Abstract
Genetic mutations causing human disease are conventionally thought to be inherited through the germ line from one’s parents and present in all somatic (body) cells, except for most cancer mutations, which arise somatically. Increasingly, somatic mutations are being identified in diseases other than cancer, including neurodevelopmental diseases. Somatic mutations can arise during the course of prenatal brain development and cause neurological disease—even when present at low levels of mosaicism, for example—resulting in brain malformations associated with epilepsy and intellectual disability. Novel, highly sensitive technologies will allow more accurate evaluation of somatic mutations in neurodevelopmental disorders and during normal brain development.
With the exception of cancer, human genetic diseases have been until relatively recently thought to reflect inherited DNA variation. Inherited mutations are present in the parent (or parents) and in all tissues of the affected individual (Fig. 1, A and B). Hence, they can be conveniently assayed in any tissue of the body, including readily available peripheral blood. Even though inherited mutations are present in essentially all cells, they may affect some tissues more than others, depending upon where and when the gene involved has its essential roles.
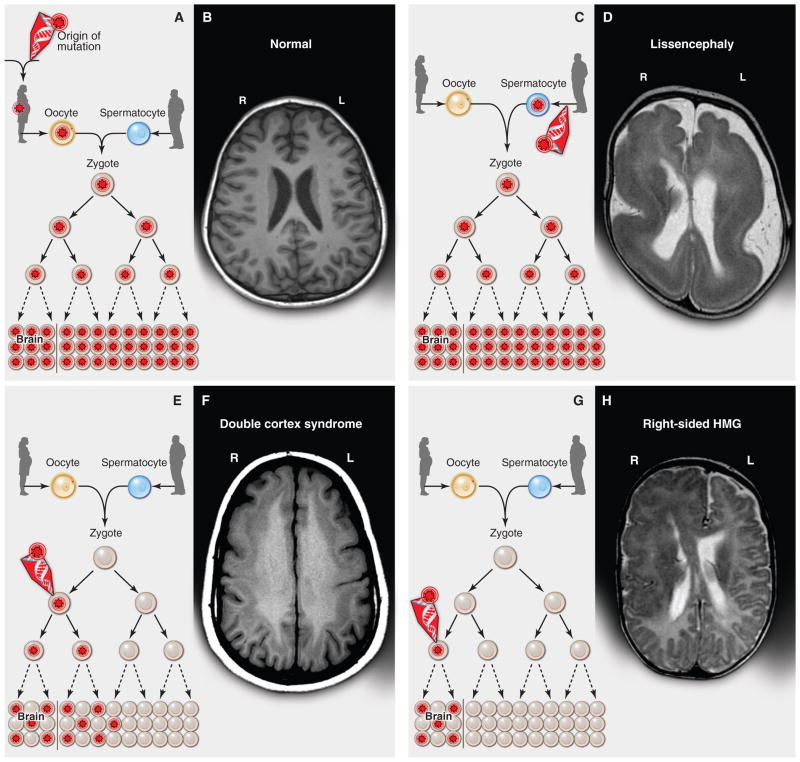
Fig. 1. Inherited, de novo, and somatic mutations causing neurological disease.
(A) A heterozygous mutation is inherited from one parent. This mechanism is typical of autosomal dominant epilepsy. In this example, the mutation originally presented in the mother, whose oocytes in turn carry the mutation. (The mutation arose during gametogenesis in one of the parents of the mother, top left.) It is present in the zygote and thus all cells of the affected child. (B) This axial T1-weighted image from a MRI study of a patient with inherited epilepsy appears normal. Individuals with dominantly inherited epilepsies caused by mutations in genes encoding ion channels, for example, have normal neuroimaging studies despite every cell carrying a mutation. (C) A de novo mutation may arise sporadically during gametogenesis, in this case spermatogenesis. This mechanism of mutation would be typical of a de novo mutation in the gene SCN1A associated with severe myoclonic epilepsy of infancy or LIS1 associated with lissencephaly. Even though every cell in the individual carries the mutation, the predominant effects of the mutation depend on the distribution of gene expression; in these examples, the brain is primarily affected. (D) An axial T2-weighted MRI image shows the severe gyral simplification—more pronounced posteriorly (the bottom of the figure)—that is associated with mutations in the gene LIS1. (E) An early post-zygotic mutation results in a mutation present in most or all tissues of the organism (including the leukocytes, which are generally assayed for clinical genetic testing) but in a mosaic fashion, with only a portion of all cells in each tissue harboring the mutation. This pattern, illustrated by the axial T1-weighted image in (F), has been observed in mosaic cases of double cortex syndrome involving the gene DCX. Visible is the extra band of gray matter underlying the normal-appearing outer aspect of the cerebral cortex. Because DCX is required for normal migration of neurons from the ventricular region deep in the brain to the superficial cortex, the cells carrying the DCX mutation only migrate about halfway to the cortex and then arrest their migration. (G) A late post-zygotic mutation will be present in only certain tissues in a mosaic fashion, in this case apparently in half of the brain. This is the pattern observed in some cases of HMG with somatic mosaic point mutations in AKT3 and other related genes and somatic mosaic increase of copy number of chromosome 1q. (H) This axial T2-weighted MRI image shows right-sided HMG, characterized here by enlargement of the right hemisphere, abnormally thick and dark-appearing gray matter anteriorly, heterotopic periventricular gray matter, and abnormal white matter signal in the right hemisphere. (R, right; L, left).
Increasing evidence shows the importance of “de novo” mutations—those present in affected offspring but not detected in the parents—in neuropsychiatric and pediatric disorders (1–8). These de novo mutations are typically present in the sperm or egg of one parent and yet are not detectable in blood taken from the parents; once transmitted to the embryo, they are present in all tissues of the offspring (Fig. 1, C and D). Whole-exome sequencing studies have shown that most individuals have one or two spontaneous mutations in the exome (the part of the genome encoding proteins) that are not present in their parents, but in individuals with neurodevelopmental and neuropsychiatric conditions [such as autism spectrum disorders (ASDs)] these de novo mutations are more likely to be damaging, suggesting that some of these de novo mutations cause disease (1, 4, 5, 8). In fact, mutations that greatly increase the risk of neurodevelopmental or neuropsychiatric disease—even when only one of the two alleles of a gene is affected (heterozygous mutations)—appear to arise de novo most of the time and are inherited relatively rarely. This is not unexpected because individuals with these disorders are less likely to bear offspring, placing the disease-causing mutations under strong negative selection. Because affected people thus rarely transmit the mutation to children, the presence of the disease reflects the ongoing appearance of new mutations.
Disease-causing mutations can also occur during the mitotic cell divisions that generate the embryo after fertilization and zygote formation. These mutations lead to individuals who are mosaic, with only a subset of their cells harboring the mutation (Fig. 1, E and F). These mutations are de novo in the sense that they are not detectable in the parents of the affected individuals but are more specifically termed somatic mutations. Somatic mutations can give rise to cancer (9), as well as noncancerous diseases. Noncancerous somatic mutations that occur during development may affect cell proliferation, as would be the case in cancer, or they may simply alter cellular function without causing a proliferative effect. There are estimates that the mutation burden in somatic cells is quite high, and estimates based on known mutation rates suggest that every cell division creates some form of genetic variation, which may or may not have an effect on cellular function (10, 11). Several recent studies have even suggested that the brain may harbor widespread somatic mutations, in the form of aneuploidy or retrotransposon insertions, perhaps as part of its “normal” development (12–14). If somatic mutations, especially in brain cells, do play a role in “complex” diseases (that is, diseases with genetic influences that are non-Mendelian), detecting them represents a substantial challenge with current sequencing strategies that mainly analyze blood DNA.
Here, we will review the present state of data about somatic mutations in human neurological disease. We highlight the recent identification of disease-associated somatic mutations present in the brain but undetectable in the blood of the same patient (Fig. 1, G and H) and discuss the challenges of identifying such rare mosaic mutations. We further discuss how emerging techniques will allow more refined study of the types and rates of somatic mutation and genomic variation in the brain.
“Obligatory” Somatic Mosaic Disorders?
Severe genetic diseases that are not compatible with survival or fertility would be expected to be preferentially or exclusively caused by either recessive mutations or dominant de novo (detectable in the affected child but not the parents) mutations. This is because recessive mutations that affect survival or fertility in the homozygous state can persist in the population in a heterozygous state, whereas severe dominant mutations cannot be passed to offspring when present in enough cells to cause severe disease in a parent. From this, we would predict that for any disease caused by a dominant mutation the ratio of sporadic cases caused by de novo mutations to cases caused by inherited mutations as seen in recurrent familial cases should correlate with the severity of the disease’s effect on survival and fertility. The most deleterious mutations that are not compatible with embryonic development might even be found only as somatic mosaic and not as inherited mutations.
Indeed, there are numerous examples of diseases spanning the spectrum of severity that follow these predictions. Proteus syndrome is a severe syndrome characterized by multiple overgrowths of the skin, bone, connective tissue, and other tissues caused by a dominant somatic mutation in the gene encoding serine-threonine protein kinase B α (PKBα), AKT1 (15). It has never been reported to be recurrent in a family or to be heritable across generations, which is consistent with its severity in the mosaic state and likely incompatibility with survival if it were inherited and present at the zygote stage. The multiple lesions of Proteus syndrome contain between 1 and 50% mutant cells, suggesting that a small fraction of abnormal cells can induce a lesion (15). McCune-Albright syndrome is often cited as an example of a severe disease caused by somatic mutation (in the gene encoding the guanine nucleotide binding protein, alpha stimulating, GNAS1) but not seen as familial inherited cases, which is likely due to the incompatibility of inherited mutations with embryonic development (16). Maffucci syndrome is another example of a severe overgrowth syndrome, characterized by multiple cartilaginous tumors, seen only as sporadic cases caused by somatic mutation in the gene encoding isocitrate dehydrogenase 1, IDH1 (17). Somatic activating mutations in the gene encoding phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha, PIK3CA, are the cause of congenital lipomatous overgrowth with vascular, epidermal, and skeletal anomalies (CLOVES) syndrome (18, 19). Similarly, somatic activating mutations in the gene encoding guanine nucleotide binding protein q polypeptide, GNAQ, are associated with the lesions seen in the neurocutaneous disease Sturge Weber syndrome (20). As with the previous examples, these latter examples do not recur in families and present as sporadic somatic disorders. Somatic mutations affecting the brain are discussed below.
The above examples resemble chromosomal aneuploidies in that many aneuploidies that cause severe disease are tolerated only as somatic mosaic mutations. Aneuploidy of chromosomes 13, 18, 21, and the sex chromosomes accounts for nearly all aneuploidy live births, in which all cells in the body carry the extra chromosome. On the other hand, aneuploidies of other chromosomes are only tolerated during development as somatic mosaics (21, 22). Down syndrome (trisomy of chromosome 21) and other chromosomal trisomies, such as trisomy 13 or 18, are associated with intellectual disability and other features of brain dysfunction caused by the abnormal chromosome number. Mosaic forms of trisomy 21 occur in a certain proportion of cases of Down syndrome in which the extra chromosome is present in some but not all cells of the body (23). Mosaic trisomies have also been observed for many other chromosomes (such as chromosomes 1, 8, 9, 16, 17, and 22) that are rarely observed constitutionally (present in all cells) because these trisomies, if present in all cells of the body, would be lethal before or soon after birth (24–30). In each of these conditions, the severity of the disorder is determined by the particular chromosome duplicated, as well as by the proportion of cells in the body carrying the abnormality.
How Do Somatic Mutations Manifest as Neurological Disease?
“Second Hit” Mutations Produce Mosaicism
The importance of somatic mutations has long been understood in the context of several dominant conditions in which patients inherit a heterozygous mutation, present in all cells, with somatic second mutations leading to overgrowth of specific tissues because of inactivation of a second allele, according to the “two-hit” model of Knudson (31). For example, neurofibromatosis type 1 (NF1)—which is associated with focal lesions of the skin, optic gliomas, and peripheral nervous system tumors called neurofibromas—is characterized by germline mutations in the gene NF1, with second mutations in the other NF1 allele causing the neurofibromas (32). A similar phenomenon occurs in the multisystem disorder tuberous sclerosis complex (TSC), a condition caused by mutations in the genes TSC1 and TSC2 (33, 34), whose gene products form a protein-protein complex together and regulate the mammalian target of rapamycin (mTOR) pathway; somatic second mutations have been shown in non-nervous system tumors of TSC (35), and “second hits” in the form of posttranslational inactivation of TSC2 have been shown in sub-ependymal giant cell astrocytomas, as well as in the noncancerous cortical tubers in patients with TSC (36). Mutation of the second TSC allele in cortical tubers has been hypothesized to occur as well but has so far been demonstrated for only a single TSC lesion (37). These neurocutaneous syndromes give us a sense of how common somatic mutations really are because the somatic mutations are revealed in the presence of hamartomatous lesions. Remarkably, typical patients with these conditions have dozens of lesions of various tissues, suggesting that deleterious somatic mutations at any single gene occur many times during normal development.
Neurodevelopmental Disorders Caused by Somatic Mutations
The role of somatic mutations has been known for many years in genetic conditions that can be diagnosed by means such as physical examination or magnetic resonance imaging (MRI). In two examples, genetic disorders of neuronal migration in the brain have been associated with somatic mutations in 5 to 10% of patients. Mutations in the gene lissencephaly-1 (LIS1) are typically associated with a “smooth brain” phenotype of lissencephaly (an example of which is shown in Fig. 1D) because of grossly abnormal neuronal migration, whereas mutations in doublecortin (DCX), located on the X chromosome, are associated with lissencephaly in males and a subcortical band heterotopia pattern (or “double cortex”) in females (an example of which is shown in Fig. 1F) (38, 39) because chromosome X-inactivation creates mosaic populations of cells in the female that have either normal or abnormal DCX function. Somatic mutations in LIS1 affecting only a portion of migrating neurons can result in the “double cortex” pattern (40), and similarly, males with somatic mutations in DCX can exhibit a “double cortex” because only some neurons carry the mutation (41). In both of these cases, mutations were detectable in mosaic form in leukocytes, suggesting that a relatively early postzygotic, somatic mutation occurred.
Somatic mutation has also been described in cases without visible focal lesions, including in epilepsy. For example, severe myoclonic epilepsy of infancy (Dravet syndrome), typically caused by de novo mutations in the gene encoding the sodium channel alpha-1 subunit, SCN1A, has recently been described as occurring not only at the gamete stage but also in the form of a somatic mutation at the postzygotic stage, including in monozygotic twins discordant for the mutation (42, 43).
“Brain-Only” Somatic Mutations
Many focal malformations of cerebral cortical development, such as focal cortical dysplasia, have been hypothesized to occur via somatic mutation in the developing brain (44), but testing this hypothesis required the availability of brain tissue that was resected from affected individuals for control of intractable epilepsy. We and others recently reported a somatic genetic explanation for a condition called hemimegalencephaly (HMG), which is characterized by enlargement and extensive malformation of an entire cerebral hemisphere (Fig. 1H) (45–47). Previously, no specific genes had been identified for isolated HMG, although there had been rare reports of HMG associated with the overgrowth syndromes TSC (48) and Proteus syndrome (49). Direct study of HMG brain tissue surgically resected in the course of treatment of the severe epilepsy led to the identification of somatic activating mutations in the gene AKT3, encoding PKBγ a serine-threonine kinase upstream from mTOR that is highly expressed in the developing brain during corticogenesis. In at least some cases, the mutation was shown to be absent from leukocytes of the affected child (46). HMG can also result from somatic mutations in other genes of the phosphoinositide 3-kinase (PI3K)–AKT3-mTOR pathway, including PIK3CA and mTOR itself (45, 47). Additional cases carry mosaic gain of copy number of chromosome 1q (the locus of AKT3) in HMG brain tissue, demonstrating a different mechanism of mutation that can occur somatically during brain development and one that would not be tolerated if it were present in the germline because of severe developmental defects in multiple organ systems (24, 46). The most common HMG-associated AKT3 mutation, Glu17Lys, is precisely paralogous to the AKT1 mutation that causes Proteus syndrome, whereas the paralogous mutation in AKT2 causes a predominantly non-neurological disorder, hypoglycemia-hemihypertrophy syndrome (15, 50). The different phenotypes presumably arise because of differences in the timing and location of expression of the three AKT genes.
In addition to some cases being caused by mutations apparently limited to brain, megalencephaly (large brain size, affecting one or both hemispheres) can also occur in the setting of somatic de novo mutations, which are detectable at low levels in leukocytes, and in AKT3 and PIK3CA as part of the megalencephaly-capillary malformation and megalencephaly-polydactyly-polymicrogyria-hydrocephalus syndromes (45). Taken together, these studies suggest that brain overgrowth syndromes can be caused by mutations in brain progenitor cells, but that some of these mutations occur early enough in development to be present in many tissues, affecting cells outside the brain as well. In contrast, other mutations might be limited to the brain because they occur after the embryonic separation of brain from nonbrain tissue.
Neurodegenerative Diseases Caused or Modulated by Somatic Mutation
Some cases of neurodegenerative diseases have been associated with somatic mutations or can be modified by somatic mutations. A fascinating case of sporadic, early-onset Alzheimer’s disease has been attributed to a somatic mosaic presenilin-1 mutation present in the brain (51). A case of sporadic Creutzfeld-Jakob disease was caused by an early embryonic somatic mutation identified by the presence of three alleles for the gene encoding the major prion protein PrP (PRNP) in the individual (52). Incontinentia pigmenti can lead to atrophy of the cortex and cerebellum, with some cases due to somatic mutation in the gene encoding the inhibitor of kappa light polypeptide gene enhancer in β cells, kinase gamma (IKBKG, also known as NEMO) (53). Some neurodegenerative diseases—such as Friedrich’s ataxia, dentatorubral-pallidoluysian atrophy, and Huntington’s disease—are caused by inheritance of expanded microsatellite repeats that are highly mutable; these loci can exhibit marked somatic heterogeneity in repeat lengths across brain regions and tissues of affected individuals (54–60). Age-related (postdevelopmental) somatic mutation, known to play a role in cancer, has also been widely postulated as playing a role in normal aging processes as well as in neurodegeneration (61, 62), but a full consideration of such age-related somatic mutation is beyond the scope of this Review.
How Does Cortical Clonal Architecture Influence Mosaic Mutations?
A specific progenitor cell in which a somatic mutation occurs transmits the mutation to its daughter cells, the extent and effects of which will depend on which cells of the developing cortex are involved. Understanding the clonal architecture of the developing human brain can therefore help us understand how somatic mutations cause neurogenetic disease. Cortical development, showing various neuronal and glial cells and their origins, is outlined in Fig. 2. Direct information about cell lineage patterns in the human brain is not available, but studies in animal models suggest substantial complexity in the brain’s clonal structure. The principal (excitatory) neurons of the cerebral cortex and hippocampus are derived from an embryonic neuroepithelium, with progenitor cells lining the ventricular surfaces, deep in the brain (63). However, unlike most embryonic epithelia, which typically produce patches of clonal cells that adjoin or remain near one another, cerebral cortical progenitors produce postmitotic neurons that migrate radially from the deep ventricle to the superficial, outer layers of the brain. This long-distance, radial migration has been shown in a number of animal models to be associated with substantial dispersion between neurons with common clonal origins (64).
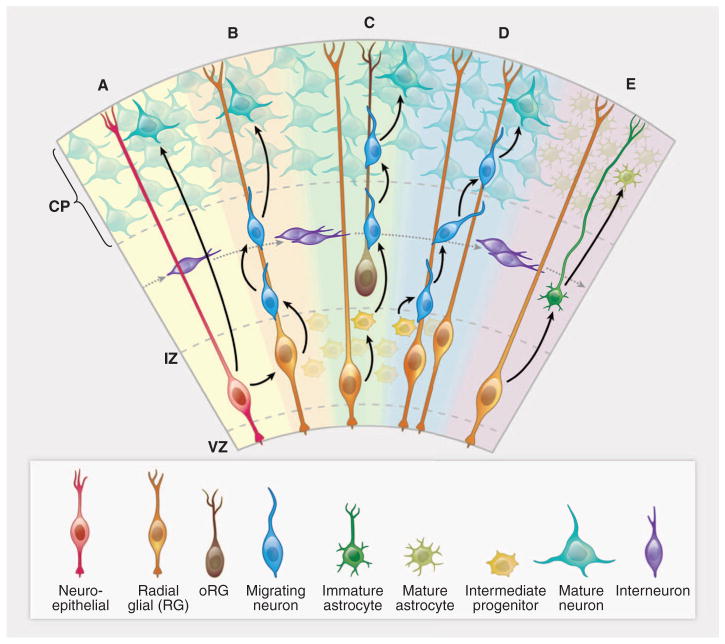
Fig. 2. Cortical development—origins of pyramidal neurons and astrocytes in the cerebral cortex.
(A) A neuroepithelial cell (red) at the ventricular zone serves as progenitor for both a pyramidal neuron (green-blue) as well as a radial glial cell (gold). (B) A newly differentiated neuron (blue) migrates along a radial glial process. (C) Neurons (blue) continue to migrate as intermediate progenitor cells (small yellow) form. (D) Intermediate progenitor cells begin to generate neurons (blue). (E) The progenitor cells in the ventricular zone begin to give rise to astrocytes (dark green). Interneurons (purple) generated elsewhere migrate tangentially. CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone. The VZ early in development has a thickness of ~10 cell bodies (50 to 100 μm). The CP ranges in thickness from two to three cell bodies at the earliest stages of development, eventually forming a mature cerebral cortex that is 2 to 4 mm thick.
Clones of related pyramidal neurons generally maintain a funnel shape but are typically highly interspersed with neurons of diverse clonal origins, so that neurons carrying a mosaic mutation would be expected to be somewhat clustered but intermingled with neurons from distinct origins (65–68). More limited studies in animals with large, gyrated brains (for example, ferret and macaque) suggest similar patterns of clustering of sibling cells, but with extensive intermingling of clones (illustrated in Fig. 3, A and B) (65, 66, 68, 69). Thus, somatic mutations could produce clones of mutant cells that might mainly concentrate in functional regions of cortex (70) but may also involve only a small proportion of the cells harboring the mutation, making such mutations difficult to detect by standard sequencing techniques. Human lesions with a funnel-shaped appearance on neuroimaging exist in the form of focal cortical dysplasias (Fig. 3, C and D). The appearance of these focal cortical dysplasias, their frequent continuity into the ventricular region of the brain even in adulthood, and their similarity to the focal cortical lesions (“tubers”) in TSC all have suggested that focal cortical dysplasias may represent mosaic mutations of these deep pyramidal-neuron progenitors, but this remains incompletely worked out. In addition to the determination of the extent of dispersion of neurons derived from progenitors at the ventricular zone, neurons undergo layer specification through a series of fate restriction and specification processes during or after mitosis (64). Any disruption in the normal sequence of layer determination and subsequent functional specification—for example, by mutations occurring during progenitor mitoses—might place an individual at risk for localized, cell type–specific or even layer-specific functional abnormalities, such as those that occur in conditions such as epilepsy.
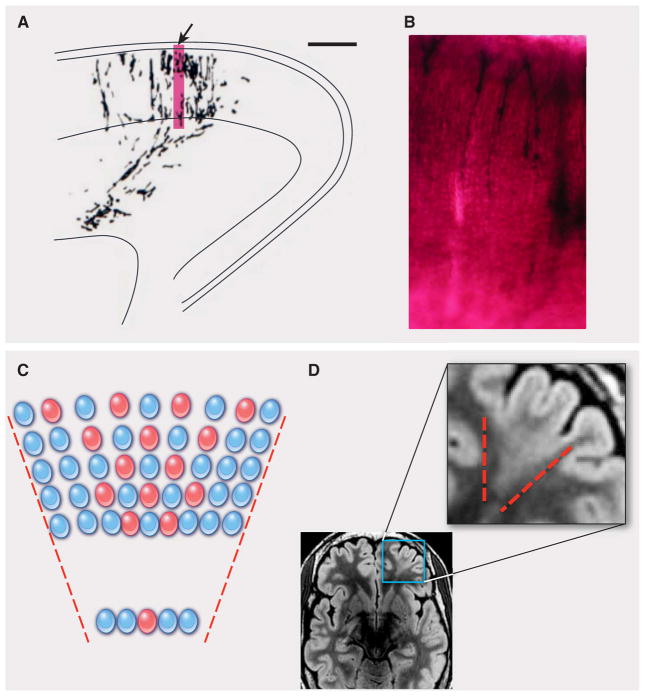
Fig. 3. Focal cortical dysplasia: a clonal-appearing brain lesion suspected to be caused by somatic mutation in a progenitor cell.
(A) A camera lucida drawing shows a radial cluster from the developing ferret brain, labeled at E29 and studied at P8 (scale bar, 500 μm) [from (68)]. The arrow indicates the region enlarged in (B). The accompanying photograph (B) shows an enlarged view of retro-virally labeled migrating pyramidal neurons at the same stage. (C) A schematic depicts the predicted effects of a somatic mutation affecting a progenitor cell in the VZ, producing a funnel-shaped lesion emanating from the ventricle. Shown are offspring (red) of the progenitor that experienced a mutation and are partially interspersed with normal cells (blue). (D) This axial T2-weighted MRI of the brain of a patient with focal epilepsy shows a focal cortical dysplasia in the left frontal region. In this image, normal gray matter appears gray, and normal white matter appears black. In the expanded view of the lesion, outlined in the smaller region by the blue box, the MRI shows a wedge-shaped abnormality roughly bounded by red dashed lines. The lesion consists of abnormal white matter signal (normally black, but in the lesion light gray in the region between the dashed lines) and thickened gray matter. The boundary between the gray matter and the white matter is blurred in this focal region as compared with the rest of the brain. In contrast, the normal regions surrounding the focal cortical dysplasia show gray matter of appropriate thickness, a sharp distinction between the gray matter and the white matter, and appropriate (black) signal in the white matter. [The MRI image is courtesy of Dr. A. James Barkovich, Department of Radiology, University of California, San Francisco]
In contrast to pyramidal neurons, other cell types of the brain show even less evidence of clonal clustering, so that sibling cells sharing a mosaic mutation would be expected to be quite widely scattered. For example, in animal models the inhibitory interneurons that populate the cerebral cortex are formed outside the cortex altogether in a second proliferative zone in the basal forebrain called the ganglionic eminence, which generates the basal ganglia. These inhibitory interneurons migrate large distances in a nonradial (tangential) direction before turning radially to enter the cortex (71). Direct lineage studies in mouse and in ferret suggest that deep progenitors of inhibitory interneurons generate clones of neurons covering broad regions of the cerebral cortex (66, 68) at very low levels of mosaicism. Astrocytic glial cells arise from several sources, including progenitors that also generate principal neurons (66, 72), whereas oligodendrocytes arise from yet a fourth source in the basal forebrain that generates cells for essentially the entire forebrain (73). Hence, cells carrying common somatic mutations would be expected to be quite dispersed, and neighboring cells in the cortex have diverse clonal origins. This complex architecture with dispersed and intermingled clones makes it difficult to detect somatic mutations through genotyping or sequencing of bulk brain tissue because of the inherent difficulty in detecting the signal of low-level mosaic mutations.
Studies in the human brain suggest the potential of somatic mutations in the cerebral cortex to create dysfunction surprisingly out of proportion to their level of mosaicism. For example, in HMG, which morphologically on MRI appears as a malformation affecting the entire affected hemisphere, as few as 8% and generally less than 35% of cells (neurons and glia) carry the disease-causing mutation yet are distributed over an entire hemisphere (46, 47, 74), which is consistent with the diverse clonal origins suggested by cell lineage studies in animal models. Even levels of mosaicism as low as 8% are sufficient to disrupt the normal architecture and function of essentially the entire hemisphere, causing motor weakness, intractable epilepsy, and cognitive dysfunction that often improves dramatically with removal or disconnection of the abnormal cerebral hemisphere. Somatic mosaic mutations in LIS1 or DCX can also cause marked cognitive dysfunction and epilepsy with levels of mosaicism detected in blood as low as 10 to 40% (40, 41); although we presume the brain is likewise mosaic, the precise level of mosaicism is such that cases can only be determined by studying cells from the brain directly, which has yet to be done. Given that complex neural circuits underlying human cognitive function are highly interconnected throughout the cortex, localized disruptions caused by somatic mutations may affect widespread networks, leading to substantial disease.
Other Neurological Diseases Attributable to Somatic Mutation?
De novo mutation has been implicated in almost all neurodevelopmental and neuropsychiatric disease, most notably intellectual disability and ASDs (2, 3, 5–8), suggesting that some disease-associated neuropsychiatric mutations may occasionally occur somatically as well—not only chromosome rearrangements, but also de novo copy number variants. Furthermore, de novo point mutations appear to be common collectively as a cause of ASD, although any given gene appears to be implicated infrequently. The example of SCN1A above demonstrates that de novo mutations may be present in some cells of a parent of an affected individual (yet not detected in parental blood) or may arise post-zygotically during development of the affected individual (somatic mutation). Similarly, de novo mutations in the X-linked methyl CpG binding protein 2 (MECP2) gene cause Rett syndrome, an ASD that is dominant in females and typically pre-natally lethal in males (75); mosaic mutations have been reported in males with both classic and atypical forms of Rett syndrome (76, 77).
Our ability to detect a pathogenic somatic mutation by using current clinical methods depends on how abundant it is in the leukocytes. The examples above suggest that at least some cases of autism, epilepsy, and perhaps other neuropsychiatric conditions such as schizophrenia may show roles for somatic mutations that have been overlooked by the usual paradigm of leukocyte DNA sequencing. Some cases of epilepsy, for example, may be due to somatic mutations affecting a specific cell lineage, such as γ-aminobutyric acid–secreting interneurons. Autism and other disorders that predominantly affect language function may be due to somatic mutations in populations of cells critical for language function in specific regions of the developing cortex. These may have been systematically missed by previous research designs that rarely sequence affected brain tissue and often do not account for the possibility of somatic mutations present at low levels in a mosaic pattern. The possibility of somatic mosaic mutations in some patients with high-functioning ASD is intriguing because it could provide a mechanism for the remarkable preservation of some abilities (“splinter or savant skills”) in some rare autistic patients: Mosaic mutations could result in a brain that is normal in some regions yet abnormal in other regions, which is analogous to HMG, in which gain-of-function mutations in the mTOR pathway result in one hemisphere that is impaired whereas the other hemisphere may functional normally. Nevertheless, studies relying on direct analysis of brain tissue are limited to autopsy studies for patients with autism or epilepsy and studies of human brain tissue removed in epilepsy surgery for patients with medically uncontrollable epilepsy. Such ongoing studies will continue to be informative, but they may or may not be generalizable to the broader group of patients with neuro-developmental disease. Thus, it is difficult to predict to what extent somatic mutation may account for conditions such as autism and epilepsy.
Contribution to Functional Cellular Diversity in the Brain?
Against the backdrop of an increasing recognition of the pathological role of somatic mutations in brain disease is an open question of whether genetic variation may generate functional diversity among cells in the brain, and if so, how this may affect brain function. In addition to single-nucleotide variation, somatic deletions and duplications have been reported in the brains of individuals without disease (12, 78). Retrotransposition of long interspersed nuclear elements (LINE-1, or L1) is a special subset of somatic mutation that has been of particular interest recently in terms of nervous system development. Retrotransposon insertion can cause gene mutation by inactivating genes by inserting into them or by changing patterns of gene expression. A number of studies made the observation that somatic L1 retrotransposition can occur during normal brain development (79, 80), and initial estimates of these events suggested that dozens of somatic L1 insertions may be present in each neuronal genome (80). Recent quantitative analysis of the rate of somatic L1 insertions in human neurons by a single-cell sequencing approach confirmed that L1 retrotransposition occurs during neurogenesis in the human brain but suggested a rate of less than one unique insertion per cell (74), which is much lower than previous estimates. This work highlights the potential of single-cell sequencing to quantify somatic mutation rates in humans. Further work remains to determine L1 retrotransposition rates across different regions of the human brain and to assess the potential role of these and other types of somatic mutations in neurological disease.
Techniques for Further Study of Somatic Variation in the Brain
As next-generation sequencing techniques become more efficient and affordable, we will be able to push the limits of detection of all types of somatic mosaic mutations and quantify somatic mutation rates across all regions, cell types, and time points of brain development. Studies of brain-only somatic mutations will still be limited by access to brain tissue. Nonetheless, the optimization of new techniques, such as single-cell sequencing and high depth sequencing, along with improved bioinformatic analyses, will allow us to address the role of somatic mosaicism in conditions such as focal epilepsy without malformations, autism, and intellectual disability in patients with mosaic mutations detectable in a small proportion of accessible cells such as leukocytes, or perhaps skin fibroblasts.
An exciting extension of the discovery of brain-only somatic mutations is the recent ability not only to characterize somatic mutations in a more refined manner but also to study the mutations as a means of understanding the lineage in which the mutation occurred. In the case of the somatic mutation of AKT3 in HMG, single-cell sorting and sequencing of neurons and glia from the surgically resected brain tissue specimen provided corroboration that the mutation was present in both neurons and glia in a mosaic fashion, in approximately one third of each cell population, indicating that the mutation occurred in a neuronal-glial progenitor (74). The detailed techniques to accurately study somatic mutation at the single-cell level are beyond the scope of this Review; we highlight some recent examples of single-cell approaches applied to tumors to evaluate their clonal evolution (81–83) and applied to gametes to evaluate spontaneous mutation rates (84, 85).
Conclusions
Ultimately, single-cell and ultra-deep genome sequencing will allow systematic measurement of somatic mutation rates in different cell types and lineages during development of the normal human brain. Deeper sequencing will enable a better understanding of (i) the prevalence of somatic mutations as an occasional cause of otherwise unexplained “complex” neuropsychiatric diseases, (ii) the extent to which somatic mutation in the brain may modify the pathogenesis of neurological diseases more broadly, and (iii) whether there are possible roles for genomic variability in normal developmental processes. Because the key to treating disease lies in understanding its underlying molecular causes, determining whether somatic mutation in the brain is responsible for relatively common conditions such as epilepsy, autism, and schizophrenia is one of the next major challenges in the field of somatic mutation. Somatic mutation sequencing also affords the exciting opportunity to perform lineage tracing that will add to our understanding of the diverse cell types and developmental processes that build the human brain.
Acknowledgments
A.P. is supported by the National Institute of Neurological Disorders and Stroke (NINDS) (K23NS069784). G.D.E. is supported in part by NIH Medical Science Training Program grant T32GM007753 and by the Louis Lange III Scholarship in Translational Research. X.C. was supported in part by NIH National Institute of General Medical Sciences grant T32GM007726-35. C.A.W. is supported by the Simons Foundation, the Manton Center for Orphan Disease Research, and grants from NINDS (R01 NS079277, RO1 NS032457, and R01 NS035129) and the National Institute of Mental Health (RO1 MH083565 and 1RC2MH089952). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
References and Notes
- 1.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 4.Awadalla P, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. Genomic variability within an organism exposes its cell lineage tree. PLOS Comput Biol. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 14.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindhurst MJ, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldred MA, Trembath RC. Activating and inactivating mutations in the human GNAS1 gene. Hum Mutat. 2000;16:183. doi: 10.1002/1098-1004(200009)16:3<183::AID-HUMU1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Pansuriya TC, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43:1256. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurek KC, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindhurst MJ, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet. 2012;44:928. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirley MD, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705:96. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Erickson RP. Somatic gene mutation and human disease other than cancer. Mutat Res. 2003;543:125. doi: 10.1016/S1383-5742(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 23.Pangalos C, et al. Understanding the mechanism(s) of mosaic trisomy 21 by using DNA polymorphism analysis. Am J Hum Genet. 1994;54:473. [PMC free article] [PubMed] [Google Scholar]
- 24.Patel C, et al. Mosaic trisomy 1q: The longest surviving case. Am J Med Genet A. 2009;149A:1795. doi: 10.1002/ajmg.a.32959. [DOI] [PubMed] [Google Scholar]
- 25.Kleczkowska A, Fryns JP, Van den Berghe H. On the variable effect of mosaic normal/balanced chromosomal rearrangements in man. J Med Genet. 1990;27:505. doi: 10.1136/jmg.27.8.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrander-Stumpel CT, et al. Mosaic tetrasomy 8p in two patients: Clinical data and review of the literature. Am J Med Genet. 1994;50:377. doi: 10.1002/ajmg.1320500416. [DOI] [PubMed] [Google Scholar]
- 27.Gérard-Blanluet M, et al. Mosaic trisomy 9 and lobar holoprosencephaly. Am J Med Genet. 2002;111:295. doi: 10.1002/ajmg.10481. [DOI] [PubMed] [Google Scholar]
- 28.Laus AC, et al. Karyotype/phenotype correlation in partial trisomies of the long arm of chromosome 16: case report and review of literature. Am J Med Genet A. 2012;158A:821. doi: 10.1002/ajmg.a.32988. [DOI] [PubMed] [Google Scholar]
- 29.Daber R, et al. Mosaic trisomy 17: Variable clinical and cytogenetic presentation. Am J Med Genet A. 2011;155:2489. doi: 10.1002/ajmg.a.34172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowe CA, Schwartz S, Black CJ, Jaswaney V. Mosaic trisomy 22: A case presentation and literature review of trisomy 22 phenotypes. Am J Med Genet. 1997;71:406. doi: 10.1002/(SICI)1096-8628(19970905)71:4<406::AID-AJMG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Linares C, et al. Dissecting loss of heterozygosity (LOH) in neurofibromatosis type 1-associated neurofibromas: Importance of copy neutral LOH. Hum Mutat. 2011;32:78. doi: 10.1002/humu.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305. doi: 10.1016/0092-8674(93)90618-Z. [DOI] [PubMed] [Google Scholar]
- 34.van Slegtenhorst M, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 35.Niida Y, et al. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am J Hum Genet. 2001;69:493. doi: 10.1086/321972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, et al. Phosphorylation of tuberin as a novel mechanism for somatic inactivation of the tuberous sclerosis complex proteins in brain lesions. Cancer Res. 2004;64:812. doi: 10.1158/0008-5472.CAN-03-3277. [DOI] [PubMed] [Google Scholar]
- 37.Qin W, et al. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol. 2010;20:1096. doi: 10.1111/j.1750-3639.2010.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleeson JG, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63. doi: 10.1016/S0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 39.Bahi-Buisson N, et al. SBH-LIS European Consortium, New insights into genotype-phenotype correlations for the doublecortin-related lissencephaly spectrum. Brain. 2013;136:223. doi: 10.1093/brain/aws323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sicca F, et al. Mosaic mutations of the LIS1 gene cause subcortical band heterotopia. Neurology. 2003;61:1042. doi: 10.1212/WNL.61.8.1042. [DOI] [PubMed] [Google Scholar]
- 41.Gleeson JG, et al. Somatic and germline mosaic mutations in the doublecortin gene are associated with variable phenotypes. Am J Hum Genet. 2000;67:574. doi: 10.1086/303043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gennaro E, et al. Somatic and germline mosaicisms in severe myoclonic epilepsy of infancy. Biochem Biophys Res Commun. 2006;341:489. doi: 10.1016/j.bbrc.2005.12.209. [DOI] [PubMed] [Google Scholar]
- 43.Vadlamudi L, et al. Timing of de novo mutagenesis—A twin study of sodium-channel mutations. N Engl J Med. 2010;363:1335. doi: 10.1056/NEJMoa0910752. [DOI] [PubMed] [Google Scholar]
- 44.Hua Y, Crino PB. Single cell lineage analysis in human focal cortical dysplasia. Cereb Cortex. 2003;13:693. doi: 10.1093/cercor/13.6.693. [DOI] [PubMed] [Google Scholar]
- 45.Rivière JB, et al. Finding of Rare Disease Genes (FORGE) Canada Consortium, De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poduri A, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cartwright MS, McCarthy SC, Roach ES. Hemimegalencephaly and tuberous sclerosis complex. Neurology. 2005;64:1634. doi: 10.1212/01.WNL.0000154465.87255.78. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths PD, Welch RJ, Gardner-Medwin D, Gholkar A, McAllister V. The radiological features of hemimegalencephaly including three cases associated with proteus syndrome. Neuropediatrics. 1994;25:140. doi: 10.1055/s-2008-1073012. [DOI] [PubMed] [Google Scholar]
- 50.Hussain K, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck JA, et al. Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum Mol Genet. 2004;13:1219. doi: 10.1093/hmg/ddh134. [DOI] [PubMed] [Google Scholar]
- 52.Alzualde A, et al. Somatic mosaicism in a case of apparently sporadic Creutzfeldt-Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1283. doi: 10.1002/ajmg.b.31099. [DOI] [PubMed] [Google Scholar]
- 53.Smahi A, et al. The International Incontinentia Pigmenti (IP) Consortium, Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. Nature. 2000;405:466. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 54.Hellenbroich Y, Schwinger E, Zühlke C. Limited somatic mosaicism for Friedreich’s ataxia GAA triplet repeat expansions identified by small pool PCR in blood leukocytes. Acta Neurol Scand. 2001;103:188. doi: 10.1034/j.1600-0404.2001.103003188.x. [DOI] [PubMed] [Google Scholar]
- 55.Hashida H, et al. Single cell analysis of CAG repeat in brains of dentatorubral-pallidoluysian atrophy (DRPLA) J Neurol Sci. 2001;190:87. doi: 10.1016/S0022-510X(01)00596-2. [DOI] [PubMed] [Google Scholar]
- 56.Kahlem P, Djian P. The expanded CAG repeat associated with juvenile Huntington disease shows a common origin of most or all neurons and glia in human cerebrum. Neurosci Lett. 2000;286:203. doi: 10.1016/S0304-3940(00)01029-6. [DOI] [PubMed] [Google Scholar]
- 57.Møllersen L, Rowe AD, Larsen E, Rognes T, Klungland A. Continuous and periodic expansion of CAG repeats in Huntington’s disease R6/1 mice. PLoS Genet. 2010;6:e1001242. doi: 10.1371/journal.pgen.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito Y, et al. Somatic mosaicism of the expanded CAG trinucleotide repeat in mRNAs for the responsible gene of Machado-Joseph disease (MJD), dentatorubral-pallidoluysian atrophy (DRPLA), and spinal and bulbar muscular atrophy (SBMA) Neurochem Res. 1998;23:25. doi: 10.1023/A:1022441101801. [DOI] [PubMed] [Google Scholar]
- 59.Ueno S, et al. Somatic mosaicism of CAG repeat in dentatorubral-pallidoluysian atrophy (DRPLA) Hum Mol Genet. 1995;4:663. doi: 10.1093/hmg/4.4.663. [DOI] [PubMed] [Google Scholar]
- 60.Montermini L, Kish SJ, Jiralerspong S, Lamarche JB, Pandolfo M. Somatic mosaicism for Friedreich’s ataxia GAA triplet repeat expansions in the central nervous system. Neurology. 1997;49:606. doi: 10.1212/WNL.49.2.606. [DOI] [PubMed] [Google Scholar]
- 61.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133:118. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 64.Franco SJ, Müller U. Shaping our minds: Stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reillo I, de Juan Romero C, García-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 66.Reid CB, Tavazoie SF, Walsh CA. Clonal dispersion and evidence for asymmetric cell division in ferret cortex. Development. 1997;124:2441. doi: 10.1242/dev.124.12.2441. [DOI] [PubMed] [Google Scholar]
- 67.Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255:434. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- 68.Ware ML, Tavazoie SF, Reid CB, Walsh CA. Coexistence of widespread clones and large radial clones in early embryonic ferret cortex. Cereb Cortex. 1999;9:636. doi: 10.1093/cercor/9.6.636. [DOI] [PubMed] [Google Scholar]
- 69.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci USA. 1998;95:1242. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 71.Pleasure SJ, et al. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727. doi: 10.1016/S0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 72.Magavi S, Friedmann D, Banks G, Stolfi A, Lois C. Coincident generation of pyramidal neurons and protoplasmic astrocytes in neocortical columns. J Neurosci. 2012;32:4762. doi: 10.1523/JNEUROSCI.3560-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 76.Pieras JI, et al. Somatic mosaicism for Y120X mutation in the MECP2 gene causes atypical Rett syndrome in a male. Brain Dev. 2011;33:608. doi: 10.1016/j.braindev.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 77.Topçu M, et al. Somatic mosaicism for a MECP2 mutation associated with classic Rett syndrome in a boy. Eur J Hum Genet. 2002;10:77. doi: 10.1038/sj.ejhg.5200745. [DOI] [PubMed] [Google Scholar]
- 78.O’Huallachain M, Karczewski KJ, Weissman SM, Urban AE, Snyder MP. Extensive genetic variation in somatic human tissues. Proc Natl Acad Sci USA. 2012;109:18018. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 80.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]