annotated in the Ingenuity Knowledge Base (Ingenuity® Systems, version 11631407, http://www.ingenuity.com; assessed at August 5, 2013). In addition, we looked for highly and significantly differentially expressed ion transport protein coding genes involved in CSF/AH production in the CPE and NPE. The ion-channels and ion transporters that are involved in CSF and AH production were, respectively, reviewed by Brown et al. [30] and Civan et al. [20]. Ingenuity was searched for corresponding genes that code for these channels and transporters. In this way, we created a list of genes involved in CSF/AH production. Next, this list was compared with the “Highly expressed CPE and NPE genes” and “Significantly differentially expressed CPE and NPE genes” to find common or different features in CPE and NPE regarding CSF/AH production. Finally, we searched for (candidate) POAG disease genes that were highly or significantly differentially expressed in the CPE and NPE. To this end we compared the “Highly expressed CPE and NPE genes” and “Significantly differentially expressed CPE and NPE genes” with a list of 65 (candidate) POAG disease genes that we recently reviewed [31].
Results
Comparison of the CPE and NPE transcriptome
There were several functions that are apparently biologically important in both epithelial layers. These were (1) neurological function and disease (migration of neuroglia, Alzheimer’s disease, Parkinson’s disease, tauopathy, Leigh syndrome), (2) immunological and infectious diseases (allergy and autoimmune disease, viral infection pathways), (3) molecular transport, (4) hematological disease (myelodysplastic syndrome, lymphosarcoma, anemia), and (5) basic cellular (dys) functions. However, it is possible that similar expression of house-keeping genes in both tissues underlie some of these shared functional annotations.
The statistical comparison between the entire gene expression datasets of 30,589 unique genes of the CPE and NPE showed that many genes were also significantly differentially expressed between the CPE and NPE. Surprisingly, with the frequently used standard ANOVA selection criteria (FC > 2.5 and p-value < 0.05; [15,16]), far more significantly differentially expressed genes (14%) were found than we had expected a-priori. To identify the most important and specific differences, we subsequently chose a more strict selection criteria (FC > 5 and p-value < 0.001). With these criteria, there were 760 (2.5%) genes significantly more highly expressed in the CPE than in the NPE. Vice versa, there were 624 (2.0%) genes significantly more highly expressed in the NPE out of a total of 30,589 unique genes. Tables 1 and 2 present the top-30 of “Significantly differentially expressed CPE and NPE genes” according to the strict criteria. The complete lists of the expressed genes in CPE and NPE can be found in Additional files 1 and 2, respectively.
Table 1.
Top-30 genes significantly higher expressed in CPE compared to NPE
| Gene name | Systematic name | FC | p -value |
|---|---|---|---|
|
TTR |
NM_000371 |
4307.5 |
2.38E-12 |
|
SLC24A4 |
NM_153646 |
425.1 |
3.49E-14 |
|
W60781 |
W60781 |
390.8 |
4.65E-09 |
|
ARMC3 |
NM_173081 |
196.7 |
7.12E-13 |
|
KL |
NM_153683 |
191.7 |
4.53E-10 |
|
SPAG6 |
NM_012443 |
171.5 |
2.74E-11 |
|
SLCO1C1 |
NM_017435 |
134.0 |
1.96E-11 |
|
KLK11 |
NM_144947 |
127.7 |
1.47E-14 |
|
IL13RA2 |
NM_000640 |
97.5 |
4.59E-12 |
|
FAM81B |
NM_152548 |
90.5 |
2.08E-11 |
|
EFCAB1 |
NM_024593 |
88.1 |
4.38E-13 |
|
THC2741529 |
THC2741529 |
82.8 |
1.86E-09 |
|
DNER |
NM_139072 |
82.6 |
2.08E-12 |
|
KIAA1456 |
NM_020844 |
77.6 |
2.09E-12 |
|
C13orf26 |
NM_152325 |
77.5 |
1.13E-12 |
|
SLC4A10 |
NM_022058 |
73.1 |
9.42E-10 |
|
FLJ23049 |
NM_024687 |
71.8 |
3.49E-12 |
|
KCNA5 |
NM_002234 |
70.4 |
1.97E-11 |
|
NAT8L |
NM_178557 |
69.9 |
5.20E-10 |
|
SPAG8 |
NM_172312 |
62.9 |
1.47E-10 |
|
BUB1B |
NM_001211 |
59.2 |
7.40E-08 |
|
AIM2 |
NM_004833 |
55.6 |
9.11E-13 |
|
THC2657259 |
THC2657259 |
55.0 |
6.28E-11 |
|
GRM8 |
NM_000845 |
54.9 |
5.38E-10 |
|
HRK |
NM_003806 |
54.6 |
8.93E-11 |
|
POLE |
AF128542 |
54.3 |
4.24E-09 |
|
FABP4 |
NM_001442 |
52.4 |
8.89E-10 |
|
C2orf39 |
NM_145038 |
50.6 |
2.09E-12 |
|
CLIC6 |
NM_053277 |
49.7 |
2.07E-11 |
| KIAA1199 | NM_018689 | 47.9 | 1.10E-12 |
Table 2.
Top-30 genes expressed significantly higher in NPE compared to CPE
| Gene name | Systematic name | FC | p-value |
|---|---|---|---|
|
AOC3 |
NM_003734 |
375.2 |
4.24E-10 |
|
FGFBP2 |
NM_031950 |
272.4 |
9.92E-11 |
|
THC2677285 |
THC2677285 |
268.8 |
9.11E-13 |
|
LOC92196 |
NM_001017920 |
250.9 |
3.32E-11 |
|
CPAMD8 |
NM_015692 |
214.9 |
9.80E-11 |
|
HSD17B2 |
NM_002153 |
195.9 |
9.23E-12 |
|
CD96 |
NM_198196 |
162.3 |
9.23E-12 |
|
AK124698 |
AK124698 |
161.5 |
7.00E-10 |
|
GRM1 |
NM_000838 |
128.9 |
2.64E-13 |
|
PCSK2 |
NM_002594 |
125.6 |
2.49E-11 |
|
CLRN1 |
NM_174878 |
118.3 |
1.09E-12 |
|
LIPC |
NM_000236 |
107.8 |
2.40E-12 |
|
ATP1A2 |
NM_000702 |
105.1 |
2.77E-08 |
|
C8orf47 |
NM_173549 |
89.4 |
2.38E-12 |
|
TFPI2 |
NM_006528 |
88.7 |
2.44E-09 |
|
DSC1 |
NM_004948 |
87.4 |
1.20E-10 |
|
GPR64 |
NM_001079858 |
86.5 |
8.89E-12 |
|
SLC5A8 |
NM_145913 |
85.6 |
5.97E-14 |
|
LHX2 |
NM_004789 |
77.8 |
2.64E-13 |
|
RAX |
NM_013435 |
77.7 |
1.70E-11 |
|
ABI3BP |
NM_015429 |
76.6 |
1.65E-09 |
|
FAM83D |
NM_030919 |
75.9 |
4.79E-12 |
|
BHLHB5 |
NM_152414 |
75.4 |
5.31E-10 |
|
SLITRK5 |
NM_015567 |
66.0 |
5.56E-10 |
|
THC2636507 |
THC2636507 |
64.2 |
1.45E-08 |
|
THC2676797 |
THC2676797 |
62.6 |
7.23E-12 |
|
DSCR8 |
NM_203429 |
61.8 |
2.38E-12 |
|
OXGR1 |
NM_080818 |
60.7 |
4.41E-11 |
|
LRRN1 |
NM_020873 |
56.7 |
9.48E-12 |
| KIAA1727 | NM_033393 | 55.6 | 3.40E-09 |
Functional annotation of the genes expressed significantly higher in CPE than in NPE
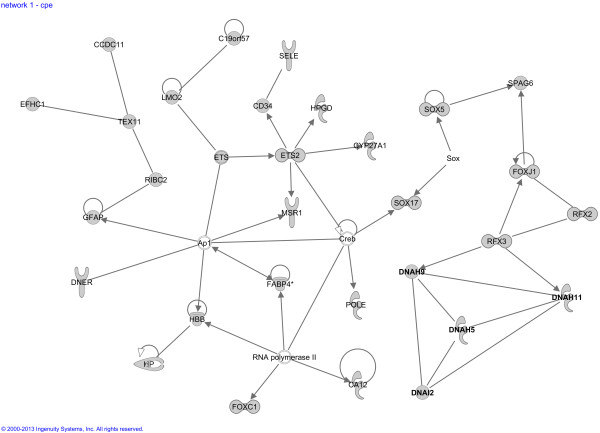
Ingenuity attributed four major statistically significant biological functions to the CPE: (1) ciliary and flagella motion and primary ciliary dyskinesia, (2) development of blood vessels and angiogenesis and related diseases (vascular disease, occlusion of artery, arteriosclerosis), (3) molecular transport, and (4) several types of cancer (adenocarcinoma, epithelial neoplasia, cancer of lung, uterine endometrium, and ovary). Using the same dataset, Ingenuity subsequently built 25 functional molecular CPE networks. Figure 1 shows as an example, a diagram of network 1 that represents the gene and protein interactions leading to ciliary movement and dyskinesia. This network is functionally annotated with “Developmental and hereditary disorders and respiratory disease”. The other 24 functional molecular network diagrams can be found in Additional file 3. After correction for multiple testing, there were no statistically significant canonical pathways specifically assigned by Ingenuity to the CPE compared with the NPE.
Figure 1.
Molecular network of genes expressed significantly more highly in the brain choroid plexus epithelium compared to the non-pigmented epithelium of the eye. Example of a molecular network generated by Ingenuity software. Grey symbols represent genes expressed more highly in the CPE than in the NPE. Transparent entries are molecules inserted by the knowledge database. Gene names are abbreviated according to those used in GenBank. Solid lines between molecules indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). The genes DNAH5, DNAH9, DNAH11 and DNAI2 indicated in bold, code for dynein proteins. Dyneins keep the microtubules of the cilia together and promote the movement of the cilia. Mutations in these genes can cause primary ciliary dyskinesia. The main functionalities given by Ingenuity for this entire molecular network are “Developmental and hereditary disorder and respiratory disease”. Reproduced with permission.
Functional annotation of the genes expressed significantly higher in NPE than in CPE
Ingenuity assigned four major statistically significant biological functions to the NPE compared to the CPE. The first and most significant biological function was (1) nervous system development, and more specifically neuronal development of sensory organ, head, body axis, eye, forebrain, brain, and telencephalon. This functionality also comprised neurological and ophthalmic diseases (seizure, epilepsy, schizophrenia, bipolar disorder, and congenital anomalies of the eye). Examples of genes underlying this NPE functionality are ALDH1A1, ATOH7, CYP1B1, MSX2, OTX1, PAX6, PVRL3, RP1, SEMA5A, SIX6, and SOX4 (eye development) and BMP2, FGFR1, GAP43, GLI3, GSX2, HES1, PAX6, RELN, SLIT1, SOX3, SOX4, and ZIC3 (brain development). For the total list, see Additional file 4. Another statistically significant function was formation of cellular protrusions and plasma membrane projections. This function was based on 47 genes expressed significantly higher in NPE, for example genes involved in actin function (ANK3, ARGHGAP24, ENC1, MARCKS, PALLD), genes involved in filopodium formation and cytoskeleton organization (MCF2, RHOU) and axonal growth and guidance (CNTN4, GAP43, LRRC4C, SEMA3A, TIAM1, TIAM2). For the total list, see Additional file 5. Finally, we identified the functions of molecular transport, and several types of cancer (adenocarcinoma, epithelial neoplasia, cancer of lung).
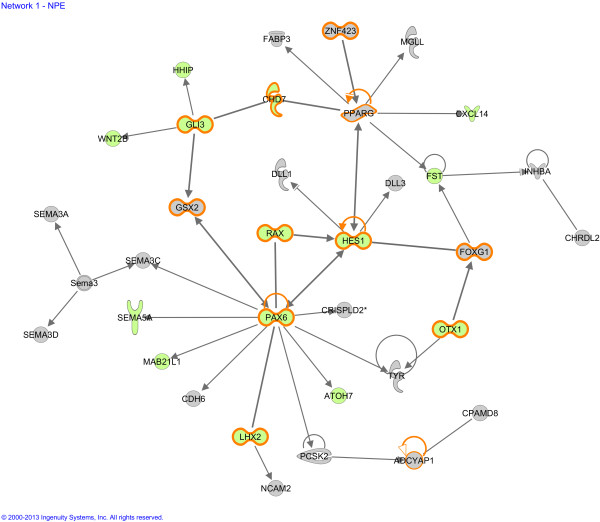
Subsequently, from the same dataset, Ingenuity built 25 functional molecular networks significantly present in the NPE, but not in the CPE. Figure 2 presents, as an example, a diagram of network 1 that shows specific neurodevelopmental functions of the NPE, annotated with top functions “Embryonic, organ and organismal development”. The other 24 functional molecular network diagrams can be found in Additional file 6. After correction for multiple testing, Ingenuity did not assign statistically significant canonical pathways specific for the NPE compared to the CPE.
Figure 2.
Molecular network of genes expressed significantly more highly in non-pigmented epithelium of the eye compared to the brain choroid plexus epithelium. Example of a molecular network generated by Ingenuity software. Gene names are abbreviated according to those used in GenBank. Solid lines between molecules indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). This network contains many genes involved in brain and nervous system development (orange bordered symbols) and eye development (green filled symbols). The main functionalities given by Ingenuity for this entire molecular network are “Embryonic organ and organismal development”. Reproduced with permission.
CPE and NPE expressed genes involved in CSF and AH production
We constructed a list of 157 coding genes corresponding to all the CPE and CBE ion channels and ion transporters proteins as described by Brown et al.[30] and Civan et al.[20] (see Additional file 7). Table 3 presents a list of ion channels and ion-transporters of which at least some of the coding genes were highly and/or significantly differentially expressed in the CPE and NPE, in order to find common or different features in CPE and NPE regarding CSF/AH production. Based on our gene expression data, all these ion channels and ion transporters were present in both epithelia. Eight coding genes were both highly expressed in CPE and NPE (SLC12A7, ATP1A1, ATP1A2, ATP1B1, ATP1B2, ATP1B3, KCNJ13, and AQP1), 13 were expressed significantly higher in the CPE compared to the NPE (ATP1B1, SLC12A2, SLC4A2, SLC4A10, KCNA5, KCNE1, KCNF1, KCNH2, KCNK1, KCNN2, KCNN3, SCN2A, and AQP4), and 10 higher in the NPE compared to the CPE (ATP1A1, ATP1B3, SLC4A4, KCNA4, KCNB2, KCNG1, KCNS1, SCN2B, SCN3B, and SCN4B).
Table 3.
Genes coding for ion channels and transporters involved in CSF and AH production in the CPE and NPE
| Ion channels and transporters | Present in | Coding genes | High gene expr. | Sign. diff. gene expr. |
|---|---|---|---|---|
| Na+/K+ ATPase |
CPE and CBE |
ATP1A1 |
Both |
NPE |
|
ATP1A2 |
Both |
|
||
|
ATP1A3 |
CPE |
|
||
|
ATP1A4 |
|
|
||
|
ATP1B1 |
Both |
CPE |
||
|
ATP1B2 |
Both |
|
||
|
ATP1B3 |
Both |
NPE |
||
| Na+/2Cl-/K+ co-transport |
CPE and CBE |
SLC12A1 |
|
|
|
SLC12A2 |
CPE |
CPE |
||
| Na+/Cl- symport |
CBE |
SLC12A3 |
|
|
| K+/Cl- symport |
CPE |
SLC12A4 |
|
|
|
SLC12A5 |
|
|
||
|
SLC12A6 |
|
|
||
|
SLC12A7 |
Both |
|
||
|
SLC12A9 |
|
|
||
| Cl-/HCO3– exchanger |
CPE and CBE |
SLC4A1 |
|
|
|
SLC4A2 |
|
CPE |
||
|
SLC4A3 |
|
|
||
| Na+/HCO3– symport |
CPE |
SLC4A4 |
NPE |
NPE |
|
SLC4A5 |
|
|
||
|
SLC4A7 |
|
|
||
|
SLC4A8 |
|
|
||
|
SLC4A9 |
|
|
||
|
SLC4A10 |
CPE |
CPE |
||
| Na+/H+ exchanger |
CPE and CBE |
SLC9A1 |
|
|
|
SLC9A2 |
|
|
||
|
SLC9A3 |
|
|
||
|
SLC9A4 |
|
|
||
|
SLC9A5 |
|
|
||
|
SLC9A6 |
|
|
||
| K+ channel |
CPE and CBE |
KCNA4 |
|
NPE |
|
KCNA5 |
CPE |
CPE |
||
|
KCNAB1 |
NPE |
|
||
|
KCNB2 |
|
NPE |
||
|
KCNE1 |
|
CPE |
||
|
KCNF1 |
|
CPE |
||
|
KCNG1 |
|
NPE |
||
|
KCNH2 |
|
CPE |
||
|
KCNJ13 |
Both |
|
||
|
KCNK1 |
CPE |
CPE |
||
|
KCNN2 |
|
CPE |
||
|
KCNN3 |
|
CPE |
||
|
KCNS1 |
|
NPE |
||
| Cl- channel |
CPE and CBE |
CLCN1 |
|
|
|
CLCN2 |
|
|
||
|
CLCN3 |
|
|
||
|
CLCN4 |
|
|
||
|
CLCN5 |
|
|
||
|
CLCN6 |
|
|
||
|
CLCN7 |
|
|
||
| Na+ channel |
CPE and CBE |
SCN1A |
|
|
|
SCN1B |
|
|
||
|
SCN2A |
|
CPE |
||
|
SCN2B |
|
NPE |
||
|
SCN3A |
|
|
||
|
SCN3B |
|
NPE |
||
|
SCN4A |
|
|
||
|
SCN4B |
NPE |
NPE |
||
|
SCN5A |
|
|
||
|
SCN7A |
|
|
||
|
SCN8A |
|
|
||
|
SCN9A |
|
|
||
|
SCN10A |
|
|
||
|
SCN11A |
|
|
||
| Water channel |
CPE and CBE |
AQP1 |
Both |
|
| |
|
AQP2 |
|
|
| |
|
AQP3 |
|
|
| |
|
AQP4 |
|
CPE |
| |
|
AQP5 |
|
|
| |
|
AQP7 |
|
|
| |
|
AQP8 |
|
|
| |
|
AQP9 |
|
|
| |
|
AQP10 |
|
|
| AQP11 |
This table lists the ion channels and ion transporters involved in cerebrospinal fluid (CSF)/aqueous humor (AH) production, their presence in choroid plexus epithelium (CPE) and/or non-pigmented epithelium (NPE) of the ciliary body epithelium according to literature [20,30] and the gene expression profiles in the CPE and NPE according to our microarray gene expression studies. Column 1: Ion channels and transporters involved in CSF/AH production. For the K+ channel, we only presented the highly and/or statistical significant different expressed genes of the CPE and NPE; Column 2 - Present in: The presence of the different ion channels and ion transporters are reviewed in [30] for the CPE and [20] for the NPE; Column 3 - Coding genes: the genes coding for the ion channels and ion transporters listed in column 1; Column 4 - High gene expr.: Here we indicate the genes that are highly expressed in the CPE and/or NPE according to our gene expression microarray data of the CPE and NPE. Highly expressed gene are genes with mean expression values >90th percentile. This means that these genes have an expression intensity that falls into the highest 10% intensity values of the microarray. Both = high expression in both CPE and NPE; Column 5 - Sign. diff. gene expr.: Here we indicate the genes that were statistically significant higher expressed in the CPE or NPE compared with the NPE and CPE, respectively, according to our gene expression microarray data of the CPE and NPE. The selection criteria were fold change > 5 and p-value < 0.001.
CPE and NPE expressed genes previously associated with POAG
We compared a recently constructed list of 65 candidate POAG genes (reviewed in [31]) with the genes that were highly and/or significantly differently expressed in the CPE and NPE. Table 4 presents the results. We found 10 candidate POAG genes that were highly expressed in both the CPE and NPE (AKAP13, C1QBP, CHSY1, COL8A2, CYP1B1, FBN1, IBTK, MFN2, TMCO1, and TMEM248), three genes that were expressed significantly higher in the CPE (CDH1, CDKN2B, and SIX1), and six genes that were expressed significantly higher in the NPE (ATOH7, CYP1B1, FBN1, MYOC, PAX6, and SIX6).
Table 4.
Gene expression profiles of candidate POAG disease genes in CPE and NPE
| Candidate POAG disease genes | High gene expr. | Sign. diff. gene expr. |
|---|---|---|
|
APOE |
CPE |
|
|
AKAP13 |
Both |
|
|
ATOH7 |
|
NPE |
|
C1QBP |
Both |
|
|
CDH1 |
CPE |
CPE |
|
CDKN2B |
|
CPE |
|
CHSY1 |
Both |
|
|
COL8A2 |
Both |
|
|
CYP1B1 |
Both |
NPE |
|
FBN1 |
Both |
NPE |
|
GSTM1 |
CPE |
|
|
IBTK |
Both |
|
|
MFN2 |
Both |
|
|
MYOC |
NPE |
NPE |
|
OPTN |
CPE |
|
|
PAX6 |
NPE |
NPE |
|
RFTN1 |
NPE |
|
|
SIX1 |
|
CPE |
|
SIX6 |
NPE |
NPE |
|
TMCO1 |
Both |
|
| TMEM248 | Both |
High gene expr: Genes that are highly expressed in the CPE and/or NPE according to our gene expression microarray data of the CPE and NPE. Highly expressed gene are genes with mean expression values >90th percentile. Both = high expression in both CPE and NPE. Sign. diff. gene expr.: Genes that are statistically significant higher expressed in the CPE or NPE compared with the NPE and CPE, respectively, according to our gene expression microarray data. The selection criteria were fold change > 5 and p-value < 0.001.
Discussion
The gene expression data of the brain CPE and the NPE of the CBE from the eye were compared. The CPE and NPE are both secretory neuroepithelia, which share a common embryological origin and important structural and functional features (fluid production, brain and eye homeostasis and pressure dynamics) but also exhibit substantial anatomical differences (location, single (CPE) or double (CBE) layered). Together, the CPE and CBE are ultimately involved in building the translaminar pressure of CSF and AH on the optic disc. A disturbed translaminar pressure is a risk factor for POAG and NTG.
Comparison of the CPE and NPE transcriptome
It had been anticipated that the transcriptomes and predicted functionalities of the CPE and NPE in the current study would be highly similar. After all, using (initially) the same criteria, methodology and platform, the neuroepithelial transcriptomes of the ocular NPE and PE differed only by 1% [16] and the transcriptomes of the macular and peripheral RPE only by 1-5% [15]. Several overlapping functionalities for the CPE and NPE most highly expressed genes were found, including neurological function and diseases, immunological properties, and molecular transport. However, unexpectedly, there were also large differences between the two neuroepithelia. In the initial analysis, in which we applied the same selection criteria (FC > 2.5 and p-value < 0.05) as used in our previous neuroepithelial microarray studies [13,15,16], there was a large, 14% difference (data not shown); in our final analysis (FC > 5 and p-value < 0.001) this was still 4.5%. Hence, although the transcriptomes of the two neuroepithelia showed substantial overlap, there were large differences as well.
Previously published data showed many molecular similarities between CPE and CBE, but also some differences: for example the ion channels and ion transporters involved in CSF and AH production. These were widely studied and extensively reviewed by Brown et al.[30] for the CPE and by Civan et al. [20] for the CBE. These reviews reported that most of the ion channels and ion-transporters involved in the CSF/AH production are found in both the CPE and CBE, for example Na+/K+-ATPase, Na+/K+/2Cl- -symporters, Cl-/HCO3– and Na+/H+ exchangers, and aquaporins. However, there were also some ion transport proteins in the CPE that have not been identified in the CBE, for example Na+/HCO3– and K+/Cl- symporters. On the other hand, Na+/Cl- exchangers were found in the CBE, but not (yet) in the CPE [20,30].
Predicted CPE functionalities not present in the NPE
Function of cilia
The most important predicted function found in the CPE, but not in the NPE, was genes for ciliary motion and primary ciliary dyskinesia. Cilia, which consist of bundles of microtubules held together by dyneins, are essential for CSF flow in the brain ventricles. Hydrocephalus is a common sign of primary ciliary dyskinesia, a disease caused by mutations in dynein proteins [32,33]. A detailed inspection of the expression datasets showed that six dynein coding genes (DNAH5, DNAH7, DNAH9, DNAH11, DNAI1, and DNAI2) were expressed significantly more highly in the CPE compared to the NPE. Vice versa, there were no dynein coding genes expressed more highly in the NPE compared to the CPE. Hence, our data are in line with the idea that ciliary function is biologically more important in CPE than in NPE.
Angiogenesis
Another important CPE function that was not present in the NPE was “angiogenesis and development of blood vessels”. Ingenuity yielded this function because of 57 relevant genes expressed significantly higher in the CPE than in the NPE, most importantly ACE2, ADRA2B, AHR, FN1, FOXC1, GATA6, HGF, HHEX, KLF2, PGF, PTHLH, SERPINF1, and TEK (for the complete list, see Additional file 8). The appearance of this function could be explained either by a technical artifact or by a genuine biological reason. The CPE samples are much more closely entangled with blood vessels than the NPE. Thus, potential contamination of CPE tissue with blood vessel cells during laser dissection microscopy may underlie this finding. We previously showed that cellular contamination of adjacent tissue is a possible limitation of (our) cellular microarray approach [13]. Alternatively, the CPE could actually have a specific function that is not present in the NPE. This function could be extramedullary hematopoiesis (EH), which previously has been assigned to the human and mouse CPE [31]. Indeed, the functional molecular network 4 (“Hematological system development and function and hematopoiesis”) built by Ingenuity of significantly higher expressed CPE genes, also points to this specific property (Additional file 3). EH is a compensatory mechanism of hematopoiesis in sites other than the bone marrow, and some case reports point to a role of EH in the CPE [34-36]. We could not find published reports of possible presence of EH in the NPE.
Predicted NPE functions not present in the CPE
Neuronal development
The first and most significant predicted function for the NPE not present in the CPE was “development of neuronal tissues and organs, including eye, retina, brain and forebrain”. The potential neural embryological properties of the CBE have been a topic of fierce recent discussion. The pars plana and the ciliary marginal zone of the CBE may, or may not, contain retinal progenitor cells, and there may even be differences between animal species. Some authors reported the presence of specific retinal stem cell or progenitor cell markers, such as NES, MITF, PAX6, SIX3, Rx, FGF2, and CHX10 [37-42]. Isolated CBE neural progenitor cells were shown to proliferate and differentiate in vitro into neural spheres and possible photoreceptor-like cells [37,43-50], although this was not found in all studies [51,52]. Interestingly, our gene expression data are primarily of the pars plicata of the human NPE, not from the ora serrata, but, even here, some embryological properties seem to be present in the adult neuroepithelium.
Cellular protrusions
The second predicted function of the NPE was “formation of cellular protrusions and plasma membrane projections”. The reason why this function is apparently more prominent in the NPE is currently not clear. After all, both the NPE and CPE contain actin and both cell types have filopodia. Possibly, the attachment of lens zonule fibers to the NPE may play a role in this finding. Future research is warranted.
Ion channels and ion transporters involved in CSF and AH production
The genes coding for the ion channels and ion-transporters involved in CSF and AH production had highly similar expression patterns in both tissues. Indeed, in the literature, some of these proteins were already identified in the CPE and CBE, such as ATP1A1, ATP1B1 and ATP1B2 [20,30], AQP1 [53,54] and SLC12A7 [55]. However, there are some significant expression differences in specific genes, which point to (subtle) differences in CSF and AH production: namely 13 specifically-expressed genes in the CPE and 10 in the NPE. To compare, a study of the molecular background of AH production in the CBE, the NPE and PE only showed four specifically expressed genes in the PE and none in the NPE [56]. At the protein level, previous studies showed expression of SLC12A2 [57], SLC4A2 [58], and SLC4A10 [59] in the CPE and expression of SLC4A4 [60] and sodium channels [61] in CBE. However, these finding were not proof that these proteins definitely differ between CPE and CBE, and comparative studies on protein level in both CPE and CPE are necessary to determine this. The differences between CPE and NPE could be a starting point for further biochemical, physiological and pharmacological research. Perhaps, these differences will open the door for specific drug delivery to the brain and the eye after systemic treatment as for example in a systemic drug that simultaneously increases ICP and lowers the IOP for POAG/NTG treatment.
Potential CPE and NPE involvement in POAG
Both the CPE and CBE may, ultimately, play a role in the development of POAG (see Background section), and especially in the development of NTG [31]. Ten (AKAP13, C1QBP, CHSY1, COL8A2, CYP1B1, FBN1, IBTK, MFN2, TMCO1 and TMEM248) of the 65 candidate POAG disease genes were highly expressed in both CPE and NPE, which might indicate overlapping pathobiological mechanisms of POAG in the two epithelia. On the other hand, we also identified three candidate POAG disease genes, CDH1, CDKN2B and SIX1, which were expressed more highly in the CPE than in the NPE. CDKN2B regulates cell cycle and growth, SIX1 is involved in developmental processes and CDH1 is a cell-cell adhesion glycoprotein. Interestingly, mutations in CDKN2B were previously implicated in NTG [62-65]. In addition, mutations in both CDKN2B and SIX1 were previously associated with the vertical-cup-disc-ratio, an endophenotype of POAG [66,67], and not, for example, with IOP. Vice versa, we found six (candidate) POAG-disease genes significantly higher expressed in the NPE than in the CPE (ATOH7, CYP1B1, FBN1, MYOC, PAX6, and SIX6). The potential involvement of these genes in POAG via the CPE and CBE is currently not clear.
As mentioned in the Background section, it seems that not only an increase in IOP but also a decrease in ICP could be a risk factor for POAG [22-29,68-70]. Thus, manipulation of either IOP or ICP, together or apart, would be a possible form of POAG treatment. For example, alpha-adrenergic receptors are present in the both NPE and CPE albeit with differences in affinity and effect upon stimulation [71-73]. Our expression data show that ADRA2B (an alpha-2-adrenergic receptor) is expressed more highly in the CPE compared to NPE and ADRA1B (an alpha-1-adrenergic receptor) is expressed more highly in the NPE compared to the CPE. Of specific interest are the alpha-2 adrenoceptors in the brain and eye. Chiou et al.[71] found that intraocular administration of an alpha-2 antagonist (yohimbine) resulted in a decreased IOP in cats, whereas McCormick et al.[73] showed that an intravenous administered alpha-2 antagonist (tolazine) resulted in an increased ICP. These findings might suggest that an intravenous alpha-2 antagonist is an ideal treatment option for POAG and NTG: it lowers the IOP and increases the ICP, thereby changing the translaminar pressure on both sides. Chiou et al.[71] also found that intraocular treatment with an alpha-2 agonist (clonidine) resulted in a decreased IOP (but less effective than alpha-2 antagonist). In the brain, intravenous alpha-2 agonist (xylazine) resulted also in a decreased ICP, making this agent less interesting for the dual, systemic treatment of POAG/NTG. However, more research is warranted, to prove these findings and hypothesis, to specify which alpha-2 antagonist does both decrease IOP and increase ICP, and if this approach is a good treatment option.
Conclusions
The gene expression patterns and predicted functions of the CPE and NPE overlap (molecular transport, immunological functions), but also show substantial differences. The ion channels and ion-transporters involved in CSF and AH production share the same expression profiles for most of their coding genes in CPE and NPE, with a few exceptions. The CPE showed specific functions in ciliary movement and angiogenesis/hematopoiesis compared to the NPE. Vice versa, the NPE analysis yielded specific neurodevelopmental properties not present in the CPE. In POAG and NTG, the CPE and NPE could act together in creating a disturbed, net posteriorly directed translaminar pressure over the optic disc, where brain and eye meet.
Abbreviations
AH: Aqueous humor; EH: Extramedullary hematopoiesis; CBE: Ciliary body epithelium; CNS: Central nervous system; CPE: Choroid plexus epithelium; CSF: Cerebrospinal fluid; ICP: Intracranial pressure; IOP: Intraocular pressure; NPE: Non-pigmented epithelium; NTG: Normal tension glaucoma; PE: Pigmented epithelium; POAG: Primary open angle glaucoma; RPE: Retinal pigment epithelium.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SJ carried out the microarray studies and drafted the manuscript; TG participated in study design and helped to draft the manuscript; JB carried out the microarray studies; NJ participated in study design and helped to draft the manuscript; AB participated in study design and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Total list of genes expressed significantly higher in CPE compared to NPE. Selection criteria were a fold change > 5 and p-value < 0.001.
Total list of genes expressed significantly higher in NPE compared to CPE. Selection criteria were a fold change > 5 and p-value < 0.001.
Molecular networks 2–25 generated by the Ingenuity software from the genes expressed significantly higher in the CPE compared to NPE. Grey symbols represent genes expressed significantly higher in the CPE. Transparent entries are molecules inserted by the knowledge database. Gene names are abbreviated according to those used in GenBank. Solid lines indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). The main functionalities given by Ingenuity for this entire molecular network are shown in the diagrams. Reproduced with permission.
Genes in the NPE expressed significantly higher than in CPE involved in the biological function of development.
Genes in the NPE expressed significantly higher than in CPE involved in the biological function of cellular protrusions.
Molecular networks generated by the Ingenuity software from the genes expressed significantly higher in the NPE compared to CPE. Grey symbols represent genes expressed significantly higher in the NPE. Transparent entries are molecules inserted by the knowledge database. Gene names are abbreviated according to those used in GenBank. Solid lines indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). The main functionalities given by Ingenuity for this entire molecular network are shown in the diagrams. Reproduced with permission.
Genes coding ion transport proteins involved in CSF and AH production. We constructed a list of 157 coding genes corresponding to all the CPE and CBE ion transport proteins as described by Brown et al. [30] and Civan et al.[20].
Genes expressed significantly higher in the CPE than in NPE involved in the biological function of angiogenesis.
Contributor Information
Sarah F Janssen, Email: s.janssen@nin.knaw.nl.
Theo GMF Gorgels, Email: t.gorgels@nin.knaw.nl.
Jacoline B ten Brink, Email: j.ten.brink@nin.knaw.nl.
Nomdo M Jansonius, Email: n.m.jansonius@umcg.nl.
Arthur AB Bergen, Email: a.bergen@nin.knaw.nl.
References
- Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Leclerc C, Neant I, Moreau M. Early neural development in vertebrates is also a matter of calcium. Biochimie. 2011;93:2102–2111. doi: 10.1016/j.biochi.2011.06.032. [DOI] [PubMed] [Google Scholar]
- Preston M, Sherman LS. Neural stem cell niches: roles for the hyaluronan-based extracellular matrix. Front Biosci (Schol Ed) 2011;3:1165–1179. doi: 10.2741/218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson PA, Irmler M, Acampora D, Beckers J, Simeone A, Gotz M. The transcription factor Otx2 regulates choroid plexus development and function. Development. 2013;140:1055–1066. doi: 10.1242/dev.090860. [DOI] [PubMed] [Google Scholar]
- Larsen KB, Lutterodt M, Rath MF, Moller M. Expression of the homeobox genes PAX6, OTX2, and OTX1 in the early human fetal retina. Int J Dev Neurosci. 2009;27:485–492. doi: 10.1016/j.ijdevneu.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Nicholson-Flynn K, Hitchcock-DeGregori SE, Levitt P. Restricted expression of the actin-regulatory protein, tropomyosin, defines distinct boundaries, evaginating neuroepithelium, and choroid plexus forerunners during early CNS development. J Neurosci. 1996;16:6853–6863. doi: 10.1523/JNEUROSCI.16-21-06853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von FJ, Wizenmann A, Gotz M. The transcription factors Emx1 and Emx2 suppress choroid plexus development and promote neuroepithelial cell fate. Dev Biol. 2006;296:239–252. doi: 10.1016/j.ydbio.2006.04.461. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kang YJ, Davies LM, Meghpara S, Lau K, Chung CY, Kathiriya J, Hadjantonakis AK, Monuki ES. BMP4 sufficiency to induce choroid plexus epithelial fate from embryonic stem cell-derived neuroepithelial progenitors. J Neurosci. 2012;32:15934–15945. doi: 10.1523/JNEUROSCI.3227-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Chen Q, Hung FC, Overbeek PA. BMP signaling is required for development of the ciliary body. Development. 2002;129:4435–4442. doi: 10.1242/dev.129.19.4435. [DOI] [PubMed] [Google Scholar]
- Whitmore SS, Braun TA, Skeie JM, Haas CM, Sohn EH, Stone EM, Scheetz TE, Mullins RF. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Mol Vis. 2013;19:2274–2297. [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Anand VN, Wang WH, Chatterton JE, Sun D, Shepard AR, Jacobson N, Pang IH, Deluca AP, Casavany TL, Scheetz TE, Mullins RF, Braun TA, Clark AF. Exon-level expression profiling of ocular tissues. Exp Eye Res. 2013;111:105–111. doi: 10.1016/j.exer.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt M, Böhm C, Tatenhorst L, Dinh V, Newrzella D, Keyvani K, Jeibmann A, Buerger H, Rickert CH, Paulus W. Identification of novel diagnostic markers for choroid plexus tumors: a microarray-based approach. Am J Surg Pathol. 2006;30:66–74. doi: 10.1097/01.pas.0000176430.88702.e0. [DOI] [PubMed] [Google Scholar]
- Booij JC, Van SS, Swagemakers SM, Essing AH, Verkerk AJ, Van Der Spek PJ, Gorgels TG, Bergen AA. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009;10:164. doi: 10.1186/1471-2164-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij JC, Ten Brink JB, Swagemakers SM, Verkerk AJ, Essing AH, van der Spek PJ, Bergen AA. A new strategy to identify and annotate human RPE-specific gene expression. PLoS One. 2010;5:e9341. doi: 10.1371/journal.pone.0009341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soest SS, de Wit GM, Essing AH, Ten Brink JB, Kamphuis W, De Jong PT, Bergen AA. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol Vis. 2007;13:1608–1617. [PubMed] [Google Scholar]
- Janssen SF, Gorgels TG, Bossers K, Ten Brink JB, Essing AH, Nagtegaal M, van der Spek PJ, Jansonius NM, Bergen AA. Gene expression and functional annotation of the human ciliary body epithelia. PLoS One. 2012;7:e44973. doi: 10.1371/journal.pone.0044973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen SF, van der Spek SJF, Ten Brink JB, Essing AHW, Gorgels TGMF, van der Spek PJ, Jansonius NM, Bergen AAB. Gene expression and functional annotation of the human and mouse choroid plexus epithelium. PlosOne. 2013;8(12):e83345. doi: 10.1371/journal.pone.0083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazielle N, Ghersi-Egea JF. Choroid plexus in the central nervous system: biology and physiopathology. J Neuropathol Exp Neurol. 2000;59:561–574. doi: 10.1093/jnen/59.7.561. [DOI] [PubMed] [Google Scholar]
- Coca-Prados M, Escribano J. New perspectives in aqueous humor secretion and in glaucoma: the ciliary body as a multifunctional neuroendocrine gland. Prog Retin Eye Res. 2007;26:239–262. doi: 10.1016/j.preteyeres.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Civan MM, Macknight AD. The ins and outs of aqueous humour secretion. Exp Eye Res. 2004;78:625–631. doi: 10.1016/j.exer.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Johanson CE, Duncan JA 3rd, Klinge PM, Brinker T, Stopa EG, Silverberg GD. Multiplicity of cerebrospinal fluid functions: New challenges in health and disease. Cerebrospinal Fluid Res. 2008;5:10. doi: 10.1186/1743-8454-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JB, Berenshtein E, Holbach L. Anatomic relationship between lamina cribrosa, intraocular space, and cerebrospinal fluid space. Invest Ophthalmol Vis Sci. 2003;44:5189–5195. doi: 10.1167/iovs.03-0174. [DOI] [PubMed] [Google Scholar]
- Morgan WH, Yu DY, Balaratnasingam C. The role of cerebrospinal fluid pressure in glaucoma pathophysiology: the dark side of the optic disc. J Glaucoma. 2008;17:408–413. doi: 10.1097/IJG.0b013e31815c5f7c. [DOI] [PubMed] [Google Scholar]
- Morgan WH, Yu DY, Cooper RL, Alder VA, Cringle SJ, Constable IJ. The influence of cerebrospinal fluid pressure on the lamina cribrosa tissue pressure gradient. Invest Ophthalmol Vis Sci. 1995;36:1163–1172. [PubMed] [Google Scholar]
- Berdahl JP, Fautsch MP, Stinnett SS, Allingham RR. Intracranial pressure in primary open angle glaucoma, normal tension glaucoma, and ocular hypertension: a case–control study. Invest Ophthalmol Vis Sci. 2008;49:5412–5418. doi: 10.1167/iovs.08-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdahl JP, Allingham RR, Johnson DH. Cerebrospinal fluid pressure is decreased in primary open-angle glaucoma. Ophthalmology. 2008;115:763–768. doi: 10.1016/j.ophtha.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Ren R, Jonas JB, Tian G, Zhen Y, Ma K, Li S, Wang H, Li B, Zhang X, Wang N. Cerebrospinal fluid pressure in glaucoma: a prospective study. Ophthalmology. 2010;117:259–266. doi: 10.1016/j.ophtha.2009.06.058. [DOI] [PubMed] [Google Scholar]
- Ren R, Wang N, Zhang X, Cui T, Jonas JB. Trans-lamina cribrosa pressure difference correlated with neuroretinal rim area in glaucoma. Graefes Arch Clin Exp Ophthalmol. 2011;249:1057–1063. doi: 10.1007/s00417-011-1657-1. [DOI] [PubMed] [Google Scholar]
- Wang N, Xie X, Yang D, Xian J, Li Y, Ren R, Peng X, Jonas JB, Weinreb RN. Orbital cerebrospinal fluid space in glaucoma: the Beijing intracranial and intraocular pressure (iCOP) study. Ophthalmology. 2012;119:2065–2073. doi: 10.1016/j.ophtha.2012.03.054. [DOI] [PubMed] [Google Scholar]
- Brown PD, Davies SL, Speake T, Millar ID. Molecular mechanisms of cerebrospinal fluid production. Neuroscience. 2004;129:957–970. doi: 10.1016/j.neuroscience.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen SF, Gorgels TG, Ramdas WD, Klaver CC, van Duijn CM, Jansonius NM, Bergen AA. The vast complexity of primary open angle glaucoma: Disease genes, risks, molecular mechanisms and pathobiology. Prog Retin Eye Res. 2013;37:31–67. doi: 10.1016/j.preteyeres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Banizs B, Pike MM, Millican CL, Ferguson WB, Komlosi P, Sheetz J, Bell PD, Schwiebert EM, Yoder BK. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 2005;132:5329–5339. doi: 10.1242/dev.02153. [DOI] [PubMed] [Google Scholar]
- Narita K, Kawate T, Kakinuma N, Takeda S. Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic. 2010;11:287–301. doi: 10.1111/j.1600-0854.2009.01016.x. [DOI] [PubMed] [Google Scholar]
- Dhechakaisaya S, Shuangshoti S, Susakares A. Extramedullary hematopoiesis of cranial dura mater and choroid plexus and terminal convulsions in a patient with thalassemia-hemoglobin E disease. J Med Assoc Thai. 1979;62:503–511. [PubMed] [Google Scholar]
- Eskazan AE, Ar MC, Baslar Z. Intracranial extramedullary hematopoiesis in patients with thalassemia: a case report and review of the literature. Transfusion. 2012;52:1715–1720. doi: 10.1111/j.1537-2995.2011.03499.x. [DOI] [PubMed] [Google Scholar]
- Tabesh H, Shekarchizadeh A, Mahzouni P, Mokhtari M, Abrishamkar S, Abbasi FS. An intracranial extramedullary hematopoiesis in a 34-year-old man with beta thalassemia: a case report. J Med Case Rep. 2011;5:580. doi: 10.1186/1752-1947-5-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Tang L, Pham H. Identification of neural progenitors in the adult mammalian eye. Biochem Biophys Res Commun. 2000;270:517–521. doi: 10.1006/bbrc.2000.2473. [DOI] [PubMed] [Google Scholar]
- Bhatia B, Singhal S, Lawrence JM, Khaw PT, Limb GA. Distribution of Muller stem cells within the neural retina: evidence for the existence of a ciliary margin-like zone in the adult human eye. Exp Eye Res. 2009;89:373–382. doi: 10.1016/j.exer.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Das AV, James J, Rahnenfuhrer J, Thoreson WB, Bhattacharya S, Zhao X, Ahmad I. Retinal properties and potential of the adult mammalian ciliary epithelium stem cells. Vision Res. 2005;45:1653–1666. doi: 10.1016/j.visres.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Transdifferentiation of pigmented epithelial cells: a source of retinal stem cells? Dev Neurosci. 2001;23:268–276. doi: 10.1159/000048710. [DOI] [PubMed] [Google Scholar]
- Lord-Grignon J, Abdouh M, Bernier G. Identification of genes expressed in retinal progenitor/stem cell colonies isolated from the ocular ciliary body of adult mice. Gene Expr Patterns. 2006;6:992–999. doi: 10.1016/j.modgep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Martinez-Navarrete GC, Angulo A, Martin-Nieto J, Cuenca N. Gradual morphogenesis of retinal neurons in the peripheral retinal margin of adult monkeys and humans. J Comp Neurol. 2008;511:557–580. doi: 10.1002/cne.21860. [DOI] [PubMed] [Google Scholar]
- Abburi C, Prabhakar S, Kalra J, Huria A, Anand A. Vascular endothelial growth factor (VEGF) induced proliferation of human fetal derived ciliary epithelium stem cells is mediated by jagged-N cadherin pathway. Curr Neurovasc Res. 2013;10:93–102. doi: 10.2174/1567202611310020002. [DOI] [PubMed] [Google Scholar]
- Ballios BG, Clarke L, Coles BL, Shoichet MS, van der Kooy D. The adult retinal stem cell is a rare cell in the ciliary epithelium whose progeny can differentiate into photoreceptors. Biol Open. 2012;1:237–246. doi: 10.1242/bio.2012027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis GC, Aruta C, Comitato A, De MA, Marigo V. Functional and molecular characterization of rod-like cells from retinal stem cells derived from the adult ciliary epithelium. PLoS One. 2012;7:e33338. doi: 10.1371/journal.pone.0033338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Cho KS, Tchedre K, Lee SW, Guo C, Kinouchi H, Fried S, Sun X, Chen DF. Ephrin-A3 suppresses Wnt signaling to control retinal stem cell potency. Stem Cells. 2013;31:349–359. doi: 10.1002/stem.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Coles BL, Dorval K, Bremner R, Bessho Y, Kageyama R, Hino S, Matsuoka M, Craft CM, McInnes RR. et al. Maximizing functional photoreceptor differentiation from adult human retinal stem cells. Stem Cells. 2010;28:489–500. doi: 10.1002/stem.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A, Pearson RA, MacLaren RE, Smith AJ, Sowden JC, Ali RR. Comparative analysis of progenitor cells isolated from the iris, pars plana, and ciliary body of the adult porcine eye. Stem Cells. 2007;25:2430–2438. doi: 10.1634/stemcells.2007-0035. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, van der Kooy D. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Xu H, Sta Iglesia DD, Kielczewski JL, Valenta DF, Pease ME, Zack DJ, Quigley HA. Characteristics of progenitor cells derived from adult ciliary body in mouse, rat, and human eyes. Invest Ophthalmol Vis Sci. 2007;48:1674–1682. doi: 10.1167/iovs.06-1034. [DOI] [PubMed] [Google Scholar]
- Cicero SA, Johnson D, Reyntjens S, Frase S, Connell S, Chow LM, Baker SJ, Sorrentino BP, Dyer MA. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc Natl Acad Sci USA. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdoni S, Baron M, Lakowski J, Decembrini S, Smith AJ, Pearson RA, Ali RR, Sowden JC. Adult ciliary epithelial cells, previously identified as retinal stem cells with potential for retinal repair, fail to differentiate into new rod photoreceptors. Stem Cells. 2010;28:1048–1059. doi: 10.1002/stem.423. [DOI] [PubMed] [Google Scholar]
- Longatti P, Basaldella L, Orvieto E. Dei Tos A, Martinuzzi A: Aquaporin(s) expression in choroid plexus tumours. Pediatr Neurosurg. 2006;42:228–233. doi: 10.1159/000092359. [DOI] [PubMed] [Google Scholar]
- Verkman AS, Ruiz-Ederra J, Levin MH. Functions of aquaporins in the eye. Prog Retin Eye Res. 2008;27:420–433. doi: 10.1016/j.preteyeres.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadsheh MF, Byun N, Mount DB, Delpire E. Localization of the KCC4 potassium-chloride cotransporter in the nervous system. Neuroscience. 2004;123:381–391. doi: 10.1016/j.neuroscience.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Janssen SF, Gorgels TGMF, van der Spek PJ, Jansonius NM, Bergen AAB. In silico analysis of the molecular machinery underlying aquoeus humor production: potential implication for glaucoma. J CLin Bioinform. 2013;3:21. doi: 10.1186/2043-9113-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin MD, Kaplan MR, Peterson LN. Gullans SRm Hebert SC, Delpire E: Expression of the Na(+)-K(+)-2Cl- cotransporter BSC2 in the nervous system. Am J Physiol. 1997;272:C173–SC183. doi: 10.1152/ajpcell.1997.272.1.C173. [DOI] [PubMed] [Google Scholar]
- Wu Q, Delpire E, Hebert SC, Strange K. Functional demonstration of Na + −K + −2Cl- cotransporter activity in isolated, polarized choroid plexus cells. Am J Physiol. 1998;275:C1565–C1572. doi: 10.1152/ajpcell.1998.275.6.C1565. [DOI] [PubMed] [Google Scholar]
- Christensen HL, Nguyen AT, Pedersen FD, Damkier HH. Na + dependent acid–base transporters in the choroid plexus; insights from slc4 and slc9 gene deletion studies. Front Physion. 2013;4:304. doi: 10.3389/fphys.2013.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D, Schibler MJ, Pushkin A, Sassani P, Abuladze N, Naser Z, Kurtz I. Immunolocalization of electrogenic sodium-bicarbonate cotransporters pNBC1 and kNBC1 in the rat eye. Am J Physiol Renal Physiol. 2001;281:F920–F935. doi: 10.1152/ajprenal.2001.281.5.F920. [DOI] [PubMed] [Google Scholar]
- Watsky MA, Cooper K, Rae JL. Sodium channels in ocular epithelia. Pflugers Arch. 1991;419:454–459. doi: 10.1007/BF00370788. [DOI] [PubMed] [Google Scholar]
- Burdon KP, Crawford A, Casson RJ, Hewitt AW, Landers J, Danoy P, Mackey DA, Mitchell P, Healey PR, Craig JE. Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmology. 2012;119:1539–1545. doi: 10.1016/j.ophtha.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. Association between genetic variants associated with vertical cup-to-disc ratio and phenotypic features of primary open-angle glaucoma. Ophthalmology. 2012;119:1819–1825. doi: 10.1016/j.ophtha.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Takamoto M, Kaburaki T, Mabuchi A, Araie M, Amano S, Aihara M, Tomidokoro A, Iwase A, Mabuchi F, Kashiwagi K. et al. Common variants on chromosome 9p21 are associated with normal tension glaucoma. PLoS One. 2012;7:e40107. doi: 10.1371/journal.pone.0040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM, Abdrabou W, Fan BJ, Wang DY, Brodeur W. et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimasi DP, Burdon KP, Hewitt AW, Fitzgerald J, Wang JJ, Healey PR, Mitchell P, Mackey DA, Craig JE. Genetic investigation into the endophenotypic status of central corneal thickness and optic disc parameters in relation to open-angle glaucoma. Am J Ophthalmol. 2012;154:833–842. doi: 10.1016/j.ajo.2012.04.023. [DOI] [PubMed] [Google Scholar]
- Ramdas WD, van Koolwijk LM, Ikram MK, Jansonius NM, De Jong PT, Bergen AA, Isaacs A, Amin N, Aulchenko YS, Wolfs RC. et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JB, Wang N. Intracranial pressure and glaucoma. J Glaucoma. 2013;22(Suppl 5):S13–S14. doi: 10.1097/IJG.0b013e31829349bf. [DOI] [PubMed] [Google Scholar]
- Killer HE, Miller NR, Flammer J, Meyer P, Weinreb RN, Remonda L, Jaggi GP. Cerebrospinal fluid exchange in the optic nerve in normal-tension glaucoma. Br J Ophthalmol. 2012;96:544–548. doi: 10.1136/bjophthalmol-2011-300663. [DOI] [PubMed] [Google Scholar]
- Wostyn P, De GV, Van DD, Audenaert K, De Deyn PP. Senescent changes in cerebrospinal fluid circulatory physiology and their role in the pathogenesis of normal-tension glaucoma. Am J Ophthalmol. 2013;156:5–14. doi: 10.1016/j.ajo.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Chiou GC. Effects of alpha 1 and alpha 2 activation of adrenergic receptors on aqueous humor dynamics. Life Sci. 1983;32:1699–1704. doi: 10.1016/0024-3205(83)90831-7. [DOI] [PubMed] [Google Scholar]
- Haywood JR, Vogh BP. Some measurements of autonomic nervous system influence on production of cerebrospinal fluid in the cat. J Pharmacol Exp Ther. 1979;208:341–346. [PubMed] [Google Scholar]
- McCormick JM, McCormick PW, Zabramski JM, Spetzler RF. Intracranial pressure reduction by a central alpha-2 adrenoreceptor agonist after subarachnoid hemorrhage. Neurosurgery. 1993;32:974–979. doi: 10.1227/00006123-199306000-00016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total list of genes expressed significantly higher in CPE compared to NPE. Selection criteria were a fold change > 5 and p-value < 0.001.
Total list of genes expressed significantly higher in NPE compared to CPE. Selection criteria were a fold change > 5 and p-value < 0.001.
Molecular networks 2–25 generated by the Ingenuity software from the genes expressed significantly higher in the CPE compared to NPE. Grey symbols represent genes expressed significantly higher in the CPE. Transparent entries are molecules inserted by the knowledge database. Gene names are abbreviated according to those used in GenBank. Solid lines indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). The main functionalities given by Ingenuity for this entire molecular network are shown in the diagrams. Reproduced with permission.
Genes in the NPE expressed significantly higher than in CPE involved in the biological function of development.
Genes in the NPE expressed significantly higher than in CPE involved in the biological function of cellular protrusions.
Molecular networks generated by the Ingenuity software from the genes expressed significantly higher in the NPE compared to CPE. Grey symbols represent genes expressed significantly higher in the NPE. Transparent entries are molecules inserted by the knowledge database. Gene names are abbreviated according to those used in GenBank. Solid lines indicate direct physical or functional relationships between molecules (such as regulating and interacting protein domains). The main functionalities given by Ingenuity for this entire molecular network are shown in the diagrams. Reproduced with permission.
Genes coding ion transport proteins involved in CSF and AH production. We constructed a list of 157 coding genes corresponding to all the CPE and CBE ion transport proteins as described by Brown et al. [30] and Civan et al.[20].
Genes expressed significantly higher in the CPE than in NPE involved in the biological function of angiogenesis.