Abstract
Transcriptome studies reveal many noncoding transcripts overlapping 3’ gene termini. The function of these transcripts is unknown. Here we characterize transcription at the progesterone receptor (PR) locus and identify noncoding transcripts that overlap the 3’ end of the gene. Small RNAs complementary to sequences beyond the 3’ terminus of PR mRNA modulate expression of PR, recruit argonaute to a 3’-noncoding transcript, alter occupancy of RNA polymerase II, induce chromatin changes at the PR promoter, and affect responses to physiologic stimuli. We find that the promoter and 3’ terminal regions of the PR locus are in close proximity, providing a potential mechanism for RNA-mediated control of transcription over long genomic distances. Computational analysis led to identification of an inhibitory miRNA that targets the 3’ downstream region and modulates PR expression. These results extend the potential for small RNAs to regulate transcription to regions beyond the 3’ termini of mRNA.
Modulation of gene expression in mammalian cells by small duplex RNAs is typically associated with recognition of mRNA1. Duplex RNAs complementary to gene promoters have been reported to either silence or activate gene expression in mammalian cells2-6. Argonaute 2 (AGO2), a key protein involved in RNAi7, is required for the action of promoter-targeted RNAs5,8, and a related protein, AGO1, has also been implicated in the mechanism9. Recent reports have suggested that the mechanism of promoter-targeted RNAs involves recognition of noncoding transcripts that overlap gene promoters10,11. Over 70 % of all genes have noncoding transcripts that overlap their promoters and these transcripts provide potential target sites for small RNA duplexes12-17.
Promoter-targeted RNAs are robust modulators of progesterone receptor (PR) transcription in T47D and MCF7 breast cancer cells4,6,8,11. We term these small RNAs antigene RNAs (agRNAs) to distinguish them from duplex RNAs that target mRNA. The main difference between activation or inhibition of gene expression by closely related agRNAs is the basal expression of PR. Gene silencing is observed in T47D cells that constitutively express PR at high basal levels, while activation of PR expression is observed in MCF7 cells that express PR at low levels6.
Both activating and inhibitory agRNAs modulate PR expression through binding to complementary target sequences within an antisense transcript that originates from inside the PR gene and is transcribed through the promoter region. agRNAs recruit AGO protein to the antisense transcript, affect levels of RNA polymerase II (RNAP2) at the promoter, and alter the mix of regulatory proteins that bind the antisense transcript and the PR promoter11.
Noncoding RNAs also overlap the 3’-untranslated region (3’-UTR) of many genes15-17. The 3’-UTR plays a major role in cellular regulation and disease pathology18 and is involved in a variety of post-transcriptional processes, including mRNA transport, localization, and stability. The function of 3’ noncoding transcripts is unclear, but their proximity to the 3’-UTR suggests that they may affect gene regulation.
There has been little investigation into the potential function of overlapping noncoding transcripts at the 3’-region of genes, and no examination of whether these noncoding transcripts might be targets for modulating gene expression by duplex RNAs. The abundance of transcripts that overlap the 3’-UTR, coupled with the ability of agRNAs to modulate gene expression by targeting overlapping 5’ transcripts, suggested that small RNAs might also influence gene expression by recognizing sequences beyond the 3’ end of genes. Here we investigate the potential for small RNAs to recognize regions beyond the 3’ termini of mRNA and regulate gene expression.
RESULTS
Characterization of the 3’ Region of PR mRNA
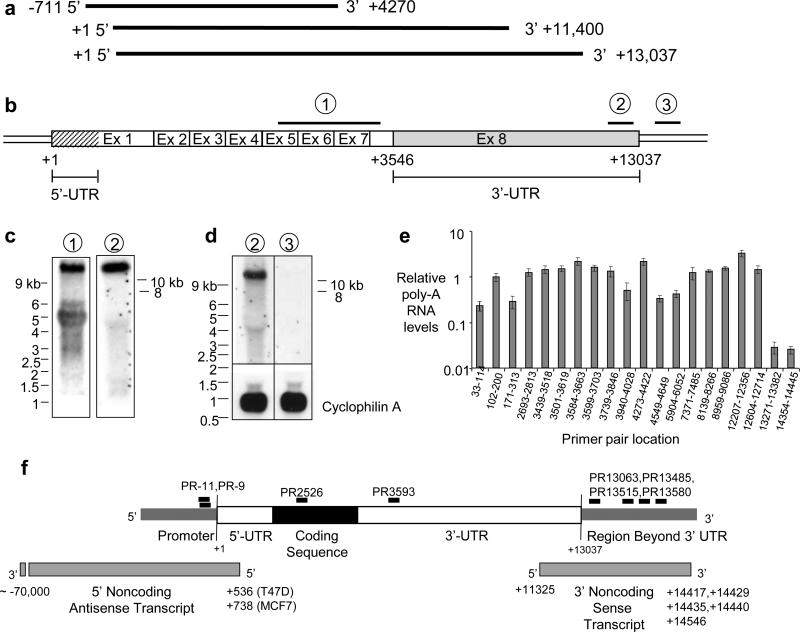
Working with agRNAs requires accurate identification of mRNA termini. Initially, however, the PR GenBank sequence had been inaccurately labeled with the 5’ end extended too far upstream and the 3’ terminus prematurely truncated (Fig. 1a, top). Northern analysis suggested lengths for PR mRNA variants19 but lacked a precise length for the largest variant (estimated to be 11.4 kB) (Fig. 1a, middle). A GenBank update based on a cluster of expressed sequence tags extended the 3’ UTR downstream to +13,037 (Fig. 1a, bottom).
Figure 1. Characterization of PR mRNA.
(a) Differing annotations of PR mRNA. Top: pre-2008 GenBank (NM_000926.3); Middle: Predicted largest transcript based on Northern analysis in published reports; Bottom: Current GenBank (NM_000926.4). (b) Schematic of PR mRNA predicted by GenBank and locations of probes for Northern analysis. (c) Northern analysis of PR mRNA comparing results using probes that detect PR mRNA (probe 1) or targeting the 3’ termini of PR mRNA (probe 2), (d) Northern analysis of PR mRNA using probe 2 or a probe immediately downstream of the potential mRNA terminus (probe 3). (e) qPCR showing levels of poly-A RNA in T47D cells detected from the PR transcription start site (+1) past the most downstream annotated terminus of PR mRNA. Notation indicates target region for PCR primers. Data is the resultant of triplicate independent experiments. (f) Location of target sequences for duplex RNAs relative to PR mRNA. The 5’ and 3’ noncoding transcripts that overlap the transcription start site and polyadenylation site are shown.
We performed northern analysis with probes complementary to i) the protein-encoding region of PR mRNA (probe 1), ii) the terminus of PR mRNA at +13,037 predicted by the recent GenBank update (probe 2), and iii) a region immediately downstream from the predicted +13,037 terminus (probe 3) (Fig. 1b). Probe 1 yielded major products at ~5.5 kb and >10 kb, similar to results observed previously19 (Fig. 1c). Probe 2 (complementary to the region at the predicted PR terminus) yielded only the >10 kb band (Fig. 1c,d), consistent with the conclusion that the band is full length PR mRNA. Probe 3 (complementary to the region immediately downstream of +13,037) did not detect transcript (Fig. 1d). Quantitative PCR (qPCR) revealed that RNA levels were relatively constant before abruptly dropping after nucleotide +13,037 (Fig. 1e, Supplementary Fig S1). 5’ and 3’ Rapid Amplification of cDNA Ends (RACE) detected polyadenylation sites near +13,037 (Supplementary Fig. S2).
Characterization of Transcripts that Overlap the 3’ Terminus of PR mRNA
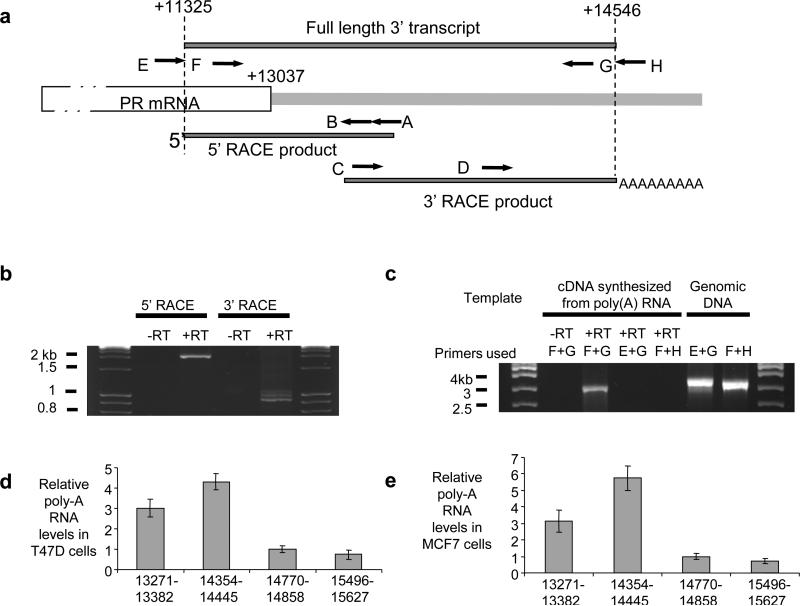
We used 3’ and 5’ RACE to search for noncoding transcripts that overlap the 3’ terminus and identified transcripts that are transcribed in sense orientation (i.e. synthesized in the same direction) relative to PR mRNA (Fig. 1f, Figs. 2a,b, Supplementary Figs. S1,S2). These 3’ transcripts share a transcription start site at +11,325 and multiple polyadenylation sites 1400 to 1500 bases downstream from to the +13,037 terminus of PR mRNA. We did not detect antisense transcripts in this region.
Figure 2. Characterization of the PR 3’ noncoding transcript.
(a) Location of RACE (A,B,C, and D) or RT-PCR (E,F,G, and H) primers relative to PR mRNA and the 3’ noncoding transcript. (b) Agarose gel analysis of RACE products. Total RNA used in RACE was treated with DNase prior to reverse transcription. (c) Agarose gel analysis of RT-PCR amplification using primers E, F, G, and H as shown. Poly(A) RNA was DNase-treated prior to reverse transcription. Amplification of genomic DNA was included as a positive control for primer function. Complete data including sequencing of amplified products is shown in Supplementary Figure S3. qPCR of relative RNA levels in (d) T47D cells and (e) MCF7 cells using primer sets upstream or downstream of the predicted +14,546 termini of the 3’ noncoding transcript (also see Supplementary Figure S1). −RT: RNA samples that were not treated with reverse transcriptase (−RT) were used as negative control. +RT: Reverse transcriptase added. Data is the result of triplicate independent experiments.
Since the 3’ transcript is transcribed in the same direction as PR mRNA, we tested whether it was long variant of PR mRNA. We performed RT-PCR using a forward primer (primer E) complementary to PR mRNA that was directly upstream of +11,325 (and therefore had no complementarity to the +11325/+14,546 transcript) and a reverse primer (primer G) upstream of +14,546 (Fig. 2a). No amplified product was detected (Fig. 2c, Supplementary Figs S2c,d). By contrast, RT-PCR using a forward primer (primer F) directly downstream of +11,325 and reverse primer directly upstream of +14,546 (primer G) detected the +11325/+14,546 product. Sequencing confirmed the identity of the amplified primer F/primer G product as the +11,325/+14,546 transcript and revealed that the transcript was unspliced (Supplementary Fig. S2e). Beyond the +14,546 terminus of the noncoding transcript, qPCR revealed that RNA levels drop abruptly in both T47D and MCF7 cells (Fig. 2d,e, Supplementary Fig. S1). RT-PCR with one primer (primer F) complementary to a sequence shared by PR mRNA and the 3’ noncoding transcript and a primer (primer H) downstream from +14,546 detected no product, suggesting that the mRNA does not extend past +13037 (Fig. 2C).
Many genes are transcribed beyond their poly-A sites and then cleaved to form the mature mRNA. Our PCR did not detect evidence for longer PR mRNA (Figure 2C), but we cannot exclude with certainty its existence or its involvement in the mechanism of 3’ agRNAs. qPCR revealed that the 3’ noncoding transcript was almost entirely in the nuclear fraction of cell lysates while PR mRNA was mostly in the cytoplasm (Supplementary Figure S3).
We verified our data using the branched DNA (bDNA) assay (Supplementary Fig. S4). The bDNA assay directly detects RNA using strand-specific probes that allow discrimination of sense and antisense transcripts. A probe set complementary to the sense transcript beyond +13,037 detected RNA at ~4 % relative to the level of PR mRNA in either T47D or MCF7 cells. Assays with a probe set designed to detect a possible antisense transcript downstream from +13,037 suggested that an antisense transcript was either not present or was present a levels too low to detect.
Choice of Target Sequences for Duplex RNAs
We chose target sequences within the +11,325/+14,546 3’ noncoding transcript but downstream from +13,037 terminus of PR mRNA. (Supplementary Table S1, Fig. 1f). Duplex RNAs are numbered by the position of the most upstream base relative to the +1 transcription start site for PR mRNA. We also tested duplex RNAs that target the PR promoter or PR mRNA to allow comparisons with other modulatory RNAs. We have previously characterized agRNAs that target the promoter region (5’ agRNAs) that modulate PR expression. PR-11 targets the region −11/+8 relative to the transcription start site and activates PR transcription in MCF7 and T47D cells6. PR-9 targets the region −9/+10 and is a robust transcriptional silencing agent in T47D cells4. PR3593 and PR2526 target PR mRNA.
Gene Inhibition by Small RNAs that Target Sequences Beyond the 3’-UTR
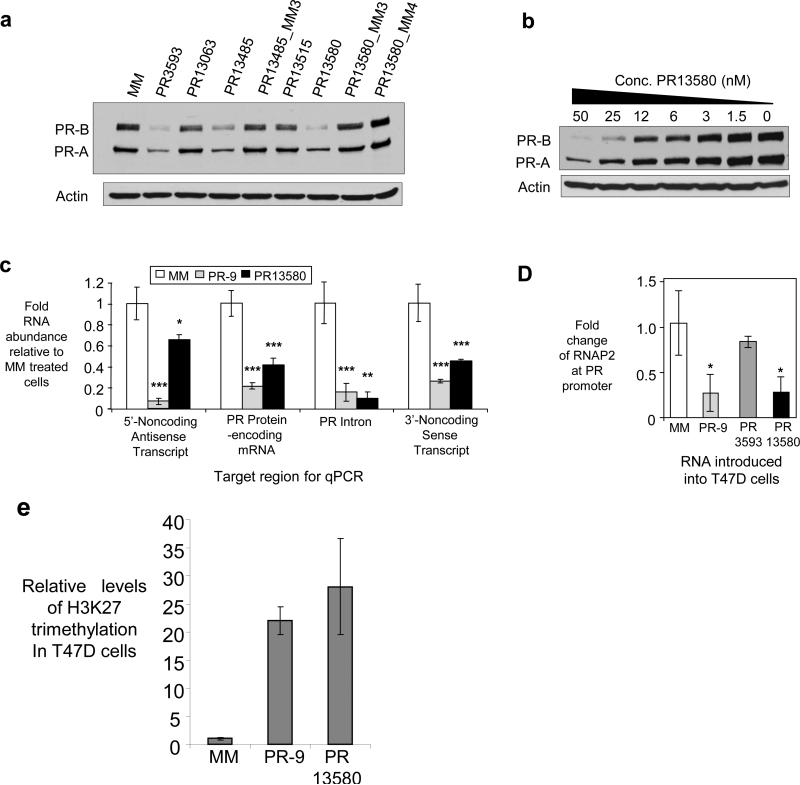
We tested several 3’ agRNAs complementary to sequences downstream from the +13,037 terminus of PR mRNA (Supplementary Table S1, Fig. 1f). agRNAs PR13485 and PR13580 inhibited PR protein expression in T47D cells (Fig. 3a). PR has two major protein isoforms, PRB and PRA19 and both of these appear during western analysis. The IC50 value for inhibition of PR13580 was 10.7 nM, similar to the value for inhibition by mRNA-targeting RNA PR2526, 11.5 nM (Fig. 3b, Supplementary Fig. S5). Mismatch-containing duplex RNAs did not inhibit gene expression. Some of the control RNAs maintained the potential for seed sequence20 recognition of the duplex (PR13485_MM3, PR13580_MM3) demonstrating that seed sequence complementarity is not sufficient to induce the observed silencing (Fig. 3a).
Figure 3. Inhibition of PR expression in T47D cells by agRNAs complementary to sequences downstream from the terminus of PR mRNA.
(a) Western analysis showing inhibition of protein expression by duplex RNAs (50 nM). (b) Dose response for PR13580. (c) qPCR showing reduction of PR mRNA levels by duplex RNAs (25 nM). Four different primer sets were used, each complementary to different regions of the PR gene (Supplementary Table 4). (d) Presence of RNAP2 at the PR transcription start site (25 nM duplex RNA) evaluated by ChIP. (e) Chromatin immunoprecipitation for the H3K27 trimethylation (H3K27me3) marker within the PR gene locus in T47D cells. ***p<0.005, **p<0.01, and *p<0.05 as compared to cells treated with a mismatch RNA. p-values were calculated using the two-tailed unpaired Student’s T-test with equal variances. All error bars represent standard deviation. Data in parts (a), (c) and (c) is the result of triplicate independent experiments. Data in part (b) is representative of duplicate experiments.
Small RNAs can induce off-target effects through induction of the interferon response21. Inhibitory 3’ agRNA PR13580 did not significantly enhance expression of interferon-responsive genes (Supplementary Fig. S6a) and was chosen for detailed investigation. Addition of poly(I:C), a potent inducer of the interferon response did not alter PR gene expression (Supplementary Fig. S6b).
We used qPCR to investigate how addition of inhibitory agRNA PR13580 affected RNA levels throughout the PR locus (Fig 3c). Our strategy was to use different primer sets, each designed to amplify a different RNA species including: i) the 5’ antisense transcript at the PR promoter previously implicated in agRNA-mediated control of transcription11, ii) the protein encoding region for PR mRNA, iii) intron 7 within PR pre-mRNA, an indicator of whether gene modulation occurs before or after splicing, and iv) the +11325/+14546 noncoding transcript overlapping the terminus of PR mRNA (Fig. 1f).
Addition of agRNA PR13580 to T47D cells reduced levels of PR mRNA (Fig. 3c). The level of PR pre-mRNA was also reduced, suggesting that modulation of RNA occurs prior to splicing. Levels of the noncoding transcript downstream from the terminus of the 3’-UTR were reduced, as were levels of the 5’ noncoding transcript overlapping the transcription start site of PR. Negative control duplex RNA PR13063 did not reduce levels of PR mRNA (Supplementary Fig. S7). The bDNA assay yielded similar results (Supplementary Fig. S8).
To test whether agRNAs targeting sequences beyond the 3’-UTR altered gene transcription, we measured RNAP2 occupancy at the PR promoter. Chromatin immunoprecipitation (ChIP) revealed that duplex agRNA PR13580 reduced occupancy of RNAP2 at the promoter (Fig. 3d). We also measured levels of the histone modification H3K27me3 which serves as a chromatin-level marker for gene silencing22. Addition of PR13580 to T47D cells led to a 27-fold increase in H3K27me3 levels within the PR gene relative to cells treated with mismatched control duplex (Fig. 3e). Addition of PR-9 also caused changes in RNAP2 occupancy and levels of H3K27me3. RNA PR3593 that targets PR mRNA did not decrease RNAP2 occupancy (Fig. 3d), emphasizing a basic difference in mechanism between agRNAs (inhibition of transcription) and duplex RNAs that function through post transcriptional gene silencing (PTGS) (inhibition of translation/destruction of mRNA).
Gene Activation by Small RNAs that Target Sequences Beyond the 3’-UTR
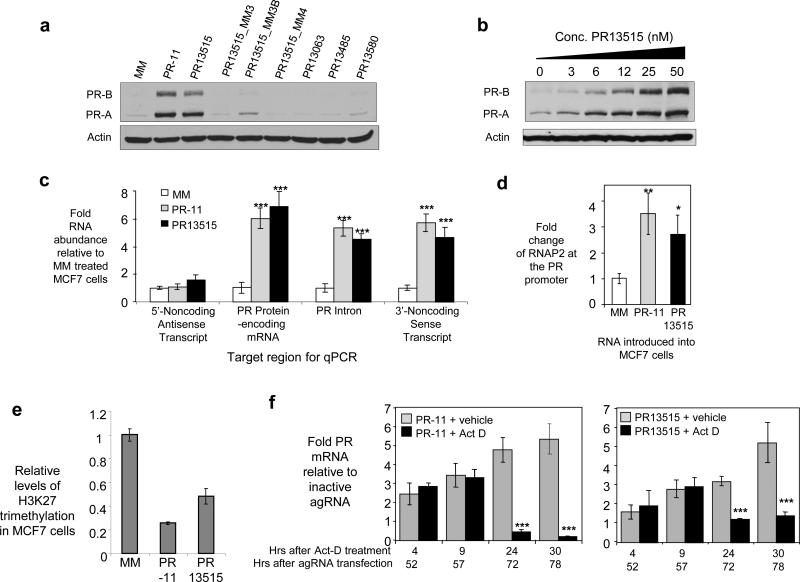
We hypothesized that agRNAs complementary to sequences beyond the 3’ terminus of PR might also yield enhanced gene expression in MCF7 cells, a cell line where PR expression is low and increased expression is easily detectable. agRNA PR13515 increased levels of PR protein and mRNA (Fig. 4a, 4b, and 4c). Levels of PR pre-mRNA were also increased (Fig. 4c), consistent with the suggestion that RNA levels increase prior to splicing. For comparison, we also measured RNA levels after addition of agRNA PR-11 that is complementary to the PR promoter/5’ noncoding RNA transcript11 and observed similar effects with all primer sets (Fig. 4c). The bDNA assay also revealed increased levels of PR mRNA and the 3’ noncoding transcript upon addition of PR13515 or PR-11 (Supplementary Fig. S8c).
Figure 4. Enhanced PR expression in MCF7 cells by an RNA complementary to a sequence downstream from the terminus of the PR 3’-UTR.
(a) Western analysis showing activation of protein expression by duplex RNAs. (b) Dose response for RNA PR13515. (c) qPCR showing effect on RNA levels. Four different primer sets were used, each complementary to different regions near the PR gene. (d) Recruitment of RNAP2 to the PR promoter upon addition of PR13515 or PR-11 evaluated by ChIP. (e) Chromatin immunoprecipitation for the H3K27 trimethylation (H3K27me3) marker within the PR gene locus in MCF7 cells. (f) MCF7 cells were transfected with either PR-11 or PR13515. After two days, actinomycin D (act D, 1 μg/ml) or vehicle was added to the media. Cells were harvested at the indicated timepoints (hours) after actinomycin D or vehicle treatment. Data were normalized to levels of 18S rRNA that did not significantly change for the duration of the experiment. Standard deviation was calculated from 4-6 samples per treatment group. ***p<0.005, **p<0.01, and *p<0.05 as compared to cells treated with RNA MM. p-values were calculated using the two-tailed unpaired Student's T-test with equal variances. All error bars represent standard deviation. Duplex RNAs were added to cells at 25 nM unless otherwise noted. Data in parts (a) - (d) are the results of triplicate experiments. Data in part (f) was the result of 4-6 independent experiments.
Addition of either PR-11 or PR13515 led to enhanced RNAP2 recruitment at the PR transcription start site (Fig. 4d) and a decrease in the silencing marker H3K27me3 (Fig. 4e). The similarity of gene activation between PR-11 and PR13515 reinforces the suggestion that modulation of gene transcription by agRNAs proceeds through similar mechanisms regardless of whether their target regions are located beyond the 5’ or 3’ boundaries of mRNA.
We tested whether increased stability of PR mRNA contributes to increased levels of PR mRNA following treatment with PR13515. Actinomycin D, a small molecule that inhibits transcription23, was added to cells that had been previously treated with PR13515 or PR-11. The half-life of PR in MCF7 cells has been established to be 7-10 hours27. In MCF7 cells treated with either PR13515 or PR-11, addition of actinomycin D reversed the agRNA-mediated increase in PR mRNA. PR mRNA levels decreased by 8-10 fold within thirty hours. (Fig. 4f). These data suggest that increased levels of PR mRNA are due to enhanced transcription.
We tested several control RNAs to evaluate specificity. Control RNA PR13515_MM4 contained mismatched bases substituted throughout the sequence of PR13515. Control RNAs PR13515_MM3 and PR13515_MM3B that had mismatched bases clustered to preserve the potential for off-target effects that might arise from seed sequence complementarity24 did not enhance protein expression (Fig. 4a). PR13515 did not increase expression of interferon-responsive genes and activation of the interferon response did not increase PR expression (Supplementary Fig. S6c,d). Off-target effects are a concern whenever duplex RNAs are introduced into cells. Our 3’ and 5’ agRNAs have partial complementarity to other genes but these matches do not suggest any source for off-target regulation of PR.
Effect of Physiological Stimuli on 3’ Transcript Expression and agRNA Action
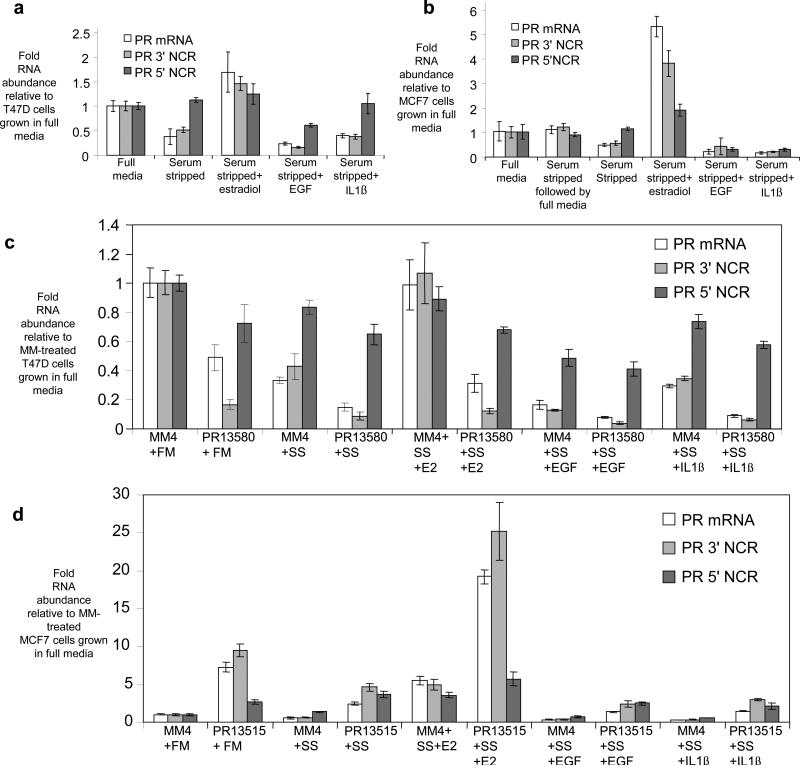
17ß-estradiol is a potent and well-characterized activator of PR gene expression in breast cancer cells24. Conversely, expression of PR is decreased in cells grown in media containing charcoal-stripped serum and supplemented with either interleukin-1ß (IL-1ß) or epidermal growth factor (EGF)25-27.
We investigated how these physiologic stimuli would affect expression of the 3’ and 5’ noncoding transcripts (Fig. 5a,b). PR mRNA levels were reduced in T47D and MCF7 cells grown using charcoal-stripped serum. Addition of EGF or IL-1ß further reduced levels of PR mRNA, while addition of 17ß-estradiol increased PR mRNA expression. In all cases, levels of the 3’ noncoding transcript varied proportionally with PR mRNA (Fig. 5a,b). By contrast, expression of the 5’ noncoding transcript did not change significantly in T47D or MCF7 cells grown in charcoal-stripped serum, or in media supplemented with 17ß-estradiol. Expression of the 5’ transcript was decreased by addition of EGF or IL1ß in MCF7 cells.
Figure 5. Effect of physiologic stimuli or effect of combining physiologic stimuli with addition of agRNAs on expression of PR mRNA, the 3’ noncoding RNA, and the 5’ noncoding RNA.
qPCR analysis showing the effect of physiologic stimuli on transcript expression in (a) T47D cells and (b) MCF7 cells. For the sample labeled “serum stripped followed by full media” cells were grown in serum stripped media. The media was replaced by full media for one day prior to harvesting. qPCR analysis of the effect of physiologic stimuli and agRNA (25 nM) addition on transcript expression in (c) T47D cells and (d) MCF7 cells. PR 3’NCR: 3’ noncoding PR RNA. PR 5’NCR: 5’ noncoding PR RNA. SS: Serum-stripped media. FM: full media. E2: 17ß estradiol treatment (100 nM). IL1ß: interleukin 1ß treatment (10 ng/mL). EGF: epidermal growth factor treatment (100 ng/mL). Results are from 3-6 independent replicates.
We then tested whether the addition of agRNAs would affect regulation of PR expression by physiologically relevant stimuli and, conversely, whether physiologic stimuli could block the action of agRNAs (Fig. 5c,d, Supplementary Fig. S9). Addition of inhibitory 3’ agRNA PR13580 to T47D cells cultured in charcoal stripped serum supplemented with IL-1ß or EGF led to a proportionate reduction in expression of PR mRNA and the 3’ noncoding transcript to levels lower than those achieved by the physiologic treatments alone. Treating cells with 17ß-estradiol increases PR mRNA expression (Fig. 5a). Inhibitory agRNA PR13580 reversed this effect, leading to low expression of PR (Fig. 5c) and almost unchanged expression of the 5’ noncoding transcript regardless of physiologic treatment.
In MCF7 cells, addition of activating agRNA PR13515 in combination with 17ß-estradiol yielded enhanced activation of PR gene expression to levels substantially above those achieved by addition of 17ß-estradiol or PR13515 alone. PR13515 reversed the repressive effects from growth in charcoal-stripped serum, EGF addition, and IL1ß addition on PR expression (Fig. 5d). These data suggest that activating and inhibitory agRNAs can counteract or supplement the effects of physiologic regulators on PR gene expression.
Argonaute Protein Recruitment by Noncoding RNAs
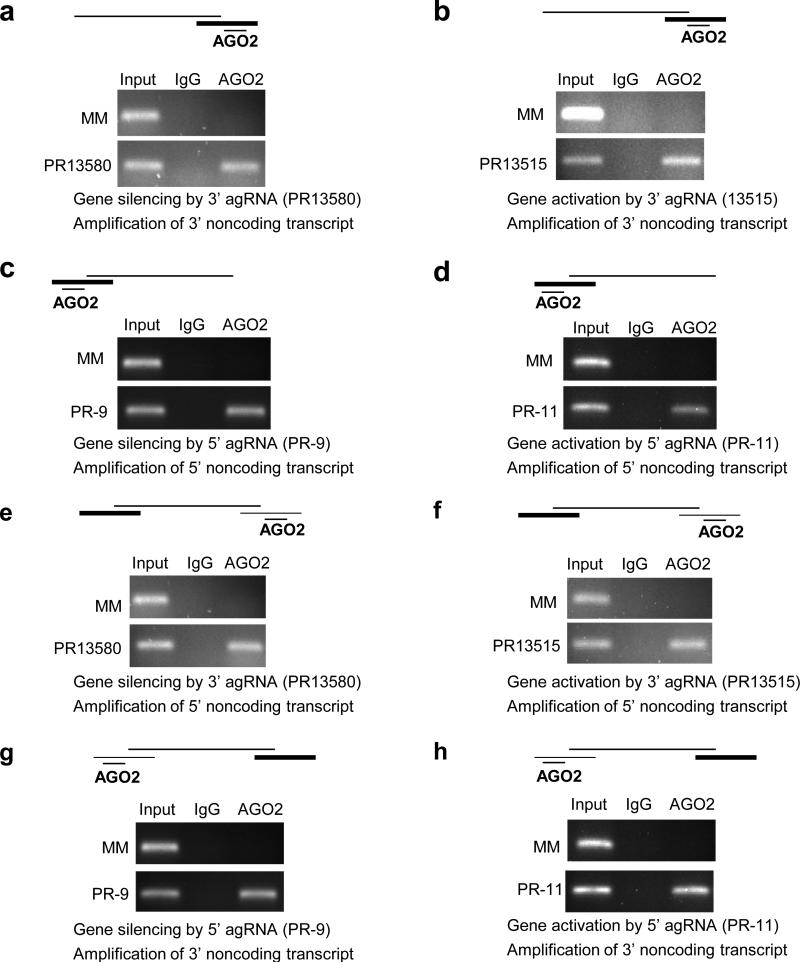
Duplex agRNAs PR-9 and PR-11 are complementary to sequences within the PR promoter and recruit AGO protein to a noncoding transcript that overlaps the PR gene promoter11. The sense noncoding transcript overlapping the 3’ terminus of the PR gene (Fig. 1f) contains complementary target sites for inhibitory or activating agRNAs PR13580 and PR13515, and is a candidate for involvement in RNA-mediated gene modulation.
We used RNA immunoprecipitation (RIP)28 from isolated nuclei with an anti-AGO2 antibody (Supplementary Fig. S10) to examine recruitment of AGO2 protein to the 3’ noncoding transcript during modulation of gene expression by agRNAs. When silencing agRNA PR13580 was added to T47D cells (Fig. 6a) or activating agRNA PR13515 was added to MCF7 cells (Fig. 6b), we observed association between AGO2 and the 3’ noncoding transcript. 5’ agRNAs targeting a noncoding RNA at the PR promoter showed similar recruitment of AGO2 to the 5’ noncoding transcript (Fig. 6c and 6d). Sequencing confirmed the identity of the RIP products (Supplementary Fig. S10). RIP with an anti-AGO1 antibody did not detect association of AGO1 (Supplementary Fig. S11). Inhibition of AGO2 expression with an anti-AGO2 siRNA reversed gene silencing by PR13580. Inhibition of AGO1, AGO3, or AGO4 expression did not reverse gene silencing (Supplementary Fig. S12). These data are consistent with a primary role for AGO2.
Figure 6. Effect of 3’ or 5’ agRNAs on recruitment of AGO2 protein to the 3’ or 5’ noncoding transcripts at the PR locus.
RNA immunoprecipitation (RIP) of 3’ noncoding RNA using an anti-AGO2 antibody after treatment with (a) inhibitory RNA PR13580 in T47D cells or (b) activating RNA PR13515 in MCF7 cells on recruitment of AGO2 protein to the 3’ noncoding transcript. Effect of adding (c) inhibitory RNA PR-9 to T47D cells or (d) activating RNA PR-11 to MCF7 cells on recruitment of AGO2 protein to the 5’ noncoding transcript. Effect of adding (e) inhibitory RNA PR13580 to T47D cells or (f) activating RNA PR13515 to MCF7 cells on co-immunoprecipitation of AGO2 protein with the 5’ noncoding transcript. Effect of adding (g) inhibitory RNA PR-9 to T47D cells or (h) activating RNA PR-11 to MCF7 cells on co-immunoprecipitation of AGO2 protein with the 3’ noncoding transcript. The scheme above each gel depicts PR mRNA, the 3’ and/or 5’ noncoding transcripts, and AGO2 bound agRNA. The heaviest line represents the transcript being amplified. Duplex RNAs were added to cells at 25 nM. Experiments are representative of two independent determinations.
We observed the same results when using a well-characterized antibody that recognizes all four AGO proteins29 (Supplementary Fig. S13A-D). Identical RIP results using two different anti-AGO antibodies supports the specificity of AGO involvement in the mechanism of 3’ agRNAs. No product was observed in the absence of reverse transcriptase (Supplementary Fig. S14) or after transfection of cells with mismatch-containing duplex RNAs. Cytoplasmic proteins GAPDH and tubulin were not detected in the nuclei, indicating that we are not detecting interactions in the cytoplasm.
Cleavage of the 3’ Noncoding Transcript is Not Detected
Small RNAs that are complementary to mRNA can induce cleavage of their target transcripts1. An important question of mechanism is whether 3’ agRNAs act by promoting cleavage of the 3’ noncoding transcript. We readily detected cleavage of PR mRNA by PR2562, a duplex RNA that is complementary to the coding region of PR mRNA (Supplementary Fig. S15).
We were unable to detect any cleavage of the 3’ noncoding RNA using silencing agRNA PR13580 in T47D cells or activating agRNA PR13515 in MCF7 cells (Supplementary Fig. S15). We note that the 3’ noncoding transcript is expressed at relatively low levels and it is possible that cleavage might occur without being detectable by 5’ RACE. Failure to detect any cleavage, however, is consistent with 1) our observation that addition of PR13515 enhances levels of the noncoding transcript (Fig. 4c) rather than reducing levels as would be predicted if transcript cleavage were occurring; 2) previous results showing that 5’agRNA PR-11 does not alter levels of the 5’ noncoding transcript and does not cause detectable cleavage of the transcript6; and 3) RIP data showing association of AGO2 because the transcript must be intact if it is to be detected during RIP.
Overexpression of 3’ Noncoding Transcript Does Not Affect PR mRNA Levels
It is possible that the 3’ noncoding transcript recruits AGO2 protein prior to its release from chromosomal DNA. Alternatively, the 3’ noncoding transcript might be released from the chromosome and subsequently return to act at the PR locus. This latter mechanism would be similar to protein transcription factors that are synthesized in the cytoplasm and return to act on their target promoters in the nucleus.
To examine the effect of altered cellular levels of the 3’ noncoding transcript and PR mRNA, we cloned and overexpressed the 3’ noncoding transcript in three breast cancer cell lines with varying basal PR expression levels (T47D-high, MCF7-low, and MDA-MB231-undetectable). Overexpression of the 3’ noncoding transcript by as much as 100-fold above endogenous levels did not yield a significant change in PR mRNA levels in any of these three cell lines (Supplementary Fig. S16). These results suggest that the cellular concentration of 3’ noncoding RNA does not affect PR expression.
Gene Looping Brings 5’ and 3’ Sequences Into Proximity
Modulation of gene transcription by small RNAs complementary to sequences beyond the PR 3’-UTR suggests that recognition of downstream sequences influences activity at gene promoters. This influence must be exerted over a long distance because the genomic locations of the PR promoter and the agRNA target sites are ~100 kB apart. Previous reports suggest that gene promoters and termini can be held in close proximity to one another30-32. Such proximity might facilitate the modulation of gene expression between otherwise distant 3’ and 5’ regions.
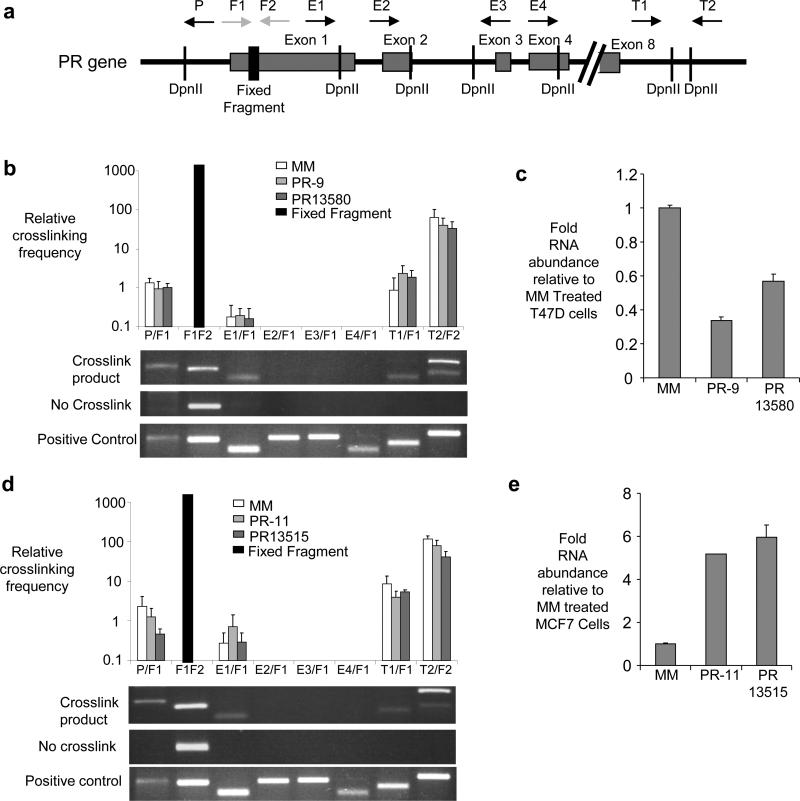
We investigated whether the promoter and terminal regions of PR might also be in proximity using chromosome conformation capture (3C) analysis31,32, a technique that examines the proximity of sequences within chromosomal DNA. In this technique chromosomal DNA is crosslinked, digested with restriction enzyme, and treated with DNA ligase to join DNA ends that are in close proximity. After reversal of crosslinks the DNA is amplified and sequenced to evaluate the proximity of target regions. We examined amplification of 3C products using multiple primer sets that vary in distance from the 5’ promoter and 3’ terminal regions of PR (Fig. 7a). For example, amplification by primer T2 (complementary to a sequence beyond the PR 3’-UTR termini) and primer F2 (complementary to sequences within PR exon 1) would only be predicted to occur if the 5’ and 3’ ends of the PR locus are held in proximity by gene looping.
Figure 7. 3C analysis of the PR locus.
(a) Schematic of the PR gene showing DpnII cleavage sites, exon boundaries, and locations of primers used for 3C analysis. The primer pairs used for 3C amplification are shown on the x axes of parts (b) and (d). (b) Top, qPCR showing the relative levels of detection of crosslinked product after treatment with a mismatch-containing RNA duplex or inhibitory duplexes PR-9 or PR13580. (c) qPCR showing reduced RNA levels in samples used for part (d). (d) Top, qPCR showing the relative levels of crosslinked product after treatment with a mismatch-containing RNA duplex or activating duplexes PR-11 or PR13515. (e) qPCR showing increased RNA levels in the samples used for part (d). Primer P amplifies a sequence at the PR promoter. Primers E1, E2, E3, and E4 amplify sequences within PR exons 1-4. Primers T1 and T2 amplify sequences beyond the terminus of PR mRNA. F1/F2= Fixed fragment. The fixed fragment is a normalization control derived from genomic DNA by primers complementary to sequences within exon 135. The bar represents performance of the normalization control, not its absolute value. Values in parts (b) and (d) are relative to amplification of sequence at the PR promoter using primer P. Duplex RNAs were added to cells at 25 nM. Two bands are observed in the T2/F2 analysis because of an alternative DpnII cleavage site. The positive control shows amplification of a synthetic DNA. Data are from three independent experiments.
We observed that the 5’ promoter region of the PR gene was ligated to the 3’ terminal regions in T47D (Fig. 7b) and MCF7 (Fig. 7d) cells. Even though expression of PR is much higher in T47D than MCF7 cells, we did not detect a difference in the relative amount of looping (Supplementary Fig. S17). Addition of inhibitory agRNAs PR13580 or PR-9 to T47D cells (Fig. 7b,c) or activating agRNAs PR13515 or PR-11 to MCF7 cells (Fig. 7d,e) did not affect the relative amount of looping at the PR locus.
We also examined the effects of physiologic stimuli on looping. We have previously described the effects of adding estrogen, growth of cells in serum-stripped media, treatment with epidermal growth factor (EGF), or adding IL1ß decreases on PR expression (Fig. 5). These treatments did not change the relative amounts of gene looping (Supplementary Figs. S18 and S19). Our data suggest that gene looping at the PR locus remains constant under a range of different cell types, environmental stimuli, and agRNA treatments. It is most likely that the looping is intrachromosomal between the termini of a single PR gene. We are unaware of evidence for interchromosomal contacts between different alleles of the same gene on different chromosomes, but we note that such contacts are possible.
Proximity of 5’ and 3’ Noncoding Transcripts
We investigated the possibility that gene looping might allow the 5’ and 3’ noncoding transcripts to form long distance associations. We used RIP with anti-AGO2 antibodies to examine the proximity of the noncoding transcript at the 3’-UTR to the noncoding transcript at the PR promoter. Addition of inhibitory 3’ agRNA PR13580 (Fig. 6e) or activating 3’ agRNA PR13515 (Fig. 6f) followed by RIP led to detection of the 5’ noncoding transcript. Similarly, addition of duplex agRNAs complementary to the PR promoter, inhibitory agRNA PR-9 (Fig. 6g) and activating agRNA PR-11 (Fig. 6h), led to recovery of the 3’-noncoding transcript. RIP uses a chemical crosslinking step that allows detection of factors within a complex. Therefore, it is not necessary for association between the 3’ and the 5’ noncoding transcripts to be direct. A more likely explanation is that the association between these noncoding RNAs is indirect. Instead, there is may be a ribonucleoprotein complex containing AGO, and the 5’ noncoding and 3’ noncoding transcripts. We also observed the same results when using a second anti-AGO antibody29 (Supplementary Fig. S13e-h).
Inhibition of BRCA1 Expression by agRNAs Targeted Beyond the 3’-UTR
To test whether gene modulation by 3’agRNAs might apply to other genes, we targeted sequences beyond the 3’-UTR of the tumor suppressor breast cancer associated gene 1 (BRCA1). BRCA1 was chosen because: 1) its 3’ termini had been well characterized33, 2) its expression is lowered in a significant percentage of human cancers34, 3) it is expressed in T47D breast cancer cells, and 4) the chromosomal loci juxtapose its promoter and termination regions31. 3’-RACE and qPCR confirmed the previously reported35 termination site for BRCA1 mRNA (Supplementary Fig. S20). 3C analysis confirmed that the BRCA1 promoter and 3’ terminal regions are in proximity (Supplementary Fig. S21).
3’ agRNAs inhibited expression of BRCA1 protein, mRNA, and pre-mRNA (Supplementary Fig. S22) (Supplementary Table 1). BRCA7851 reduced levels of RNAP2 at the BRCA1 promoter. An siBRCA complementary to BRCA1 mRNA (Ma et al., 2006) did not decrease recruitment of RNAP2 to the BRCA1 promoter. The 3’ agRNAs did not activate interferon responsive genes (Supplementary Fig. S6e,f). These data suggest that 3’ agRNAs can also regulate expression of BRCA1.
Potential miRNA Target Sites Occur Downstream from 3’-UTRs
One explanation for the robust action of 3’ agRNAs is that they might be exploiting endogenous regulatory mechanisms that involve microRNAs (miRNAs). To determine whether miRNAs are candidates for recognition of sequences beyond the 3’ termini of mRNA we investigated the complementarity of miRNAs with sequences downstream from 3’-UTRs of known genes (Supplementary Figure S23). We had previously reported a similar examination for complementarity of miRNAs to gene promoters that revealed substantial sequence matches36.
Gene silencing by miRNAs requires complementarity between the target and the miRNA seed sequence (bases 2-8)37. Seed sequence matches downstream from gene termini occurred at the same relative frequency as within 3’-UTRs (Supplementary Figure S23b). We measured the frequency of seed matches relative to sequence matches that resulted from comparisons with randomizing downstream regions and observed a ~20 % increase in seed sequence matches relative to randomized sequences (Supplementary Figure S23c). Examination of highly ranked matches revealed many miRNAs with good complementarity to regions downstream from gene termini (Supplementary Figure S23a).
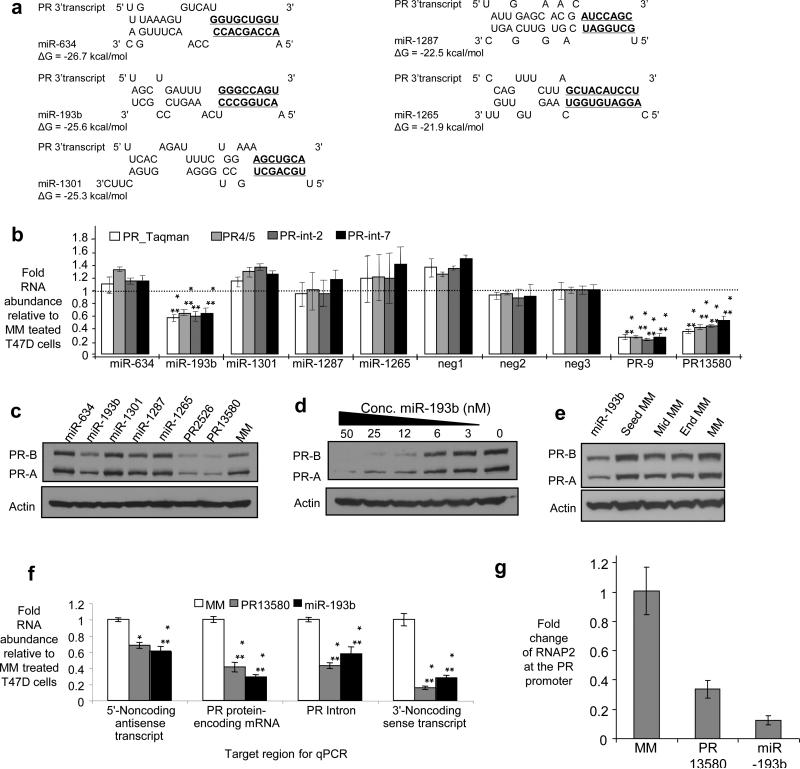
We used our computational algorithm to identify miRNAs with complementarity to the portion of the PR 3’ noncoding transcript that is downstream from the termination site for PR mRNA (Fig. 8a and Supplementary Fig. S24). Synthetic miRNAs were synthesized and introduced into T47D cells. miR-193b inhibited expression of PR mRNA when assayed using four different PCR primer sets targeting different regions of PR mRNA (Fig. 8b). miR-193b is encoded within an intergenic region of chromosome 16. It is present in both the nucleus (~40-50 %) and cytoplasm of the cell lines used in these studies. The potency of miR-193b to T47D was similar to agRNA PR13580 (Fig. 3b, Fig. 8d). Introduction of mismatches into the region of potential seed sequence complementarity between miR-193b and the 3’ noncoding transcript prevented inhibition (Fig. 8e). These results are consistent with a direct interaction at the predicted target site but do not rule out an indirect effect from a seed sequence interaction with some other gene. Comparison of miR-193b and 3’ agRNA PR13580 revealed similar effects on levels of the 5’ noncoding transcript, PR pre-mRNA, and the 3’ noncoding transcript (Fig. 8f). ChIP analysis of cells transfected with miR-193b revealed decreased levels of RNAP2 at the PR promoter (Fig. 8g). These data suggest that miR-193b can act as an inhibitory agRNA by a mechanism similar to designed agRNA PR 13580.
Figure 8. Inhibition of PR expression by a miRNA complementary to the PR 3’ noncoding RNA.
(a) Sequences for five computational matches between miRNAs and the region of PR 3’ noncoding RNA beyond +13,037 (Supplementary Fig. S27). Potential seed sequence matches (bases 2-8) are in boldface and underlined. (b) qPCR using four different primer sets showing the effects of adding miRNA mimics on PR expression. (c) Western analysis showing the effects of adding miRNA mimics on PR protein expression. (d) Dose response analysis showing effects of inhibitory miRNA miR-193b. (e) Western analysis showing inhibition of PR expression by MiR-193b and mismatch-containing analogs. (f) qPCR comparing the effects of inhibitor agRNA PR13580 or miR-193b on levels of the 5’ noncoding transcript, PR mRNA, PR pre-mRNA, and the 3’ noncoding transcript. (g) ChIP showing the effect of addition of miR-193b or inhibitory agRNA PR13580 on the presence of RNAP2 at the PR promoter. Experiments were performed in T47D cells. ***p<0.005, **p<0.01, and *p<0.05 as compared to cells treated with RNA MM. p-values were calculated using the two-tailed unpaired Student's T-test with equal variances. All error bars represent standard deviation. Duplex RNAs were added at 25 nM unless otherwise noted. Data are from triplicate independent experiments.
DISCUSSION
RNA Recognition Beyond the 3’-UTR
Our data shows that duplex RNAs can modulate transcription by targeting sequences beyond the 3’ UTR. These results 1) expand the pool of RNA transcripts that can be targeted by small RNAs; 2) suggest that interactions encoded beyond the 3’ UTR can be important; 3) show that RNA-mediated recognition can control transcription over a 100,000 base distance; 4) are consistent with gene looping as an explanation for the long distance control of transcription by RNA; 5) demonstrate that agRNAs can counteract or supplement the effects of physiologic stimuli; 6) suggest that endogenous miRNAs complementary to sequences beyond the 3’ termini of genes can regulate gene transcription; and 7) uncover additional layers of regulation and gene structure at the PR locus. agRNAs that target the 5’ promoter region have similar effects on gene expression and involve the essential RNA binding protein AGO, suggesting that the mechanisms of 3’ agRNAs and 5’ agRNAs are related.
Mechanism of 3’ agRNAs
Overexpression of the 3’ noncoding RNA does not affect expression of PR (Supplementary Fig. S16), consistent with the suggestion that the 3’ noncoding RNA species becomes involved in the complex prior to dissociation from chromosomal DNA. Proximity of the newly synthesized RNA to the proteins controlling gene transcription would simplify the challenge of initiating gene specific inhibition of transcription by increasing the effective concentration of the RNA relative to genomic DNA. Once noncoding RNA has left the target chromosomal locus it would face greater obstacles returning and forming sequence-selective interactions upon addition of complementary agRNA.
RNA immunoprecipitation data suggest that noncoding RNAs are the direct molecular targets of agRNAs, not sequences within chromosomal DNA. AGO2 appears to play a critical role in the mechanism by promoting binding of the agRNA to the noncoding transcript. After being recruited to the noncoding transcript, AGO2 is available to form interactions with other proteins or disrupt existing interactions. We have previously shown that agRNAs modulate recruitment or displacement of other proteins such as HP1γ and hnRNPk, and also alter histone modifications11. A scheme showing a potential orientation of noncoding transcripts, PR genomic DNA, AGO2, and 3’ agRNA is presented in Fig. 9. While AGO2 is the best candidate for involvement and we do not observe cleavage of the target transcript, additional experiments will be needed to fully investigate involvement of AGO1, AGO3, or AGO4 and whether or not the target transcript is cleaved.
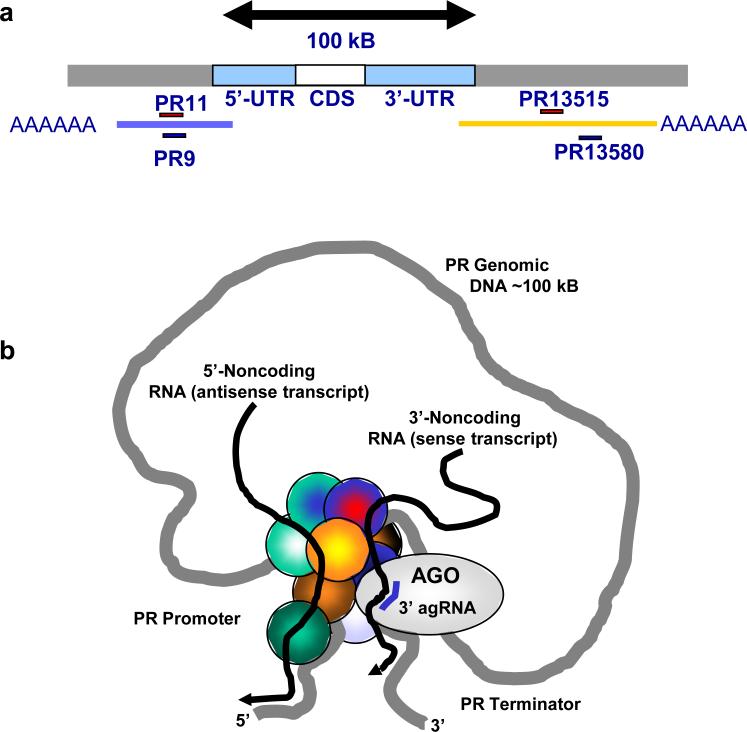
Figure 9. Model for modulation of transcription by 3’-agRNAs.
(a) 100,000 bases separates the genomic locations of the promoter and 3’ terminal regions of the PR gene. Gene looping juxtaposes the 5’ promoter and 3’ terminator, bringing DNA sequences into close proximity. Addition of the 3’ agRNA recruits AGO2 to the 3’ noncoding transcript. The arrival of AGO2 may affect other proteins (here shown as unlabeled circles) at the gene promoter and alter regulation of transcription. The proximity of 3’ and 5’ noncoding transcripts allows them to co-immunoprecipitate during RIP with anti-AGO antibodies.
How can transcription be affected by an RNA-mediated binding event across 100,000 bases (Fig. 9a)? Three lines of evidence support the conclusion that the PR gene “loops” (i.e. juxtaposes its 3’ and 5’ termini) (Fig. 9b): 1) striking similarity in the properties of 3’ and 5’ agRNAs, 2) 3C analysis demonstrating proximity of 5’ promoter and 3’ terminus regions, and 3) RNA immunoprecipitation showing association of the 3’ and 5’ noncoding transcripts. Gene looping has been observed at the X-inactivation center38. The X-inactivation center controls transcriptional inactivation of the X chromosome in females and is regulated by noncoding RNAs Xist, Tsix, and Xite. 3C analysis indicates the loci for these noncoding RNAs are positioned close enough for interactions during the inactivation process and the molecular mechanism may be related to the mechanism of agRNA-mediated gene regulation that we observe.
Similarities to Other Regulators of Transcription
While the molecular basis for the action of nuclear hormone receptors and other protein regulators is still not completely understood after many years of study, insights into how proteins control transcription provide a framework for broadly understanding the mechanism of agRNAs. Like our agRNAs, nuclear hormone receptors recognize specific sequences. For nuclear hormone receptors, ligand binding controls recognition of a coactivator or corepressor protein, while agRNAs recruit AGO2. Endogenous RNA coactivators and transcriptional activators have already been identified39-41 and it is possible that agRNAs and the agRNA/AGO2 complex may make similar interactions at gene promoters. More generally, many transcription factors have domains that contain RNA binding motifs42-45. HIV-1 Tat protein is an especially compelling example because it binds a viral transcript and interacts with other proteins to control transcription44,45.
Regulation of gene expression by 3’ agRNAs, both activation and inhibition, is robust, suggesting that the mechanism of agRNA-mediated regulation of PR expression shares components with endogenous PR regulation. Consistent with this hypothesis, silencing of CARM-1 or SRC3, proteins known to be critical for regulation of PR by estrogen46, blocked the activity of activating or inhibitory agRNAs PR-11 (Janowski, unpublished).
Both 3’ and 5’ agRNAs regulate the levels of PR transcripts. Why do we observe activation under some conditions and inhibition under others? Expression of PR is poised to respond to in vivo signals such as hormones, cytokines, and growth factors47. The primary difference between agRNA-mediated activation and repression is that activation is more easily observed in cells that express low basal levels of PR (i.e. cells poised to increase expression) while inhibition is observed in cells that express relatively high amounts of PR (i.e. cells poised to decrease expression). We note that relatively greater PR activation in cells with low basal expression is not restricted to agRNAs. 17ß-estradiol also activates PR expression much more robustly in low-PR expressing MCF7 cells than in higher-PR expressing T47D cells.
There are many examples of proteins acting as either activators or repressors depending on: 1) context48,49, 2) binding to accessory proteins50-54, or 3) antagonist/agonist binding55. Noncoding RNAs can also either activate or repress transcription through interactions with proteins at promoters possibly through regulating transcriptional coactivator and corepressor complexes56.
The transcriptional control machinery can adapt to different stimuli to produce different effects, and this versatility probably underlies the observed mechanism of agRNA action. Conversely, the ability of 3’ agRNAs to modulate gene expression over seemingly large genomic distances emphasizes the remarkable diversity of macromolecules and mechanisms affecting transcription and the power of RNA as a genetic regulator.
METHODS
Cell culture
T47D, MCF7, and MDA-MB-231 cells were cultured at 5 % CO2 in RPMI media (Sigma) supplemented with 10 % FBS (Atlanta Biologicals), 10 mM HEPES, 0.5 % NEAA, 10 μg/mL insulin, and 1 mM sodium pyruvate (Sigma). For some experiments, T47D or MCF7 cells were cultured in media containing 5 % dextran/charcoal-stripped FBS (Atlanta Biologicals) three days prior to treatment with either 17ß-estradiol (E2) (Steraloids), epidermal growth factor (EGF) (Sigma), or interleukin-1ß (Sigma).
Cellular delivery of duplex RNAs and expression assays
We used RNAi-max (Invitrogen) to deliver duplex RNAs (Supplementary Table S1) into cells. Primary antibodies (Ab) included PR-Ab (6A1, Cell Signaling) and BRCA1-Ab (MS110, Calbiochem). β-actin-Ab (Sigma) was used as an internal control and for quantitation. Protein was visualized using HRP-tagged secondary anti-mouse antibody (Jackson Immunolabs) and Super Signal developing solution (Pierce). Primers used to evaluate RNA levels of treated cells are shown in Supplementary Table S8.
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP)
ChIP was performed using monoclonal anti-RNAP2 antibody (Millipore 05-623) or mouse IgG negative control antibody (Millipore 12-371). Primers used for ChIP are shown in Supplementary Table S7. RIP was performed using the general anti-AGO antibody provided by Z. Mourelatos, University of Pennsylvania, anti-AGO1 antibody (Millipore, 07-599) or anti-AGO2 antibody (Millipore 07-590). Samples were treated with DNase I, reverse transcribed, and amplified using primers complementary to 5’ or 3’ noncoding transcripts (Supplementary Table S9).
Rapid amplification of cDNA ends (RACE)
5’-RACE and 3’-RACE were performed according to the manufacturers protocol using the GeneRacer kit (Invitrogen). This kit includes enzymatic treatments that select for full length RNA with intact 5’ caps rather than truncated products. For 5’-RACE, RNA was treated with phosphatase prior to removal of the cap to prevent cloning of truncated transcripts. For 3’-RACE, cDNA was made using oligo dT primers to allow cloning of the polyadenylated 3’ ends. Multiple primer sets (Supplementary Table S4) were used to maximize detection of transcripts and reduce the likelihood of bias from any one primer set. We used the Platinum Taq High Fidelity kit (Invitrogen) to produce product for cloning. We sequenced multiple clones from at least two independent experiments to confirm results. Both 3’ and 5’ determinations used techniques optimized for identification of full length transcripts rather than truncated products.
Chromatin conformation capture (3C)
20 million cells were grown and crosslinked in 1% formaldehyde. Cells were recovered by scraping (5 μg genomic DNA). Nuclei were purified using hypotonic lysis and distributed into 1 million nuclei aliquots. Aliquots were stored at −80°C. An additional 10 million nuclei were recovered without the use of formaldehyde for a no crosslink control.
Aliquots were removed from −80°C storage and resuspended in 500 μL of 1x restriction buffer (RB) and 3% SDS and incubated for 1.5 hr at 37°C with shaking at 1000 rpm to loosen chromatin. Then Triton-X was added to 1.8% and samples were incubated 1 hr more at 37°C with shaking to sequester SDS. 300 units of restriction enzyme DpnII were added and incubated overnight at 37°C with shaking. Next day, SDS was added to 1.6% and samples were incubated at 65°C for 30 min to inactivate restriction enzyme. 150 μL of sample was saved to check restriction enzyme efficiency.
Samples were diluted to 2 mL volume with 1.2x final concentration of ligase buffer and 1% final concentration of Triton-X. Samples were incubated at 37°C for 1 hr. Samples were placed on ice and either 40 units of T4 ligase were added and incubated overnight at 16°C. The next day samples were incubated for 30 min at room temp. Crosslinks were reversed by adding NaCl to 200 mM and incubating at 65°C for 2 hrs with proteinase K. Samples were then incubated with RNase A for 45 minutes at 41°C. Finally DNA was purified by phenol:chloroform extraction and ethanol:sodium acetate precipitation. DNA was resuspended in 50 μL of nuclease free water and 1 μL was used in each PCR reaction.
Primers were designed to span several DpnII cut sites at the 5’ end, 3’ end, and internal sites (Supplementary Table 10). Positive control templates for primers were synthesized as single strand DNA oligonucleotides (Sigma). Product was cloned and sequenced to ensure product was specific. Sequenced products aligned with their respective sites in the genome with the GATC consensus sequence for DpnII between them.
Computational methods
Methods for detecting potential matches between miRNAs and 3’ gene regions were adapted from our previously described protocol (Younger). RNA sequences were obtained from miRBase (Release 13.0) and gene sequences were obtained from Genome Browser (Mar. 2006 genome assembly). Sequences overlapping gene termini were defined as the 1000 bases downstream of the annotated 3’ end of the gene.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIGMS GM77253 to DRC, GM85080 to BAJ, and NIBIB EB 05556 to JCS) and the Robert A. Welch Foundation (I-1244 to DRC). BAJ was also supported by the High Impact/High Risk Grants Program, UT Southwestern Medical Center, and an Institutional Research Grant from the American Cancer Society (IRG-02-196-04). We gratefully acknowledge Dr. Zissimos Mourelatos (University of Pennsylvania) for providing anti-argonaute antibody. We thank Amritha Bhat for technical assistance, Dr. Alex Pertsemlidis for helpful discussions, and Dr. Jonathan Watts for comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTION
All authors designed experiments and analyzed data. All authors except DRC performed experiments. DRC wrote the manuscript. The authors recognize that the contributions of JCS, YC, and STY would have been sufficient to merit separate publications.
Supplementary Figures, Tables, and a complete descriptions of methods is available on the Nature Chemical Biology Website.
REFERENCES
- 1.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 3.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Gen. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski BA, et al. Inhibiting gene expression at transcription start sites in chromosomal DNA by antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 5.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Janowski BA, et al. Involvement of Ago1 and Ago2 link mammalian transcriptional silencing. Nat. Struc. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struc. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. USA. 2007;2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz JC, et al. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucl. Acids Res. 2005;33:5533–5543. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingeras TR. Origin of phenotypes: Genes and Transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 16.Wahlstedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 18.Chen J-M, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′ UTR variants. Hum. Genet. 2006;120:301–333. doi: 10.1007/s00439-006-0218-x. [DOI] [PubMed] [Google Scholar]
- 19.Kastner P, et al. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–14. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birmingham A, et al. 3′-UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Meth. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 21.Hornung V, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 22.Barski A, et al. High resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Keen JC, et al. Protein phosphatase 2A regulates estrogen receptor α (ER) expression through modulation of ER mRNA stability. J. Biol. Chem. 2005;280:29519–29524. doi: 10.1074/jbc.M505317200. [DOI] [PubMed] [Google Scholar]
- 24.Nardulli AM, Greene GL, O'Malley BW, Katzenellenbogen BS. Regulation of progesterone receptor messenger ribonucleic acid and protein levels in MCF-7 cells by estradiol: Analysis of estrogen's effect on progesterone receptor synthesis and degradation. Endocrinology. 1988;122:935–944. doi: 10.1210/endo-122-3-935. [DOI] [PubMed] [Google Scholar]
- 25.Cho H, Aronica SM, Katzenellenbogen BS. Regulation of progesterone receptor gene expression in MCF-7 breast cancer cells: A comparison of the effects of cyclic adenosine 3′,5′-monophosphate, estradiol, insulin-like growth factor-1, and serum factors. Endocrinology. 1994;134:658–664. doi: 10.1210/endo.134.2.7507831. [DOI] [PubMed] [Google Scholar]
- 26.Stoica A, Saceda M, Doraiswamy VL, Coleman C, Martin MB. Regulation of estrogen receptor-α gene expression by epidermal growth factor. J. Endocrinology. 2000;165:371–378. doi: 10.1677/joe.0.1650371. [DOI] [PubMed] [Google Scholar]
- 27.Zaragoza DB, Wilson RR, Mitchell BF, Olson DM. The interleukin 1beta-induced expression of human prostaglandin F2α receptor messenger RNA in human myometrial-derived ULTR cells requires the transcription factor, NFkappaB. Biol. Reprod. 2006;75:697–704. doi: 10.1095/biolreprod.106.053439. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–64. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 29.Nelson PT, et al. A novel monoclonal antibody against human argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Sullivan JM, et al. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 31.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiwari VK, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLOS Biology. 2008;12:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith TM, et al. Complete genomic sequence and analysis of 117kb of human DNA containing the Gene BRCA1. Genome Res. 2009;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 34.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol. Endocinol. 2006;20:14–34. doi: 10.1210/me.2004-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg. Med. Chem. Lett. 2009;19:3791–3794. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNAs targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 38.Tsai C-L, Rowntree RK, Cohen DE, Lee JT. Higher order chromatin structure at the X-inactivation center via looping DNA. Dev. Biol. 2008;319:416–425. doi: 10.1016/j.ydbio.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 40.Colley SM, Leedman PJ. SRA and its binding partners: an expanding role for RNA-binding co-regulators in nuclear receptor-mediated gene regulation. Crit. Reviews Biochem. Mol. Biol. 2009;44:25–33. doi: 10.1080/10409230802661719. [DOI] [PubMed] [Google Scholar]
- 41.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 42.Cassiday LA, Maher LJ. Having it both ways:transcription factors that bind RNA and DNA. Nucl. Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurokawa R, Rosenfeld MG, Glass CK. Transcriptional regulation through noncoding RNAs and epigenetic modifications. RNA Biol. 2009;6:233–236. doi: 10.4161/rna.6.3.8329. [DOI] [PubMed] [Google Scholar]
- 44.D'Orso I, Frankel AD. Tat acetylation modulates assembly of a viral host RNA-protein transcription complex. Proc. Natl. Acad. Sci. USA. 2009;106:3101–3106. doi: 10.1073/pnas.0900012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southgate C, Zapp ML, Green MR. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- 46.Naeem H, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM-1 dependent methylation. Mol. Cell. Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange CA, Gioeli D, Hammes SR, Marker PC. Integration of rapid signaling events with steroid hormone receptors action in breast and prostate cancer. Annu. Rev. Physiol. 2007;69:171–199. doi: 10.1146/annurev.physiol.69.031905.160319. [DOI] [PubMed] [Google Scholar]
- 48.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–984. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 49.Chahrour M, et al. MeCP2, a key contributor to neurologic disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scsucova S, et al. The repressor DREAM acts as a transcriptional activator on Vitamin D and retinoic acid response elements. Nucl. Acids. Res. 2005;33:2269–2279. doi: 10.1093/nar/gki503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubnicoff T, et al. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glass CK, Lipkin SM, Devary OV, Rosenfeld MG. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. Cell. 1989;59:697–708. doi: 10.1016/0092-8674(89)90016-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 54.Herschberger PA, et al. Regulation of endogenous gene expression in human non-small cell lung cancer cells by estrogen receptor ligands. Cancer Res. 2005;65:1598–1605. doi: 10.1158/0008-5472.CAN-04-2694. [DOI] [PubMed] [Google Scholar]
- 55.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodrich JA, Kugel JF. From Bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit. Rev. Biochem. Mol. Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.