Abstract
Context
The growth inhibitory effect of tamoxifen, which is used for the treatment of hormone receptor–positive breast cancer, is mediated by its metabolites, 4-hydroxytamoxifen and endoxifen. The formation of active metabolites is catalyzed by the polymorphic cytochrome P450 2D6 (CYP2D6) enzyme.
Objective
To determine whether CYP2D6 variation is associated with clinical outcomes in women receiving adjuvant tamoxifen.
Design, Setting, and Patients
Retrospective analysis of German and US cohorts of patients treated with adjuvant tamoxifen for early stage breast cancer. The 1325 patients had diagnoses between 1986 and 2005 of stage I through III breast cancer and were mainly postmenopausal (95.4%). Last follow-up was in December 2008; inclusion criteria were hormone receptor positivity, no metastatic disease at diagnosis, adjuvant tamoxifen therapy, and no chemotherapy. DNA from tumor tissue or blood was genotyped for CYP2D6 variants associated with reduced (*10, *41) or absent (*3, *4, *5) enzyme activity. Women were classified as having an extensive (n = 609), heterozygous extensive/intermediate (n = 637), or poor (n = 79) CYP2D6 metabolism.
Main Outcome Measures
Time to recurrence, event-free survival, disease-free survival, and overall survival.
Results
Median follow-up was 6.3 years. At 9 years of follow-up, the recurrence rates were 14.9% for extensive metabolizers, 20.9% for heterozygous extensive/intermediate metabolizers, and29.0%for poor metabolizers, and all-cause mortality rates were 16.7%, 18.0%, and 22.8%, respectively. Compared with extensive metabolizers, there was a significantly increased risk of recurrence for heterozygous extensive/intermediate metabolizers (time to recurrence adjusted hazard ratio [HR], 1.40; 95% confidence interval [CI], 1.04–1.90) and for poor metabolizers (time to recurrence HR, 1.90; 95% CI, 1.10–3.28). Compared with extensive metabolizers, those with decreased CYP2D6 activity (heterozygous extensive/intermediate and poor metabolism) had worse event-free survival (HR, 1.33; 95% CI, 1.06–1.68) and disease-free survival (HR, 1.29; 95% CI, 1.03–1.61), but there was no significant difference in overall survival (HR, 1.15; 95% CI, 0.88–1.51).
Conclusion
Among women with breast cancer treated with tamoxifen, there was an association between CYP2D6 variation and clinical outcomes, such that the presence of 2 functional CYP2D6 alleles was associated with better clinical outcomes and the presence of nonfunctional or reduced-function alleles with worse outcomes.
Tamoxifen has been the gold standard for the last25years for endocrine treatment of breast cancer. It is estimated that the lives of half a million women have been saved with adjuvant tamoxifen therapy.1 Although long-term experience showed that tamoxifen is safe and effective,2 de novo and acquired drug resistance is a major issue. Several lines of evidence indicate that most of the tamoxifen antiproliferative effects in breast cancer models are mediated by the metabolites 4-hydroxytamoxifen and endoxifen,3–10 with a different mode of action for endoxifen being suggested.11 Endoxifen is predominantly formed by the polymorphic enzyme cytochrome P450 (CYP) 2D6.12 Approximately 100 CYP2D6 genetic variants have been identified, which manifest in the population in 4 distinct phenotypes, extensive (normal activity), intermediate (reduced activity), poor (no activity), and ultrarapid (high activity) metabolism,13–17 and a gene-dose effect with respect to endoxifen plasma concentrations has been demonstrated.18 Thus, it can be speculated that genotype-related differences in the formation of active metabolites influence therapeutic response to tamoxifen. Recent clinical studies in Europeans and Asians showed that non-favorable outcome with tamoxifen was associated with poor or intermediate metabolizer genotypes19–24; however, 3 studies showed discrepant results.25–27
Data from clinical trials comparing the efficacy of aromatase inhibitors (AIs) and tamoxifen showed an advantage of AIs in the adjuvant endocrine treatment of early stage breast cancer.28–30 However, because the absolute difference in recurrence rates comparing tamoxifen and AIs is less than 5%,29 it has been hypothesized that treatment outcomes following tamoxifen in patients with normal CYP2D6 enzyme function would be similar to treatment outcomes following an AI.31
While CYP2D6 tamoxifen pharmacogenetics have generated considerable interest and have been subject to extensive review,17,32–35 the clinical relevance has been questioned because of discrepant results and limited sample sizes. Therefore, we conducted an adequately powered multicenter study including retrospectively and prospectively collected patient data based on strict inclusion criteria.
METHODS
Study Population
The study included patients recommended to receive 5 years of adjuvant tamoxifen. Inclusion criteria were a histologically proven diagnosis of primary breast cancer (stage I, II, or III), no previous chemotherapy or endocrine treatment other than tamoxifen, no metastatic disease at diagnosis, and hormone receptor positivity (estrogen receptor– positive and/or progesterone receptor– positive) assessed by locally performed immunohistochemistry.
A total of 1580 patients, a consecutively collected retrospective German breast cancer cohort (Stuttgart, Karlsruhe, Erlangen, and Mainz) and prospectively collected patients from the US North Central Cancer Treatment Group (NCCTG) 89-30-52 trial (Mayo Clinic, Rochester, Minnesota), were included. Patients in the German cohort were treated according to standard hospital practice, and inclusion followed the defined inclusion criteria. The NCCTG 89-30-52 trial is a randomized phase 3 clinical trial in post-menopausal women comparing tamoxifen alone vs tamoxifen in combination with the androgen fluoxymesterone, of which patients from the tamoxifen-only group were included.36
Last follow-up was between August and December 2008. The present study included 350 patients from previous reports19–21 with extended follow-up and extensive genotyping; 219 patients were noneligible (123 had chemotherapy, 60 had metastatic disease, and/or 47 had negative or unknown hormone receptor status or inconsistent follow-up data). Thus, 1361 patients underwent pharmacogenetic analysis. Ethical approval was obtained for all participating institutions. The need for additional informed consent beyond the NCCTG trial was waived by institutional review boards. For the majority of German patients, written informed consent was obtained. Among cases for whom informed consent could not be obtained, inclusion in the study is in agreement with German ethical standards and approved by ethical committees.
Study Design and End Points
The primary objective was to determine the association between CYP2D6 genetic variants and tamoxifen outcome. End points were as follows: (1) Time to recurrence was defined as time from diagnosis or randomization to documentation of a breast event, any local, locoregional, or distant recurrence of breast cancer or a contralateral breast cancer. (2) Event-free survival was defined as time to the first occurrence of a breast event or death from any cause. (3) Disease-free survival was defined as time to first occurrence of a breast event, a second nonbreast primary cancer, or death from any cause. (4) Overall survival was estimated as the time from registration to death from any cause. The secondary objective was to perform an exploratory analysis of clinical outcomes of tamoxifen-treated patients within CYP2D6 genotype subgroups vs the outcome stratification derived from the Arimidex, Tamoxifen, Alone or in Combination (ATAC) 100-monthanalysis of patients randomized to tamoxifen and anastrozole.29
DNA Source, Genotyping, and Definition of Phenotypes
Genomic DNA was extracted from whole blood (n = 601), fresh frozen tumor (n = 101), and formalin-fixed, paraffin-embedded tumor tissues (n = 659) according to standard procedures. Genotyping of German samples was at Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology (IKP), Stuttgart, and of NCCTG samples was at Mayo Clinic (eAppendix, available at http://www.jama.com). The CYP2D6 polymorphisms refer to the CYP Allele Nomenclature Committee (http://www.cypalleles.ki.se) (eTable).
The CYP2D6 pharmacogenetic analysis uses defined genotype-phenotype relationships based on known biochemical and pharmacological effects and included major CYP2D6 alleles within a population of European descent.15,16 We refer to the following phenotypes: those with poor metabolism (PM), lacking active enzyme function, are homozygous or compound heterozygous for CYP2D6*3, *4, or *5 alleles. Intermediate metabolizers (IM) have reduced enzyme activity and carry *10 and *41 alleles either homozygous or in combination with a PM allele. Since heterozygous carriers of PM or IM alleles (hetEM) have no distinct phenotype, this group was combined with IM to define a patient group associated with intermediate impairment of CYP2D6 activity (hetEM/IM). Patients with extensive metabolism (EM) have normal enzyme function and were characterized by the absence of PM and IM alleles. Ultrarapid metabolizer (UM) patients, with high enzyme activity, have duplicated gene copies without a PM or IM allele.
Statistical Analyses
Clinical and genotyping data were analyzed at the IKP central database. Because outcome effects were previously described to be strongest between PM and EM phenotypes, the sample size was estimated with respect to those 2 patient groups, assuming a ratio of 1 in 10. To detect a hazard ratio (HR) of 1.85 between CYP2D6 phenotypes PM and EM (originally reported by Goetz et al19), with a statistical power of 90% and a 1-sided α level of .05, the number of required events is 166, resulting in a minimum sample size of 1279.
Log-rank tests were used to compare 3 genotype-phenotype states: 2 functional alleles (EM), heterozygous EM or homozygous IM alleles (hetEM/IM), and homozygous PM alleles (PM), and Kaplan-Meier estimates were calculated (patients with the UM phenotype with duplicated gene copies were included in the EM group). Cox proportional hazards modeling with adjustment for tumor size, node status, and histological grade was used to adjust for clinical prognostic factors. In all Cox models, menopause status and subcohort assignment (ie, mode of patient recruitment— retrospective vs prospective) were used as stratum variables to account for population heterogeneity. To decide whether the consideration of CYP2D6 phenotypes improves the prognostic accuracy, Cox models with and without CYP2D6 genotype status were compared by means of concordance probability estimates and analysis of deviance including a likelihood ratio test.37
To model the differences in outcome between AIs and tamoxifen, HRs between CYP2D6 phenotypes in the present study were compared with HRs assigned to tamoxifen in the ATAC trial29 using a 1-sided Mantel-Haenszel log-rank test. Supposing the Cox proportionality hazard assumption, a hypothetical anastrozole survival curve was calculated (eAppendix) based on the HR from the ATAC trial and compared with survival curves of the present study. We used R statistical software, version 2.6.2, including libraries survival and Hmisc (http://www.r-project.org), as well as SPSS, version 16 (SPSS Inc, Chicago, Il-linois). Unless stated otherwise, all statistical tests were 2-sided and statistical significance was defined as P < .05.
RESULTS
Pharmacogenetic analyses were performed for 1325 of 1361 eligible patients for whom genotypes could be assigned. Baseline and tumor characteristics were similar among CYP2D6 phenotype subgroups (Table 1). Overall median age at diagnosis was 66 years and median follow-up was 76.1 months.
Table 1.
Baseline Patient and Tumor Characteristics and Frequencies of CYP2D6 Variants
| Characteristics | Overall (N = 1325) |
CYP2D6 Extensive Metabolism (n = 609)a |
CYP2D6 Decreased Metabolism (n = 716)a |
|
|---|---|---|---|---|
| Age at diagnosis, median (range), y | 66.0 (36.5–93.1) | 66.0 (36.5–93.1) | 66.0 (40.2–91.8) | |
| Follow-up, median (range), mo | 76.1 (2.1–243.6) | 75.2 (2.1–238.3) | 76.4 (2.2–243.6) | |
| Menopausal status, No. (%) | ||||
| Premenopausal | 54 (4.1) | 24 (3.9) | 30 (4.2) | |
| Postmenopausal | 1271 (95.9) | 585 (96.1) | 686 (95.8) | |
| Tumor size, cm, No. (%) | ||||
| ≤2 | 686 (51.8) | 318 (52.2) | 368 (51.4) | |
| >2 | 624 (47.1) | 284 (46.6) | 340 (47.5) | |
| Unknown | 15 (1.1) | 7 (1.1) | 8 (1.1) | |
| Node status, No. (%) | ||||
| N0 | 850 (64.1) | 407 (66.8) | 443 (61.8) | |
| N1 | 392 (29.6) | 165 (27.1) | 227 (31.7) | |
| N2 + N3 | 57 (4.3) | 24 (3.9) | 33 (4.6) | |
| Unknown | 26 (1.9) | 13 (2.1) | 13 (1.8) | |
| Grade, No. (%) | ||||
| 1 | 161 (12.1) | 72 (11.8) | 89 (12.4) | |
| 2 | 918 (69.3) | 423 (69.4) | 495 (69.1) | |
| 3 | 201 (15.2) | 87 (14.3) | 114 (15.9) | |
| Unknown | 45 (3.4) | 27 (4.4) | 18 (2.5) | |
| Estrogen receptor status, No. (%) | ||||
| Positive | 1282 (96.7) | 592 (97.2) | 690 (96.4) | |
| Negative | 38 (2.8) | 15 (2.5) | 23 (3.2) | |
| Unknown | 5 (0.4) | 2 (0.3) | 3 (0.4) | |
| Progesterone receptor status, No. (%)b | ||||
| Positive | 984 (74.3) | 449 (73.7) | 535 (74.7) | |
| Negative | 123 (9.3) | 56 (9.2) | 67 (9.3) | |
| Unknown | 218 (16.4) | 104 (17.1) | 114 (15.9) | |
| Radiotherapy, No. (%)b | ||||
| Yes | 769 (58.0) | 354 (58.1) | 415 (57.9) | |
| No | 456 (34.4) | 206 (33.8) | 250 (34.9) | |
| Unknown | 100 (7.5) | 49 (8.0) | 51 (7.1) | |
| CYP2D6 variants by genotype (allele %)c | ||||
| 2D6*3 (PM) | 917/31/1 (1.7) | |||
| 2D6*4 (PM) | 854/400/71 (20.0) | |||
| 2D6*5 (PM) | 646/33/0 (2.4) | |||
| 2D6*10 (IM) | 1175/40/4 (2.0) | |||
| 2D6*41 (IM) | 1080/206/15 (9.0) | |||
| 2D6 duplication (UM) | (2.3) | |||
Abbreviations: CYP, cytochrome P450; EM, extensive metabolism; IM, intermediate metabolism; PM, poor metabolism; UM, ultrarapid metabolism.
CYP2D6 phenotypes were assigned according to genotypes with 2 functional alleles (extensive) vs genotypes with at least 1 intermediate or 1 nonfunctional allele (decreased).
Progesterone receptor status and radiotherapy information were not available for all patients.
Genotype frequencies shown in the following order: homozygous EM/heterozygous/homozygous variant. Allele frequencies in percentages are in parentheses.
Genotype-Phenotype Frequencies
Numbers of CYP2D6 genotype frequencies (given as homozygous EM/heterozygous/homozygous variant with allele frequencies in parentheses) were as follows: CYP2D6*3: 917/31/1 (1.7%); CYP2D6*4: 854/400/71 (20.0%); CYP2D6*5: 646/33/0 (2.4%); CYP2D6*10: 1175/40/4 (2.0%); and CYP2D6*41: 1080/206/15 (9.0%). For technical reasons, gene duplication (UM) and deletion of allele *5 could be assessed only in blood- or fresh tumor–derived samples (n = 679) but not in paraffin-derived DNA samples. The prevalence of the UM phenotype was 2.3% (20/679), and prevalence of deletion of allele*5was 4.9% (33/679). Among the 1325 patients with genotyping were 609 EM (45.9%), 637 hetEM/IM (48.1%), and 79 PM (5.9%).
End-Point Analyses According to CYP2D6 Phenotypes
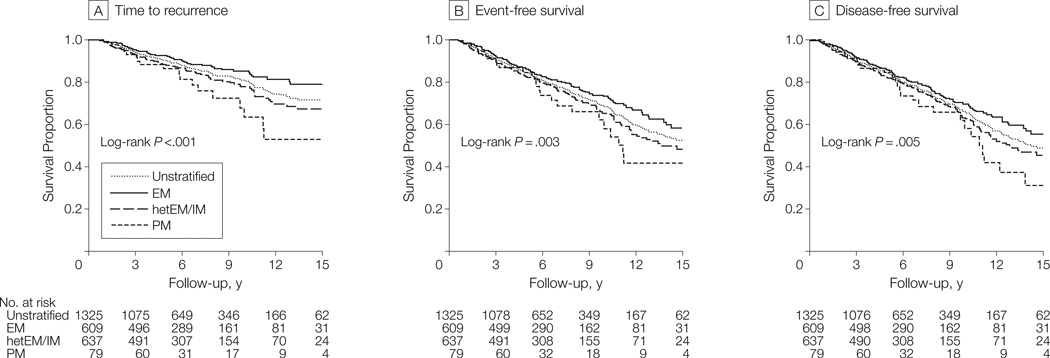
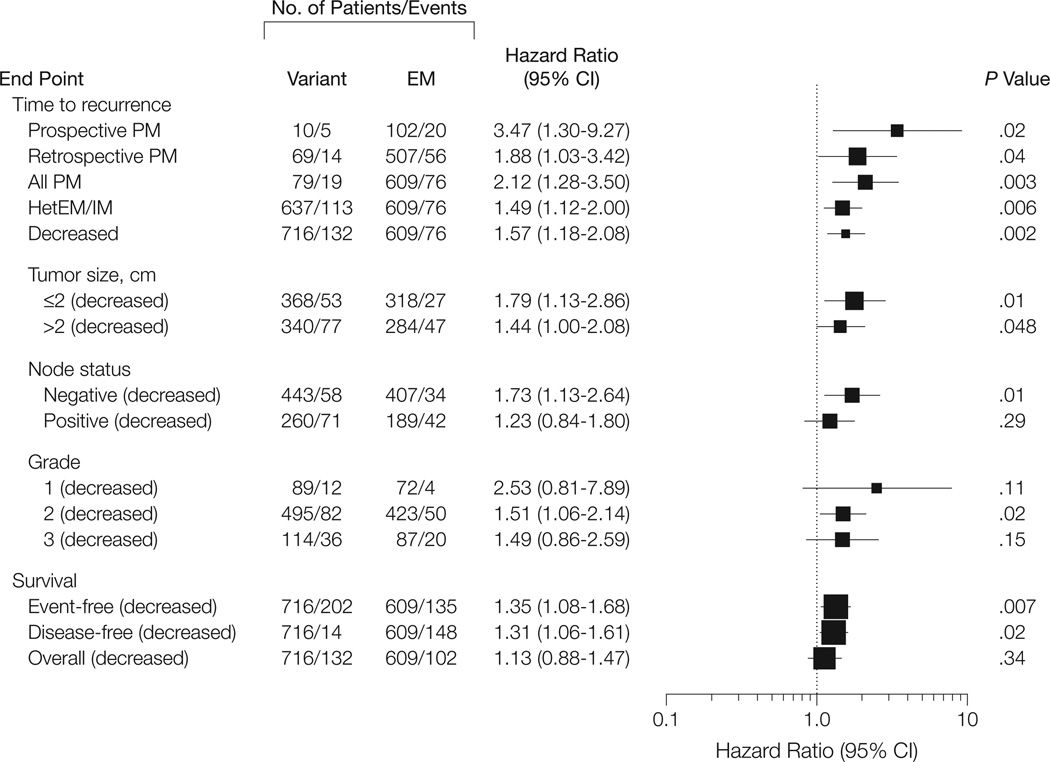
There was a variant-allele dose-dependent increase of the incidence of recurrence and death events among CYP2D6 phenotypes. Patients with EM had the lowest event probabilities, followed by hetEM/IM patients, and PM patients had the highest rates (Table 2). Kaplan-Meier estimates showed significantly higher breast cancer event rates in patients with reduced or absent CYP2D6 function vs EM patients (time to recurrence: P < .001; event-free survival: P = .003; and disease-free survival: P = .005) (Figure 1). The recurrence rates at 9 years of follow-up were 14.9% for EM, 20.9% for hetEM/IM, and 29.0% for PM. The unadjusted HR for PM was highest at 2.12 (95% confidence interval [CI], 1.28–3.50; P = .003) and for hetEM/IM was somewhat less at 1.49 (95% CI, 1.12–2.00; P = .006) (Figure 2). High enzyme activity in UM patients was associated with nonsignificantly fewer recurrences vs patients with 1 to 2 functional CYP2D6 gene copies. (eFigure).
Table 2.
Incidence of End-Point Events for CYP2D6 Phenotypes Predicted by Genotype
| Event | No. (% in Subgroup) | ||
|---|---|---|---|
| EM (n = 609)a | hetEM/IM (n = 637)a | PM (n = 79)a | |
| Local recurrence including second primary breast cancer | 40 (6.6) | 66 (10.4) | 12 (15.2) |
| Distant recurrence | 48 (7.9) | 57 (8.9) | 10 (12.7) |
| Overall recurrences | 76 (12.5) | 113 (17.7) | 19 (24.1) |
| Death from any cause | 102 (16.7) | 114 (18.0) | 18 (22.8) |
| Event-free survival eventb | 135 (22.2) | 176 (27.6) | 26 (32.9) |
Abbreviations: CYP, cytochrome P450; EM, extensive metabolism; IM, intermediate metabolism; PM, poor metabolism; UM, ultrarapid metabolism.
CYP2D6 phenotypes are defined as EM with 2 functional alleles (EM/EM, including UM), hetEM/IM with intermediate or 1 poor metabolism alleles (EM/IM, EM/PM, IM/IM, or IM/PM), and PM homozygous for poor metabolism alleles (PM/PM).
Event-free survival event was defined as the first event of local, regional, or distant recurrence; a second primary breast tumor; or death from any cause.
Figure 1. Kaplan-Meier Estimates of Time to Recurrence, Event-Free Survival, and Disease-Free Survival.
EM indicates extensive metabolism (ie, patients with 2 functional CYP2D6 alleles, including patients with ultrarapid metabolism); hetEM/IM, patients with intermediate or 1 poor metabolism allele; PM, patients homozygous for poor metabolism alleles. The curves were truncated at 15 years after diagnosis and calculations included overall time (median follow-up, 6.3 years). Percentage differences vs unstratified are as follows: for time to recurrence, at 9 years, 3.4% for EM and 10.7% for PM, and at 15 years, 7.2% for EM and 19.0% for PM; for event-free survival, at 9 years, 3.3% for EM and 6.4% for PM, and at 15 years, 5.7% for EM and 10.9% for PM; and for disease-free survival, at 9 years, 2.3% for EM and 4.5% for PM, and at 15 years, 6.4% for EM and 17.9% for PM.
Figure 2. Cox Proportional Hazards Model Estimates of CYP2D6 Phenotypes for Various End Points and Patient Subgroups.
CYP indicates cytochrome P450; EM, extensive metabolism (ie, patients with 2 functional alleles, including patients with ultrarapid metabolism); hetEM/IM, patients with intermediate or 1 poor metabolism allele; PM, patients homozygous for poor metabolism alleles; decreased, combined group of PM and hetEM/IM. Non-adjusted hazard ratios were calculated for the entire patient cohort and subgroups. Subgroups were analyzed with respect to mode of patient sampling and clinical prognostic factors. All comparisons refer to CYP2D6 EM as the reference. The size of the data markers is inversely proportional to the standard error of the hazard ratio.
The effect size for impaired CYP2D6 metabolism was greater in the prospectively collected group (HR, 3.47 [n = 212]) than in the retrospectively collected cohort (HR, 1.88 [n = 1113]); however, there was no statistical difference between the HRs (P = .15). We further evaluated the effect of decreasedCYP2D6 metabolism within each prognostic subgroup and demonstrated that the HR was consistently greater than 1 within each subgroup (Figure 2), with no significant interactions being detectable.
Because tumor size, node status, and histological grade were independently associated with outcome, we estimated gene variant–associated risks by Cox proportional hazards models adjusted for prognostic factors, conditional on menopausal and subcohort status. The adjusted HR for time to recurrence in PM relative to EM was 1.90 (95% CI, 1.10–3.28; P = .02) and was approximately half of this effect size in hetEM/IM patients (HR, 1.40; 95% CI, 1.04–1.90; P = .03). Decreased CYP2D6 activity was associated with worse event-free survival (HR, 1.33; 95% CI, 1.06–1.68; P = .01) and disease-free survival (HR, 1.29; 95% CI, 1.03–1.61; P = .02). Genotypes did not show a significant effect on overall survival (Table 3 and Figure 2). For all end points, we compared the Cox models that incorporated CYP2D6 with corresponding models without CYP2D6 genotypic information. Analysis of deviance and likelihood ratio tests showed that the predictive accuracy of the Cox models for time to recurrence (deviance, 7.51; P = .02), event-free survival (deviance, 5.1; P = .01), and disease-free survival (deviance, 6.19; P=.02) improved significantly with the incorporation of CYP2D6 genotypic information. Concordance probability estimates were 0.658 (model with genotypic status) vs 0.648 (model without genotypic status) for time to recurrence, 0.646 vs 0.639 for event-free survival, and 0.642 vs 0.636 for disease-free survival. For overall survival, no significant increase of discriminatory power by genotypic information was found (P = .31).
Table 3.
Cox Proportional Hazard Models for CYP2D6 Metabolizer Phenotypes at Different End Points
| Variablea | Hazard Ratio (95% Confidence Interval)b |
P Value |
|
|---|---|---|---|
| Time to Recurrence | |||
| CYP2D6 | |||
| EM | 1 [Reference] | ||
| hetEM/IM | 1.40 (1.04–1.90) | .03 | |
| PM | 1.90 (1.10–3.28) | .02 | |
| Tumor size | 1.52 (1.20–1.91) | <.001 | |
| Node status | |||
| 1 vs 0 | 1.76 (1.30–2.39) | <.001 | |
| 2/3 vs 0 | 2.38 (1.37–4.13) | <.001 | |
| Grade | 1.76 (1.35–2.30) | <.001 | |
| Event-Free Survival | |||
| CYP2D6 | |||
| EM | 1 [Reference] | ||
| Decreased | 1.33 (1.06–1.68) | .01 | |
| Tumor size | 1.44 (1.20–1.73) | <.001 | |
| Node status | |||
| 1 vs 0 | 1.88 (1.48–2.38) | <.001 | |
| 2/3 vs 0 | 2.59 (1.61–4.15) | <.001 | |
| Grade | 1.56 (1.26–1.93) | <.001 | |
| Disease-Free Survival | |||
| CYP2D6 | |||
| EM | 1 [Reference] | ||
| Decreased | 1.29 (1.03–1.61) | .02 | |
| Tumor size | 1.41 (1.18–1.69) | <.001 | |
| Node status | |||
| 1 vs 0 | 1.89 (1.50–2.38) | <.001 | |
| 2/3 vs 0 | 2.60 (1.63–4.16) | <.001 | |
| Grade | 1.53 (1.24–1.88) | <.001 | |
| Overall Survival | |||
| CYP2D6 | |||
| EM | 1 [Reference] | ||
| Decreased | 1.15 (0.88–1.51) | .32 | |
| Tumor size | 1.66 (1.33–2.08) | <.001 | |
| Node status | |||
| 1 vs 0 | 2.13 (1.60–2.82) | <.001 | |
| 2/3 vs 0 | 2.89 (1.54–5.42) | .001 | |
| Grade | 1.67 (1.30–2.15) | <.001 | |
Abbreviations: CYP, cytochrome P450; EM, extensive metabolism; IM, intermediate metabolism; PM, poor metabolism; UM, ultrarapid metabolism.
CYP2D6 phenotypes are defined as EM with 2 functional alleles (EM/EM, including UM), hetEM/IM with intermediate or 1 poor metabolism alleles (EM/IM, EM/PM, IM/IM, or IM/PM), and PM homozygous for poor metabolism alleles (PM/PM). Decreased refers to combined hetEM/IM and PM patients and was used in Cox models if either 1 of the 2 subphenotypes did not reach significance.CYP2D6 phenotype status was categorical, tumor size was coded as 1 (tumors ≤2 cm; reference), 2 (2–5 cm), or 3 (≥5 cm).Grade was coded as1 (low grade; reference), 2, and 3 (high grade). Node status was coded categorically with factor levels N0, N1, and combined N2/N3 (Table 1).
Hazard ratio estimates for CYP2D6 phenotypes were adjusted for tumor size, node status, and histological grade and stratified by menopause status and subcohort.
CYP2D6 Stratification and Endocrine Therapy Outcome
Patients with moderately impaired enzyme activity (hetEM/IM) had similar time-to-recurrence Kaplan-Meier survival curves vs unselected patients (Figure 1A). In contrast, recurrence risk for EM was lower vs unselected patients (P = .03) and was markedly increased for PM (P = .02). In an analogous way, Kaplan-Meier curves for CYP2D6-associated event-free and disease-free survival were compared with the entire cohort (Figure 1, B and C). Compared with the entire cohort unselected by genotype, after 9 years the absolute risk of CYP2D6 PM was increased by 10.7% (time to recurrence) and 6.4% (event-free survival), and was reduced for EM patients by 3.4% (time to recurrence) and 3.3% (event-free survival).
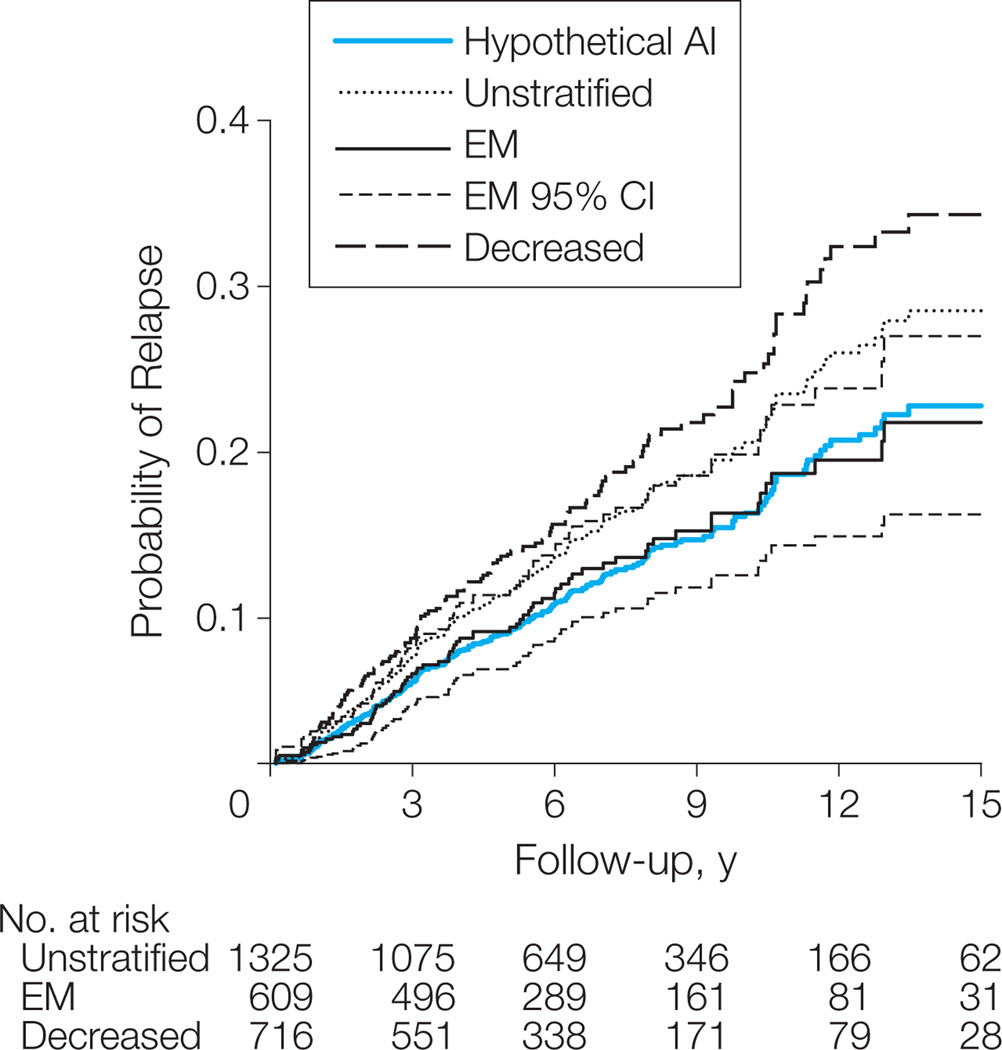
We evaluated whether CYP2D6 genotype stratification of tamoxifen outcome in the present study was more pronounced than the difference between tamoxifen vs anastrozole in the ATAC trial. The absolute difference in the proportion of patients with relapse between EM and PM was 6.7% at 5 years and 13.4% at 9 years. This effect size corresponds to an unadjusted HR of 2.12, which was significantly larger than the HR assigned to tamoxifen relative to anastrozole of 1.31 (P = .01) (Table 4).29 Supposing the proportional hazard assumption, a hypothetical survival function for anastrozole was calculated on the basis of a recurrence HR of 0.76 (1/1.31) for anastrozole vs tamoxifen29 and the Kaplan-Meier estimate for time to recurrence of the entire cohort from the present study. This hypothetical AI curve was plotted onto the survival curves of the CYP2D6 phenotypes (Figure 3). The hypothetical anastrozole survival function is located within the 95% CI of the Kaplan-Meier estimate for CYP2D6 EM patients, suggesting similar outcomes for patients treated with an AI vs patients with 2 functional CYP2D6 alleles receiving tamoxifen.
Table 4.
Optimization of Endocrine Therapy
| Current Study | ATAC Trial | |||
|---|---|---|---|---|
| CYP2D6 Stratificationa |
Hazard Ratio (95% CI)b |
Treatment Mode |
Hazard Ratio (95% CI)c |
P Valued |
| Time to Recurrence | ||||
| EM | 1 [Reference] | Aromatase inhibitors | 1 [Reference] | .19 |
| hetEM/IM | 1.40 (1.04–1.90) | Tamoxifen | 1.31 (1.15–1.49) | .01 |
| PM | 2.12 (1.10–3.28) | .11 | ||
| Decreased | 1.57 (1.18–2.08) | |||
| Event-Free Survivale | ||||
| EM | 1 [Reference] | Aromatase inhibitors | 1 [Reference] | |
| Decreased | 1.35 (1.08–1.68) | Tamoxifen | 1.18 (1.06–1.31) | .10 |
Abbreviations: CI, confidence interval; CYP, cytochrome P450; EM, extensive metabolism; IM, intermediate metabolism; PM, poor metabolism; UM, ultrarapid metabolism.
CYP2D6 phenotypes are defined as EM with 2 functional alleles (EM/EM, including UM), hetEM/IM with intermediate or 1 poor metabolism alleles (EM/IM, EM/PM, IM/IM, or IM/PM), and PM homozygous for poor metabolism alleles (PM/PM). Decreased refers to combined hetEM/IM and PM patients.
Hazard ratios are unadjusted and population heterogeneity was accounted for.
Hazard ratios from the Arimidex, Tamoxifen, Alone or in Combination (ATAC) 100-month analysis29 shown as reciprocals: 1/0.76 for time to recurrence and 1/0.85 for event-/disease-free survival.
P Values of 1-sided, 1-sample Mantel-Haenszel log-rank tests, testing the alternative hypotheses that the hazard ratio of hetEM/IM, PM, or decreased vs EM group exceeds the hazard ratio of tamoxifen vs aromatase inhibitor group in the ATAC 100-month analysis.29
Event-free survival in this study corresponds to disease-free survival in the ATAC definition.
Figure 3. Kaplan-Meier Estimates of Recurrence Probabilities Comparing Tamoxifen With a Hypothetical AI Curve.
EM indicates extensive metabolism (ie, patients with 2 functional cytochrome P450 (CYP) 2D6 alleles, including patients with ultrarapid metabolism); decreased, patients with any intermediate or poor metabolism alleles. Nonadjusted, heterogeneity-corrected Kaplan-Meier estimates for the CYP2D6 decreased and EM phenotypes as well as the entire tamoxifen cohort unselected by genotype; 95% confidence interval (CI) is shown for EM patients. Assuming the Cox proportional hazards assumption, a hypothetical aromatase inhibitor (AI) survival curve (blue) was estimated based on a hazard ratio of 0.76 for anastrozole relative to tamoxifen29 and the Kaplan-Meier estimate of the entire tamoxifen cohort40 (eAppendix).
COMMENT
This study, for the first time to our knowledge, provides sufficiently powered evidence for an association between CYP2D6 genetics and clinical outcome of tamoxifen. Since tamoxifen is standard of care for premenopausal women with estrogen receptor–positive breast cancer, and tamoxifen as well as AIs are valid treatment options for postmenopausal patients, our findings provide a powerful argument for refined endocrine treatment. Unequivocal conclusions have not been possible to date because there have been contradictory reports with respect to this association25–27 and data from prospective clinical trials are lacking. To address this shortcoming, we took advantage of accessible data and biological materials from patients with long-term follow-up. We defined strict inclusion criteria to exclude confounding effects from unclear hormone receptor status and combinatorial chemotherapy. We recruited a large number of patients to reach sufficient statistical power and compensated for sampling heterogeneity known to cause bias in retrospective studies.
Patients lacking CYP2D6 enzyme function (PM) had an almost 2-fold increased risk of developing breast cancer recurrence compared with patients with 2 functional CYP2D6 alleles (EM). The lower effect in patients with intermediate impairment of enzyme function (hetEM/IM) is indicative of a variant allele dose-dependent associated risk and, thus, underscores the primary role of CYP2D6-mediated tamoxifen activation to its active metabolite; ie, endoxifen.6,12,18 A relationship between the number of functional alleles and clinical outcome is further evident fromCYP2D6 UM phenotypes, accounting for 2.3% of all patients, who may have the best tamoxifen outcome.
CYP2D6 is not known to be associated with breast cancer biology but is thought to be a predictive factor specific to tamoxifen treatment outcomes. While a direct comparison between tamoxifen-treated and non- or AI-treated patients was not possible within the context of this study, several lines of evidence suggest that the observed association between the constitutional CYP2D6 genotype and tamoxifen outcome is a valid hypothesis. In our recent study, a control group of 280 patients not treated with tamoxifen showed that CYP2D6 genetic variants had no effect on the outcome. 21 Together with the strong pharmacological evidence of CYP2D6 being a key enzyme in tamoxifen bioactivation,6,12 the observed association in the present study is best explained in relation to tamoxifen rather than treatment outcome per se. This is supported by subgroup analyses and analysis of deviance in Cox models indicating a role of CYP2D6 to predict tamoxifen outcome independent of prognostic factors.
Although the clinical end points of event-free and disease-free survival showed lower effect sizes than in previous smaller studies,20,21,23 the gain of statistical power stresses the significance of defective CYP2D6 activity for nonfavorable tamoxifen outcome. Overall survival was not significantly associated, but a median follow-up of 6.3 years prevents this study from being conclusive because of the limited number of deaths or more common non–breast cancer deaths.38
Our findings were not shown in 3 other studies.25–27 This discrepancy can be explained by a number of reasons. As demonstrated by our sample size calculation, the number of patients/events required for CYP2D6 analysis is much higher than in any of the previously published studies. The reason why Goetz et al19,20 and Schroth et al21 observed significant associations between PM status and poor outcome may be explained by the prospective patient recruitment19,20 and the comprehensive coverage of PM and IM predisposing alleles.21 In contrast, the patient cohort characteristics of other studies25–27 appear to be different with respect to treatment modes, estrogen receptor status, and CYP2D6 genotyping. Of note, an incomplete coverage of PM genotypes likely results in missing up to 30% of PMs in a given patient cohort.39
Recently, a modeling analysis questioned the superiority of AI treatment to tamoxifen31 based on assumptions made from theCYP2D6*4 variant and assumed effect sizes from a small data set.19 Based on a more comprehensive CYP2D6 phenotype definition, our data fit well with the model predictions by Punglia et al.31 Accordingly, EM patients have the best benefit from tamoxifen compared with the unstratified tamoxifen cohort, with an absolute difference in recurrence risk of 3.4% at 9 years. While Punglia et al used 5-year follow-up comparisons from the Breast International Group Trial 1–98 data,31 we performed a 9-year follow-up comparison between our data and results from the ATAC study.29 We superimposed a hypothetical survival curve for AIs based on the reported HR of 0.76 for time to recurrence between anastrozole and tamoxifen29 onto the time-to-recurrence curves for tamoxifen of the present study. Of note, within this comparison, the outcome of the hypothetical AI curve is within the 95% CI of the CYP2D6 EM survival curve. For CYP2D6 EM, which accounts for 46% of patients, this suggests that CYP2D6 stratification attains the therapeutic benefit of AIs. Notably, we regard this approach as hypothesis-generating, yet this has been the best option, considering that a direct comparison of recurrence rates between both studies has not been possible. In support of the strong signal inherent to CYP2D6 stratification and by use of a statistical test independent of modeling, we demonstrated that the effect size of tamoxifen outcome between PM and EM patients in the present study was significantly larger than the HR assigned to tamoxifen relative to anastrozole in the ATAC study (Table 4). This amounts to an absolute risk increase of greater than 10% for PM patients compared with unselected tamoxifen patients, a finding that has implications for many thousands of women each year. Final conclusions must await confirmation from pharmacogenetic analysis in the studies comparing tamoxifen with AIs.
There are potential limitations to our study. The retrospective patient recruitment may have been prone to suboptimal documentation of events, insufficient control of adherence, lack of data regarding the coprescription of CYP2D6 inhibitors with tamoxifen, and differences in the length of follow-up. Also, because of difficulties inherent in the analysis of paraffin-derived DNA, we may have missed 15% to 20% of expected PM phenotypes, as it was apparent from comparison with PM frequency in blood-derived DNA. Although these limitations must be appreciated, our final conclusions are likely to have underestimated the effect size rather than overestimated the CYP2D6-related outcome effect. Importantly, we demonstrated by subgroup cross-comparison between prospectively and retrospectively collected patients that a CYP2D6 PM associated risk of breast cancer recurrence was detectable irrespective of patient sampling schemes. However, different degrees of effect size as well as large CIs indicate that both follow-up time and sample size are critical factors in the assessment of robust CYP2D6-associated risk calculations, once more underscoring the need for high powered data sets and possibly explaining spurious results in underpowered previous studies.
Our findings indicate that CYP2D6 PM patients have a substantially higher risk of tamoxifen treatment failure. Our analysis indicating that EM patients treated with tamoxifen had an outcome similar to patients treated with AIs in the ATAC trial should provide new impetus to the medical and scientific community to revisit the issue of relative efficacy of these 2 approaches in women with early breast cancer. Genotyping has the potential for identification of women who have the CYP2D6 PM phenotype and for whom the use of tamoxifen is associated with poor outcomes, thus indicating consideration of alternative forms of adjuvant endocrine therapy.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by the Robert Bosch Foundation, Stuttgart, Germany (W.S., M.S., and H.B.) and by National Institutes of Health grants CA 90628 (M.P.G.), Mayo Clinic Breast Cancer Specialized Program of Research Excellence CA116201 (J.N.I., M.P.G., V.J.S., and R.M.V.), CA 15083: U-01 GM61388 (R.M.W., M.P.G., and J.N.I.), and CA-25224 (North Central Cancer Treatment Group).
Role of the Sponsor: None of the funders had any input in the design and conduct of the study, collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs Schroth, Winter, and Brauch had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Schroth and Goetz contributed equally to this article.
Study concept and design: Schroth, Goetz, Winter, Suman, Weinshilboum, Ingle, Eichelbaum, Schwab, Brauch.
Acquisition of data: Schroth, Goetz, Hamann, Fasching, Schmidt, Fritz, Simon, Suman, Ames, Safgren, Kuffel, Ulmer, Boländer, Strick, Beckmann, Koelbl, Ingle, Brauch.
Analysis and interpretation of data: Schroth, Goetz, Hamann, Fasching, Winter, Suman, Ames, Safgren, Kuffel, Ingle, Schwab, Brauch.
Drafting of the manuscript: Schroth, Goetz, Winter, Ingle, Eichelbaum, Schwab, Brauch.
Critical revision of the manuscript for important intellectual content: Schroth, Goetz, Hamann, Fasching, Schmidt, Winter, Fritz, Simon, Suman, Ames, Safgren, Kuffel, Ulmer, Boländer, Strick, Beckmann, Koelbl, Weinshilboum, Ingle, Eichelbaum, Schwab, Brauch.
Statistical analysis: Schroth, Goetz, Fasching, Winter, Fritz, Suman.
Obtained funding: Goetz, Ames, Beckmann, Ingle, Schwab, Brauch.
Administrative, technical, or material support: Goetz, Hamann, Fasching, Schmidt, Simon, Ames, Safgren, Kuffel, Ulmer, Boländer, Strick, Beckmann, Koelbl, Eichelbaum, Schwab, Brauch.
Study supervision: Schroth, Goetz, Ames, Schwab, Brauch.
Financial Disclosures: Drs Goetz, Suman, Ames, Weinshilboum, and Ingle report that they are named inventors (along with Mayo Clinic) in regard to nonprovisional patent applications regarding tamoxifen and CYP2D6. The technology is not licensed, and no royalties have accrued. Dr Schmidt reports that he is named inventor on patent applications regarding prediction of chemotherapeutic response in breast cancer and molecular markers for breast cancer prognosis. Drs Goetz and Ingle report that they have been consultants for Roche; Dr Schmidt reports that he has been a consultant for Pfizer and Sanofi Aventis; and Dr Simon reports that he has been a consultant for AstraZeneca. Dr Goetz reports that he has received honoraria from Pfizer, Roche, and DNA Direct; Dr Fasching reports that he has received honoraria from AstraZeneca and Novartis; Dr Schmidt reports that he has received honoraria from AstraZeneca, Novartis, Pfizer, Essex, GlaxoSmithKline, Lilly, SanofiAventis, Roche, and Amgen; Drs Ulmer and Ingle report that they have received honoraria from AstraZeneca, Pfizer, and Novartis; Dr Beckmann reports that he has received honoraria from AstraZeneca, Novartis, Sanofi Aventis, Glaxo Smith Kline, and Roche; Drs Eichelbaum and Schwab report that they have received honoraria from Roche Molecular Diagnostics; and Dr Brauch reports that she has received honoraria from AstraZeneca. Drs Schwab and Brauch report that they have initiated scientific collaborations in 2009 with Roche Molecular Diagnostics and Siemens Healthcare Diagnostics Products GmbH. Dr Schmidt reports that he received previous funding from Siemens Healthcare Diagnostics Products GmbH.
Additional Contributions: We thank V. Craig Jordan, DPhil, DSc, MD, Lombardi Comprehensive Cancer Center at Georgetown University Medical School, Washington, DC, for helpful critical comments and discussion on the manuscript. Dr Jordan did not receive any compensation.
Additional Information: The eAppendix, eTable, and eFigure are available at http://www.jama.com.
REFERENCES
- 1.Jordan VC. Tamoxifen: catalyst for the change to targeted therapy. Eur J Cancer. 2008;44(1):30–38. doi: 10.1016/j.ejca.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75(2):305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 4.Jordan VC. Metabolites of tamoxifen in animals and man: Identification, pharmacology, and significance. Breast Cancer Res Treat. 1982;2(2):123–138. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 5.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49(8):2175–2183. [PubMed] [Google Scholar]
- 6.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coad-ministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95(23):1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 7.Allen KE, Clark ER, Jordan VC. Evidence for the metabolic activation of non-steroidal antioestrogens: a study of structure-activity relationships. Br J Pharmacol. 1980;71(1):83–91. doi: 10.1111/j.1476-5381.1980.tb10912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55(5):471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 9.Buck MB, Coller JK, Murdter TE, Eichelbaum M, Knabbe C. TGFβ2 and TβRII are valid molecular biomarkers for the antiproliferative effects of tamoxifen and tamoxifen metabolites in breast cancer cells. Breast Cancer Res Treat. 2008;107(1):15–24. doi: 10.1007/s10549-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 10.Robertson DW, Katzenellenbogen JA, Long DJ, Rorke EA, Katzenellenbogen BS. Tamoxifen antiestrogens: a comparison of the activity, pharmacokinetics, and metabolic activation of the cis and trans isomers of tamoxifen. J Steroid Biochem. 1982;16(1):1–13. doi: 10.1016/0022-4731(82)90137-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC. The tamoxifen metabolite, endoxifen, is a potent anti-estrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res. 2009;69(5):1722–1727. doi: 10.1158/0008-5472.CAN-08-3933. [DOI] [PubMed] [Google Scholar]
- 12.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro. J Pharmacol Exp Ther. 2004;310(3):1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 13.Griese EU, Zanger UM, Brudermanns U, et al. Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998;8(1):15–26. doi: 10.1097/00008571-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348(6):529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 15.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 16.Raimundo S, Toscano C, Klein K, et al. A novel intronic mutation, 2988G>A, with high predictivity for impaired function of cytochrome P450 2D6 in white subjects. Clin Pharmacol Ther. 2004;76(2):128–138. doi: 10.1016/j.clpt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies. Pharmacol Ther. 2007;116(3):496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism. Clin Pharmacol Ther. 2006;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23(36):9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 20.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101(1):113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 21.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25(33):5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 22.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25(25):3837–3845. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19(8):1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 24.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99(5):995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 26.Wegman P, Elingarami S, Carstensen J, et al. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9(1):R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7(3):R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell A, Cuzick J, Baum M, et al. ATAC Trialists’ Group. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 29.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists’ Group. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer. Lancet Oncol. 2008;9(1):45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 30.Thürlimann B, Keshaviah A, Coates AS, et al. Breast International Group (BIG) 1–98 Collaborative Group. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353(26):2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 31.Punglia RS, Burstein HJ, Winer EP, Weeks JC. Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst. 2008;100(9):642–648. doi: 10.1093/jnci/djn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flockhart D. CYP2D6 genotyping and the pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol. 2008;6(7):493–494. [PubMed] [Google Scholar]
- 33.Ingle JN. Pharmacogenomics of tamoxifen and aromatase inhibitors. Cancer. 2008;112(3) suppl:695–699. doi: 10.1002/cncr.23192. [DOI] [PubMed] [Google Scholar]
- 34.Weinshilboum R. Pharmacogenomics of endocrine therapy in breast cancer. Adv Exp Med Biol. 2008;630:220–231. doi: 10.1007/978-0-387-78818-0_14. [DOI] [PubMed] [Google Scholar]
- 35.Brauch H, Schroth W, Eichelbaum M, Schwab M, Harbeck N. Clinical relevance of CYP2D6 genetics for tamoxifen response in breast cancer. Breast Care. 2008;3(1):43–50. doi: 10.1159/000114642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingle JN, Suman VJ, Mailliard JA, et al. Randomized trial of tamoxifen alone or combined with fluoxymesterone as adjuvant therapy in postmenopausal women with resected estrogen receptor positive breast cancer: North Central Cancer Treatment Group Trial 89-30-52. Breast Cancer Res Treat. 2006;98(2):217–222. doi: 10.1007/s10549-005-9152-1. [DOI] [PubMed] [Google Scholar]
- 37.McCullagh P, Nelder J. Generalized Linear Models. 2nd ed. Chapman & Hall: CRC; 1989. [Google Scholar]
- 38.Chapman JA, Meng D, Shepherd L, et al. Competing causes of death from a randomized trial of extended adjuvant endocrine therapy for breast cancer. J Natl Cancer Inst. 2008;100(4):252–260. doi: 10.1093/jnci/djn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroth W, Haman U, Fasching PA. Comparison of CYP2D6 genotyping methods in breast cancer patients treated with tamoxifen–MALDI TOF-MS and microarray-based by Amplichip CYP450 test. Presented at: Joint Cold Spring Harbor Laboratory Wellcome Trust Conference; September 12–16, 2009; Hinxton, England. [Google Scholar]
- 40.Andersen PK, Borgan Ø, Gill RD, Keiding N. Statistical Models Based on Counting Processes. New York, NY: Springer; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.