Abstract
The Mayo Clinic Center for Individualized Medicine (CIM) is designed to discover and integrate the latest in genomic, molecular, and clinical science into personalized care for patients across a multiple-site academic medical center. Despite a highly integrated structure, fully electronic medical record, and strong administrative support, achievement of this goal has had challenges. This article will describe the activities of the CIM, with emphasis on the strategy being used to clinically implement genomics.
BACKGROUND
The Mayo Clinic is an integrated, not-for-profit academic medical center with origins in Rochester, Minnesota, with other major sites in Jacksonville, Florida, and Phoenix–Scottsdale, Arizona. The Mayo Clinic is large, with 58,300 employees, including 3,800 staff physicians and scientists in 2011. A total of 1,113,000 individual patients were seen at Mayo sites that year. The Mayo Clinic is governed by a single public board of trustees that is responsible for the medical practice and for the clinic’s research and educational programs, including the Mayo Medical School, the Mayo Graduate School, and large residency and fellowship programs at all three major locations.1 It is within this structure that the CIM is tasked to implement clinically relevant genomic science into as many aspects of the clinic’s activities as possible, especially into the daily medical practice.
CIM: OPPORTUNITIES, STRUCTURE, AND PROGRAMS
There is increasing agreement that medicine is on the verge of a potentially transformational event as a result of the application of genomics and other “omic” sciences to patient care, an event comparable with the transformation that occurred in the late nineteenth century as a result of the discovery of anesthesia and aseptic surgery. The Mayo Clinic CIM was created to catalyze movement of genomic science to the bedside. It focused initially on creating the infrastructure required to achieve that goal, beginning with enhanced centralized research genotyping and DNA sequencing core facilities but including the recruitment of staff with expertise in bioinformatics and bioethics, and the expanded use of newer technologies, e.g., next-generation DNA sequencing. However, in 2011 it became clear that a more ambitious and significantly expanded effort would be required to achieve the goal of the medical implementation of genomics. As a result, an expanded effort consisting of a series of complementary “Translational” and “Infrastructural” programs was created (Table 1). This effort can be compared with programs at other academic medical centers that have similar goals.2,3 Each of the Translational programs has initiated a series of multidisciplinary “key projects,” and each has also issued “Requests for Applications” (RFAs) focused on projects likely to have an effect on the clinical practice, including clinical trials and the development of novel organizational approaches for clinical decision making using genomic data. These proposals, supported by either institutional funds or benefactors, were peer reviewed with an emphasis on both scientific merit and potential impact on the clinical practice. These projects also served to highlight areas of need for faculty recruitment as well as organizational structures that required modification to facilitate the clinical implementation of genomics, for example, realization of the need to establish an “Individualized Medicine Clinic.”
Table 1.
Mayo Center for Individualized Medicine Translational and Infrastructural programs
| Translational programs | Infrastructural programs |
|---|---|
| Pharmacogenomics | Biomedical Informatics |
| Biomarker Discovery | Biobank and Biorepository |
| Clinomics | Bioethics |
| Epigenomics | Medical Genome Facility |
| Microbiome | Information Technology |
| Administration |
We will use the CIM Pharmacogenomics Program to illustrate the approach being taken, in part, because this area of genomic medicine will be familiar to readers of Clinical Pharmacology & Therapeutics. For example, during the past year, the CIM Pharmacogenomics Program has funded six internally peer-reviewed RFAs led by faculty members in five different clinical departments or divisions. These pilot projects involved genotyping or DNA sequencing to discover or test genomic biomarkers for drug response phenotypes. In many cases, the principal investigators for these RFAs have been junior faculty members attempting both to answer questions related to clinical practice and to obtain preliminary data that might be used to apply for extramural peer-reviewed funding for more definitive studies. The use of the RFA mechanism has served both to engage a broad cross section of our faculty and to reveal clinical areas that are ripe for the application of genomic science.
The CIM Pharmacogenomics Program also sponsors a series of larger projects that range from the clinical implementation of the US Food and Drug Administration–reviewed genomic markers for “drug–gene pairs” related to variation in efficacy or adverse drug reactions (see the “Table of Pharmacogenomic Biomarkers in Drug Labels” at the FDA.gov website), tumor and germline DNA sequencing projects involving breast cancer and prostate cancer designed to identify “signatures” for variation in drug response, and a large genotyping study designed to test the clinical utility of genomic biomarkers for cardiovascular drug response. For example, the breast cancer study offers the opportunity to women with high-risk breast cancer to undergo germline and tumor exome sequencing before, during, and after standard chemotherapy in the neoadjuvant setting. All of the Translational programs listed in Table 1 have developed RFAs and a portfolio of projects—each emphasizing both discovery and clinical implementation. In October 2012, the CIM structure was expanded to include Individualized Medicine Clinics at all three Mayo Clinic sites. These Individualized Medicine Clinics are focused initially on the application of whole-exome and/or whole-genome sequencing to two groups of patients, patients with end-stage cancer and “diagnostic odyssey” patients, patients who clearly suffer from disease but without a diagnosis or a defined therapeutic program. Creation of the Individualized Medicine Clinics was necessary to overcome challenges associated with the need to have operational responsibility outside of traditional departmental structures, and the needs to focus expertise, to have strong selection criteria, and to concentrate therapeutic expertise.
Creation of the CIM, including the Individualized Medicine Clinics, required significant institutional resources but was also made possible by a preexisting foundation created over decades by extramural peer-reviewed support, primarily from National Institutes of Health grants, that helped make it possible to recruit the faculty and build the infrastructure that made the creation of the CIM possible. Specifically, the decades-long support provided for the National Cancer Institute– funded Mayo Comprehensive Cancer Center, a grant from the National Institute of General Medical Sciences and other National Institutes of Health institutes supporting the Mayo Pharmacogenomics Research Network, the National Human Genome Research Institute–funded eMERGE grant, and one of the first Clinical and Translational Science Award grants are just a few examples of the extramural funding vehicles that created the infrastructure on which the CIM rests. However, it should be emphasized that, critical as this support from the National Institutes of Health and other funding sources was, it was also critically important that institutional leadership provide substantial resources and, of equal importance, vision and a determination to make genomic implementation a success.
PROGRESS AND CHALLENGES
The creation of the CIM and the center’s expansion in 2011 have substantially advanced the implementation of genomic medicine at the Mayo Clinic, but achievement of the CIM mission remains a work in progress. Substantial challenges are associated with attempting to achieve that mission in any medical center, challenges that are mirrored and magnified by the size and geographic dispersion of the Mayo Clinic. In an earlier publication in this journal,4 we listed a few of the steps required to implement even relatively straightforward “drug–gene pairs”; that list included evidence of clinical utility, the existence of objective practice guidelines, the availability of DNA genotyping, or sequencing in a Clinical Laboratory Improvement Amendments (CLIA)–approved environment, a mechanism for incorporating genomic data into the electronic medical record, a mechanism for pharmacy “alerts,” user-friendly decision-support tools, insurance coverage of genomic testing, and broad acceptance by both patients and the medical staff. To this list, we also need to add challenges associated with the generation, interpretation, visualization, and storage of “clinical-grade” whole-exome and whole-genome sequencing data. To achieve these additional goals involves informatics challenges unlike any that medicine has faced previously. We have now accomplished all or most of these goals for an initial set of drug–gene pairs, specifically, “TPMT–thiopurines,” “HLA-B*5071–abacavir,” “ HLA-B*1502– carbamazepine,” and “IL28B–ribavirin–pegylated interferon.” Testing for these particular drug–gene pairs is usually reimbursed, but this is variable. Genotyping usually occurs when the ordering physician requests it in response to an alert, but we are also testing “preemptive” genotyping. This initial process, led by a Pharmacogenomics Task Force, took nearly a year, but we now plan to add four to six additional drug–gene pairs to the list each year, using the same process. The Pharmacogenomics Task Force is part of the Mayo Pharmacy Formulary Committee but is sponsored by the CIM Pharmacogenomics Program (see Figure 1).
Figure 1.
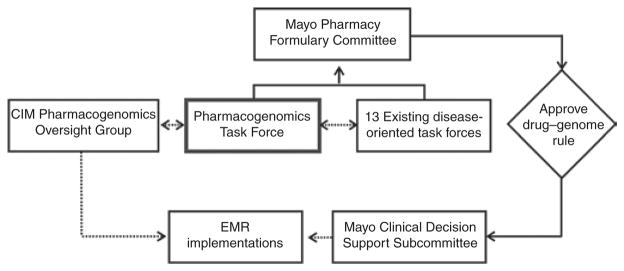
The position of the “Pharmacogenomics Task Force” within the structure of the multiple-site Mayo Pharmacy Formulary Program is depicted graphically. This task force is part of the Mayo Center for Individualized Medicine Pharmacogenomics Program and is led by the director of the Department of Laboratory Medicine and Pathology Nucleotide Polymorphism Laboratory. The task force selects, in consultation with existing disease-oriented task forces, clinically relevant drug–gene pairs for which there is evidence of clinical utility for inclusion in the electronic medical record, as shown in the figure.
CONCLUSION
In summary, we are privileged to live at a time in which biomedical science is advancing so rapidly that our profession is passing through a singularity; if we rise to the challenges, it will make the curriculum that most of us were taught in medical school seem old-fashioned. The founders of the Mayo Clinic, the two surgeon brothers William and Charles Mayo, lived at a time in which the development of anesthesia and aseptic surgery made it possible for a clinic in a small town in Minnesota to grow to the size of today’s Mayo Clinic. We may be living through a similar time of dramatic change for medicine as a result of rapid advances in biomedical science and their application to medical practice. We believe that the creation of practice-based centers similar to the Mayo Clinic CIM will accelerate the evolution of the “promise” of genomics to truly become the “practice” of genomic medicine.
Acknowledgments
This work was supported by National Institutes of Health grants R01 DK57061, R01 DK52766, U01 DK74008, P01 DK68055, U19 GM61388 (The Pharmacogenomics Research Network), U01 HG005137, R01 GM28157, and R01 CA132780, as well as by the Mayo Center for Individualized Medicine.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Weinshilboum RM. The Gordon Wilson Lecture. The Mayo model: one path to an academic medical center. Trans Am Clin Climatol Assoc. 2002;113:91–103. discussion 103. [PMC free article] [PubMed] [Google Scholar]
- 2.Kitzmiller JP, et al. Program in pharmacogenomics at the Ohio State University Medical Center. Pharmacogenomics. 2012;13:751–756. doi: 10.2217/pgs.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulley JM, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira NL, Weinshilboum RM. The impact of pharmacogenomics on the management of cardiac disease. Clin Pharmacol Ther. 2011;90:493–495. doi: 10.1038/clpt.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]


