Abstract
We evaluated a sequential elution protocol from immobilized metal affinity chromatography (SIMAC) employing gallium-based immobilized metal affinity chromatography (IMAC) in conjunction with titanium-dioxide-based metal oxide affinity chromatography (MOAC). The quantitative performance of this SIMAC enrichment approach, assessed in terms of repeatability, dynamic range, and linearity, was evaluated using a mixture composed of tryptic peptides from caseins, bovine serum albumin, and phosphopeptide standards. While our data demonstrate the overall consistent performance of the SIMAC approach under various loading conditions, the results also revealed that the method had limited repeatability and linearity for most phosphopeptides tested, and different phosphopeptides were found to have different linear ranges. These data suggest that, unless additional strategies are used, SIMAC should be regarded as a semi-quantitative method when used in large-scale phosphoproteomics studies in complex backgrounds.
Reversible phosphorylation of serine (Ser), threonine (Thr), and tyrosine (Tyr) residues is critical for the regulation of many biological processes and is a highly dynamic aspect of the proteome. In recent years, mass spectrometry (MS) based phosphoproteomics has emerged as a useful tool to survey the phosphorylation state of a complex protein mixture in a large-scale and high-throughput fashion. However, given the fact that most phosphoproteins are in low abundance with phosphorylation in low stoichiometry, enrichment technique(s) before MS analysis become a necessary step to analyze phosphopeptides from a complex background such as a total cell lysate. Immobilized metal affinity chromatography (IMAC) based on ferric ions has long been used to capture phosphopeptides non-specifically [1]. Over the years, new IMAC chemistries based on various multivalent metal cations, including gallium [2], zirconium [3] and titanium [4], have been introduced with varying selectivity and efficiency. Much attention has also been drawn to the use of metal oxide affinity chromatography (MOAC) for phosphopeptide enrichment due to its reported higher recovery rate and selectivity compared to IMAC [5-14]. Numerous MOAC protocols based on different multivalent metal oxides such as titanium dioxide (TiO2) [15], zirconium dioxide (ZrO2) [16] and aluminum oxide (Al2O3) [17] have been widely adopted. Interestingly, it was reported recently that IMAC is less efficient for enrichment of mono-phosphorylated peptides than for multiply-phosphorylated species [18-20]. In contrast, MOAC was shown to be more efficient for capturing mono-phosphorylated peptides [21]. This is probably due to the fact that mono-phosphorylated peptides have poor retention on IMAC material while MOAC provides interactions that are strong enough to capture mono-phosphorylation but make it difficult to elute multiple-phosphorylated peptides. Recognizing this phenomenon, Thingholm et al. introduced a novel sequential elution protocol from IMAC (SIMAC) using MOAC as the secondary enrichment step to capture mono-phosphorylated peptides that were not retained by IMAC enrichment [21; 22]. The application of this SIMAC protocol on whole cell lysate from human mesenchymal stem cells provided more phosphopeptide identifications than using MOAC or IMAC alone [21]. Since then, this sequential combination of IMAC and MOAC enrichment has gained popularity in various large-scale phosphoproteomics studies [23-25].
Despite the fact that IMAC- or MOAC-based protocols have been used in large-scale phosphoproteomics studies in recent years, questions about the reliability of these methods remain. In particular, there has been very limited knowledge on whether metal-based affinity enrichment techniques can be used in quantitative phosphoproteomics scenarios. Attention has usually been given to test the selectivity and sensitivity of the enrichment methods but not of quantitative performances such as the repeatability, dynamic range, and linearity. In a typical large-scale phosphoproteomics study, a liquid chromatographic separation step (e.g., SCX, HILIC or ERLIC) is performed as a peptide fractionation procedure to reduce the sample complexity prior to the isolation of phosphopeptides from each fraction using metal-based affinity chromatography. However, each fraction usually contains peptide subsets with different total peptide amounts and complexity. Such dynamic sample characteristics have made the estimation of the quantitative performance of metal-based affinity chromatography even more difficult in a real large-scale phosphoproteomics application.
In this study, we evaluated the repeatability, dynamic range, and linearity of metal-based affinity chromatography for quantitative phosphoproteomics applications. The testing protocol was modified from the SIMAC procedure in which IMAC and MOAC were performed sequentially as described by Thingholm et al. [21]. In the first step, a gallium-based IMAC method was selected because gallium has been shown to have higher selectivity and sensitivity than other metal-based IMAC methods [26]. The subsequent MOAC procedure was based on the most widely used TiO2-MOAC protocol as described by Jensen and Larsen. [18]. Glycolic acid was used to prevent non-specific binding of non-phosphorylated peptides with acidic amino acid residues. Jensen and Larsen have shown glycolic acid to be an effective alternative to 2,5-dihydroxybenzoic acid (DHB) as an acidic quenching agent [18]. Two experiments were performed. First, to test the enrichment repeatability of SIMAC from varying backgrounds, we constructed a series of peptide mixtures with a variety of loading conditions and complexity to mimic sample characteristics of peptide mixture as the result of LC pre-fractionation. In the second experiment, a complex background was spiked with a series of phosphopeptide standard mixtures of different concentrations to estimate the linearity and dynamic range of the SIMAC method.
Materials and Methods
Materials
HPLC-grade acetonitrile (ACN), water and acetic acid were obtained from Thermo Fisher (Waltham, MA, USA). Urea, dithiothreitol (DTT), iodoacetamide (IAA), sodium dodecyl sulfate (SDS), and 2DE Ready Prep clean up kits were purchased from Bio-Rad (Hercules, CA, USA). Bovine serum albumin (BSA), α-casein and β-casein, ammonium bicarbonate (ABC), formic acid (FA), trifluoroacetic acid (TFA), glycolic acid, and ammonium hydroxide were purchased from Sigma (St. Louis, MO, USA). Sep-Pak SPE columns with 200 mg C18 resin were obtained from Waters (Milford, MA, USA). Spin columns with filters (Cat# M105010S) were purchased from Boca Scientific (Boca Raton, FL, USA). Ga(II)-IMAC Nutip (part No.TT2GAA) from Glygen (Columbia, MD, USA) and Titansphere TiO2 beads from GL Science (Tokyo, Japan) were used as enrichment media in all SIMAC experiments. Trypsin was purchased from Promega (Fitchburg, WI, USA). One phosphopeptide standard mixture (P33357) was purchased from Invitrogen (Carlsbad, CA, USA), and another phosphopeptide standard mixture (PHOSPHOSTD01) was from Glygen. Table 1 provides a detailed description of these peptide standards.
Table 1.
List of phosphopeptide standards used in this study. The actual m/z species used in LC-MS label free quantitation are shown in bold.
| Name | Sequence | Number of Phosphorylations | (M+H)1+ | (M+2H)2+ | (M+3H)3+ |
|---|---|---|---|---|---|
| P33357 mixture standard (Invitrogen) | |||||
| NP1 | 4 DRVYIHPF | 0 | 1046.54 | 523.77 | 349.52 |
| NP2 | 5 DRVYIHPFHL | 0 | 1296.69 | 648.85 | 432.90 |
| NP3 | 6 GKGRGLSLSRFSWGA | 0 | 1578.85 | 789.93 | 526.95 |
| P1 | 7 DHTGFL[pT]E[pY]VATR | 2 | 1669.67 | 835.34 | 557.23 |
| P2 | 8 TRDI[pY]ETDYYRK | 1 | 1702.75 | 851.87 | 568.25 |
| P3 | 9 VPIPGRFDRRV[pT]VE | 1 | 1720.89 | 860.95 | 574.29 |
| P4 | 10 DLDVPIPGRFDRRV[pS]VAAE | 1 | 2192.09 | 1096.53 | 731.37 |
| PHOSPHOSTD01 standard (Glygen) | |||||
| P5 | 11 WWGSGPSGSGG[pS]GGGK | 1 | 1500.60 | 750.80 | 500.87 |
| P6 | 12 WWGSGPSG[pS]GG[pS]GGGK | 2 | 1580.58 | 790.78 | 527.53 |
| P7 | 13 WWGSGP[pS]G[pS]GG[pS]GGGK | 3 | 1660.53 | 830.77 | 554.18 |
| P8 | 14 WWG[pS]GP[pS]G[pS]GG[pS]GGGK | 4 | 1740.45 | 870.75 | 580.82 |
Protein preparation and digestion
The protein mixtures in each experiment were dissolved in 25 mM ABC with 0.1% SDS, then directly reduced by 10 mM DTT for 1 h and alkylated by 40 mM IAA for 30 min in the dark. Alkylation was quenched by adding DTT to the final concentration of 20 mM. Protein was then precipitated using 2DE Ready Prep cleanup kits according to the manufacturer’s protocol. The resulting protein pellet was reconstituted in 25 mM ABC and digested with trypsin at a 30:1 protein:protease ratio. Digestion was carried out at 37 °C for 5 h and stopped by acidification using TFA. Tryptic peptides were purified using a 200 mg C18 Sep-Pak SPE column and dried with a Speed Vac (Thermo Electron).
Experiment 1
The goal of this experiment was to estimate the repeatability of the SIMAC procedure with different loading backgrounds. BSA was chosen to create a non-phosphopeptide background as it is rich in acidic amino acids such as Asp and Glu that compete with phosphopeptides during SIMAC enrichment. For practical purposes, three different loading amounts (100, 200, and 500 μg) were tested. These values were selected because most large-scale phosphoproteomics studies start with 1-5 mg of total lysate and most LC-prefractionation procedures generate 10-20 fractions. In each loading test, two different levels of sample complexity were created by mixing tryptic peptides from at caseins:BSA ratios of 1:49 or 1:99 w/w, corresponding to the low stoichiometry of phosphoproteins typically seen in real samples such as cell lysate. In all experiments, caseins consist of equal amount of α and β isoforms. All six tests were repeated in triplicate.
Experiment 2
To test the linearity and dynamic range of phosphopeptides enrichment using SIMAC, a series of phosphopeptide mixtures were spiked into four samples, each containing 200 μg of tryptic peptides from the 1:49 caseins:BSA matrix. The amounts of spiked phosphopeptides are summarized in Table 2. All phosphopeptides were spiked prior to IMAC enrichment.
Table 2.
Composition of spiked peptide mixtures used in Experiment 2 to test the linearity and dynamic range of the SIMAC enrichment protocol. Peptides are listed in Table 1. These tests used 200 μg of casein:BSA tryptic peptides (1:49) spiked with phosphopeptide standards as shown. Phosphopeptide mixture p33357 contains an equimolar mixture of P1-P4. The mixtures were used in a randomized order in the spiking tests.
| Peptide mixture composition (pmol) | |||||
|---|---|---|---|---|---|
| Sample | P33357 | P5 | P6 | P7 | P8 |
| #1 | 10 | 5 | 50 | 1 | 500 |
| #2 | 80 | 10 | 500 | 50 | 100 |
| #3 | 20 | 1 | 10 | 5 | 50 |
| #4 | 40 | 50 | 100 | 10 | 10 |
Phosphopeptide enrichment by SIMAC
In all tests, the sequential elution protocol was carried out by employing the Ga(II)-IMAC Nutip as the first stage of enrichment and then using Titansphere TiO2 beads as the second enrichment step to further enrich phosphorylated peptides from the flow-through of IMAC as summarized in Figure 1. Solution components in each step during SIMAC enrichment can be found in Supplementary Materials I. During the IMAC enrichment, the Nutips were first equilibrated twice with 150 μL IMAC Binding Solution and then loaded with peptide mixture in 150 μL IMAC Binding Solution. Eluate was collected and reloaded again for complete binding. Two steps of washing, each with 150 μL IMAC Washing Solution 1 and 2, were performed followed by a 100 μL water wash to remove acid. Phosphopeptides were then eluted by 100 μL Elution Solution 1 and 2 sequentially. Flow-through fractions from IMAC loading and all washing steps were combined and dried by Speed Vac for MOAC enrichment. For the MOAC enrichment, 2 mg of TiO2 beads were aliquoted into a spin column filter, equilibrated with 300 μL MOAC Binding Solution twice, and then loaded with dried IMAC flow-through in 300 μL of MOAC Binding Solution. Eluate was collected and reloaded again to achieve a greater extent of binding. Washing steps with 300 μL MOAC Washing Solution 1 and 200 μL MOAC Washing Solution 2 were performed followed by 100 μL water wash to remove acid. Phosphopeptides were then eluted with 100 μL of Elution Solution 1 and 2 sequentially. Enrichment fractions from both IMAC and MOAC were then combined, dried and re-dissolved in 10 μL ACN:water (3:97) with 0.1% formic acid for LC-MS analysis.
Figure 1.
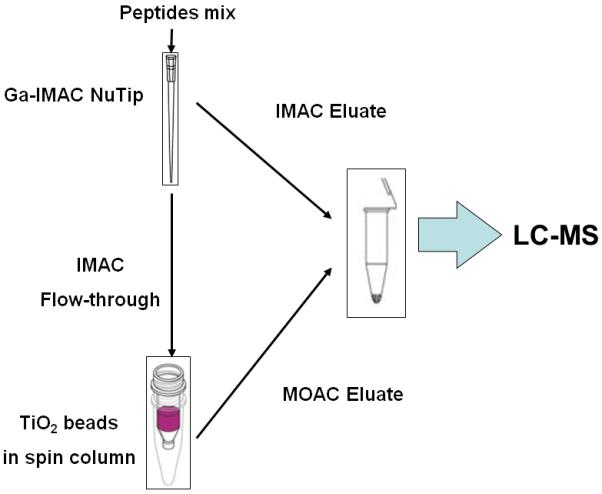
Schematic diagram of sequential IMAC workflow for phosphopeptides enrichment using Ga-IMAC and TiO2-MOAC. Eluates from both enrichment steps were combined prior to LC-MS analysis.
Casein peptide identification by LC-MS/MS
The tryptic peptides mixture from α-casein and β-casein were injected in triplicate onto a G4240-62001 C-18 HPLC-Chip (40 nL enrichment column, 75 μm × 43 mm analytical column, 5 μm C-18SB-ZX, 300 Å, Agilent) hyphenated with a 6150 hybrid ESI-Q-TOF (Agilent). Nano-LC was performed with a 30-min gradient of 16–90% ACN in 0.1% FA at 0.5 μL/min. Mass spectrometric analysis was performed under positive mode with nanoelectrospray generated at 2.1 kV. The m/z response of the instrument was calibrated regularly with standards from manufacturer. The precursor scan and five subsequent product ion spectra were acquired in centroid mode using data-dependent acquisition in MassHunter (Agilent) with mass ranges for MS and MS/MS at m/z 250–2400 and 59–3000, respectively. The switching from TOF-MS to MS/MS is triggered by precursors with ion intensity >1000 counts with dynamic exclusion for 30 sec. The collision energy was set with 5 V/100 Da slope offset with 2.5 V.
Spectra were searched by Spectrum Mill 3.3.084 (Agilent) on local server at Colorado State University against a SwissProt protein database under the taxonomy of Bos taurus. The search parameters were set to allow for up to two missed cleavages, carboxyamidomethylation on Cys as fixed modification, oxidation on Met, phosphorylation on Ser/Thr/Tyr as dynamic modification and 100 ppm for both MS and MS/MS mass tolerance with decoy search mode. Autovalidation was performed using default setting within the Spectrum Mill to qualify confident identifications.
LC-MS quantitation of phosphopeptides
For LC-MS analysis, 2 μL of samples of the tryptic peptides mixture from both experiments were injected in triplicate into the Q-TOF instrument. The LC setting and LC-gradient were the same as described in the previous section except that the Q-TOF was operated in the MS scan-only mode. Data were collected in centroid mode with MS m/z ranges set to 250–2400. Raw LC-MS data were then convert to mzData format using MassHunter (Agilent) for further analysis.
Data analysis
All mzData files from both Experiment 1 and 2 were loaded into MZmine 2 [27] to quantify phosphopeptide abundance based on the LC-MS peak area. Briefly, compounds from each LC-MS run were recognized by their unique m/z and retention time values, isotopic peaks of each compound were then grouped to give reliable quantitation. Chromatograms of each compound were then aligned across samples/injections for comparison. Phosphopeptide species were searched using unique m/z values, and their peak areas from each sample/injection were exported to Excel for further analysis. The detailed procedure can be found in Supplemental Materials. We note that some casein phosphopeptides, such as CP3-1, CP3-2, CP5-1, CP5-3, and CP5-4, contain coeluting isoforms that are indistinguishable in the LC-MS analysis and thus were treated as one identity for quantitation.
For Experiment 1, the repeatability of SIMAC in each loading condition was evaluated based on each enriched casein phosphopeptide across the SIMAC repeats with peptide mixtures at different starting amounts and mixing ratios. For Experiment 2, the signal linearity of each spiked phosphopeptides was analyzed individually. For all phosphopeptides, the relative abundance was generated based on the peak area normalized by the total ion chromatogram in each LC-MS run.
Results
Phosphopeptides identification
As expected, most qualified peptide IDs using LC-MS/MS were from BSA or the caseins. Non-phosphorylated peptides from both BSA and caseins were observed in relatively high abundance in all experiments (data not shown), indicating that non-specific binding is still an issue with the SIMAC procedure. Phosphopeptides from caseins with confident identification (see Supplemental Materials II) were used for the LC-MS quantitative analysis in Experiment 1. Some phosphopeptide isoforms such as DIGSE[pS][pT]EDQAMEDIK and DIG[pS]ES[pT]EDQAMEDIK, which have the identical molecular weight and nearly the same retention times owing to their very similar sequences, were grouped into one m/z species for LC-MS quantitation. Phosphopeptides containing oxidized Met or additional missed cleavage sites were considered as separate m/z species for LC-MS quantitation. As a result, five, three, and one phosphorylation sites from casein αSI, αSII and β were identified from which 13 different m/z species were generated. All identified casein phosphopeptides are summarized in Table 3.
Table 3.
List of phosphopeptides identified from the casein digest. The actual m/z species used in LC-MS label free quantitation are shown in bold.
| Name | Sequence | Number of Phosphoryl- ations |
Casein | Retention time (min) |
(M+H)1+ | (M+2H)2+ | (M+3H)3+ |
|---|---|---|---|---|---|---|---|
| CP1 | (K)VNEL[pS]K(D) | 1 | αS1 | 3.4 | 769.35 | 385.18 | 257.12 |
| CP2-1 | (K)VPQLEIVPN[pS]AEER(L) | 1 | 6.9 | 1660.79 | 830.90 | 554.27 | |
| CP2-2 | (K)YKVPQLEIVPN[pS]AEER(L) | 1 | 7.4 | 1951.95 | 976.48 | 651.32 | |
| CP3-1 | (K)DIG[pS]ESTEDQAMEDIK(Q) (K)DIGSE[pS]TEDQAMEDIK(Q) (K)DIGSES[pT]EDQAMEDIK(Q) |
1 | 5.1 | 1847.71 | 924.37 | 616.58 | |
| CP3-2 | (K)DIG[pS]E[pS]TEDQAMEDIK(Q) (K)DIG[pS]ES[pT]EDQAMEDIK(Q) (K)DIGSE[pS][pT]EDQAMEDIK(Q) |
2 | 6.3 | 1927.69 | 964.35 | 643.24 | |
| CP4 | (K)NMAINP[pS]KENLCSTFCK(E) | 1 | αS2 | 6.5 | 2093.88 | 1047.45 | 698.64 |
| CP5-1 | (K)TVDME[pS]TEVFTK(K) (K)TVDMES[pT]EVFTK(K) |
1 | 5.9 | 1466.61 | 733.81 | 489.54 | |
| CP5-2 | (K)TVD[oxiM]ES[pT]EVFTK(K) | 1 | 4.5 | 1482.61 | 741.81 | 494.88 | |
| CP5-3 | (K)TVDME[pS]TEVFTKK(T) (K)KTVDME[pS]TEVFTK(T) |
1 | 5.1 | 1594.70 | 797.85 | 532.24 | |
| CP5-4 | (K)TVD[oxiM]E[pS]TEVFTKK(T) (K)KTVD[oxiM]E[pS]TEVFTK(T) |
1 | 3.5 | 1610.69 | 805.85 | 537.57 | |
| CP5-5 | (K)KTVDME[pS]TEVFTKK(T) | 1 | 10.5 | 1722.81 | 861.91 | 574.94 | |
| CP6-1 | (K)FQ[pS]EEQQQTEDELQDK(I) | 1 | β | 4.3 | 2061.83 | 1031.42 | 687.95 |
| CP6-2 | (K)IEKFQ[pS]EEQQQTEDELQDK(I) | 1 | 5.9 | 2432.05 | 1216.53 | 811.35 |
SIMAC repeatability
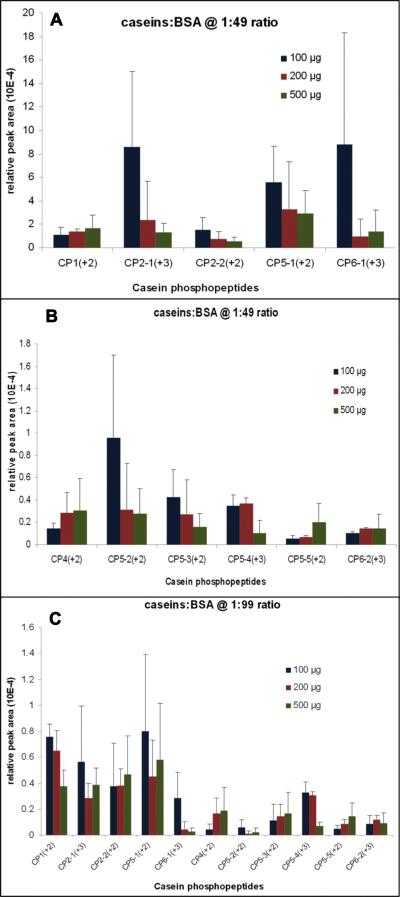
In our hands, for most casein phosphopeptides that were quantified, the SIMAC procedure demonstrated overall limited repeatability for quantitative purposes (Figure 2). The biggest variance came from the difference between individual phosphopeptides. For example, phosphopeptide ions such as CP12+, CP2-13+, CP5-12+, CP6-13+ had significantly higher MS responses than other phosphopeptide ions. Moreover, CP3-1 and CP3-2 were not detectable in most LC-MS runs and were not included in the quantitation analysis. Significant signal variances for most casein phosphopeptides were observed regardless of the changes in loading amount and sample complexity. The median coefficient of variation (CV) of the normalized MS signals from triplicate SIMAC procedures across all peptides under different loading conditions is 81.5%. In contrast, the median CV of the normalized MS signals of triplicate LC-MS injections across all peptides under different loading conditions is 9.7%. Therefore, we conclude that the majority of the MS signal variance was introduced by SIMAC enrichment, and that this SIMAC protocol may generate unreliable phosphopeptide quantitation. Poor correlation between the amount of loading and the mean relative MS signal was observed for most casein phosphopeptides been investigated in Experiment 1. Only a few phosphopeptides such as CP12+, CP42+, CP5-52+ and CP6-23+ in the 1:49 casein:BSA test and CP2-22+, CP42+, CP5-32+, CP5-52+, and CP6-23+ in the 1:99 casein:BSA test showed incremental MS signal differences as expected when an increased amount of peptide mixture was enriched via SIMAC. However, due to the large CV of SIMAC replication, ANOVA tests for each of those peptides did not correlate with the significant increase of the average MS signal (p-value ranging from 0.097 to 0.79) with increasing loading amount. Interestingly, there was a negative correlation between the amount of loading and the mean relative MS signal of phosphopeptides CP2-13+, CP2-22+, CP5-12+, CP5-22+ and CP5-32+ in the 1:49 casein:BSA test and CP12+, CP5-43+, and CP6-13+ in the 1:99 casein:BSA test. With the same loading amount, the MS signals of all casein phosphopeptides investigated were higher when the SIMAC workflow was loaded with 1:49 casein:BSA than with 1:99 casein:BSA.
Figure 2.
Repeatability test of SIMAC in Experiment 1. Plot A/B and C/D shown enrichment results from the 1:49 and 1:99 casein:BSA backgrounds, respectively. Each cluster of bars represents one casein phosphopeptide been quantified using LC-MS. Peak areas were normalized against the TIC and then averaged across injection replicates. The height of each bar represents the average relative peak areas of three enrichment replicates with standard deviation. Enrichment with 100, 200, and 500 μg tryptic peptides are indicated by blue, red, and green, respectively.
SIMAC linearity and dynamic range
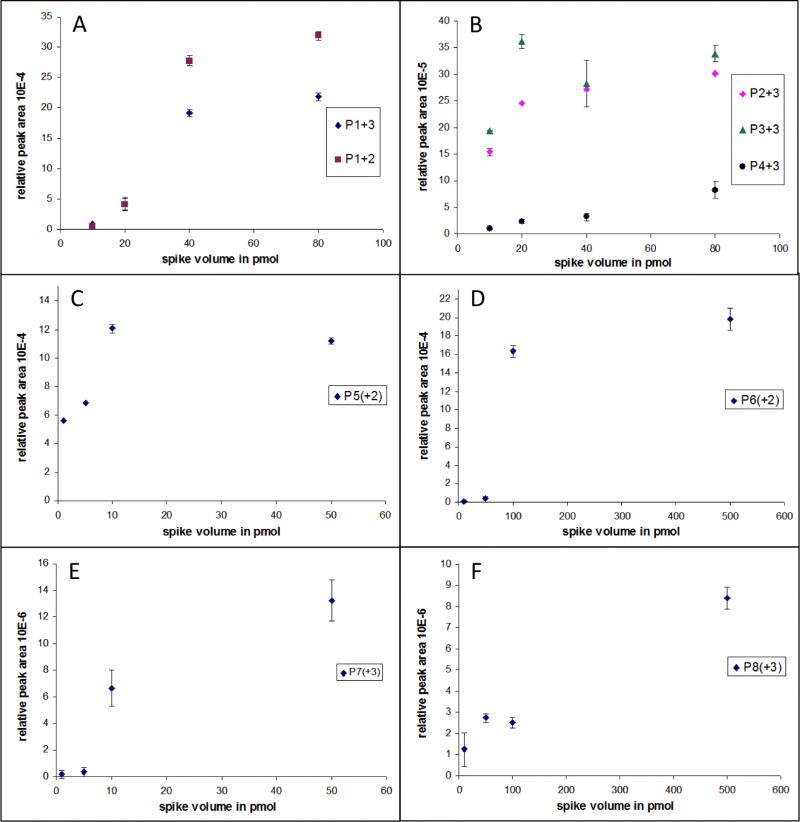
In the second experiment, the four Invitrogen phosphopeptide standards (P1-P4) were mixed and spiked at 10-80 pmol, while peptides P5 and P7 were spiked at 1-50 pmol, and peptides P6 and P8 were spiked at 1-500 pmol into four samples, respectively. Satisfactory linear response was observed from the MS signal of P43+ (Figure 3B, R2=0.975) when the spiked concentrations ranged from 10-80 pmol prior to SIMAC enrichment. However, none of the other standard phosphopeptides investigated here had a linear response over the spiked range, despite the overall trends of increasing MS signal intensity with increasing concentrations. The MS signals of P12+, P13+ (Figure 3A), P23+ (Figure 3B) and P52+ (Figure 3C) only showed linear trends in the first three concentration tiers but the signal became saturated at higher spiked concentrations. MS signals of standard P33+ (Figure 3B) leveled off after only 20 pmol of loading amount. The triply-phosphorylated peptides P73+ (Figure 3E) and P83+ (Figure 3F) had significantly lower MS responses than did other species.
Figure 3.
Results of SIMAC linearity test at different dynamic ranges. Four phosphopeptides (P1-P4) from Invitrogen P33357 were spiked from 10-80 pmol (panel A/B) and Glygen PHOSPHOSTD01 standard P5 (panel C), P7 (panel E) were spiked from 1-50 pmol and P6 (panel D), P8 (panel F) were spiked from 10-500 pmol into the 200 μg 1:49 casein:BSA backgrounds before SIMAC enrichment. Peak areas were normalized against the TIC and then averaged across injection replicates.
Discussion
Metal-based phosphopeptide enrichment techniques such as IMAC and MOAC have been successfully employed for large-scale phosphoproteomics studies. The enrichment preferences of IMAC and MOAC have been observed previously [18-20] and gave rise to separation strategies for the enrichment of multi-phosphorylated and then mono-phosphorylated peptides, in series [21]. However, most method development of metal affinity based enrichment techniques has focused primarily on the selectivity and sensitivity rather than the quantitative attributes of these methods. In a typical large-scale phosphoproteomics study, LC-based peptide fractionation generates multiple peptide mixture fractions with varying peptide amounts and complexity from which phosphopeptides are enriched.
In our first experiment, we evaluated whether dynamic loading conditions such as loading amount and sample complexity can affect the performance of metal-based affinity chromatography. Overall, the SIMAC method demonstrated limited repeatability between technical replicates, regardless of loading amount and sample complexity. Thus, it is recommended that technical replicates be included at enrichment step to prevent detection of false changes. It is still unknown if the poor repeatability was the result of competitive binding of non-phosphorylated peptides to the SIMAC materials, significant ion suppression during electrospray with a complex background, or a combination of both reasons. Nevertheless, to alleviate the adverse effects from both mechanisms, prefractionation is recommended to simplify the sample before SIMAC enrichment and MS analysis. It is also notable that for phosphopeptide species such as CP4-22+ and CP5-52+ in the repeatability test, the difference of a MS signal due to 2-fold and 5-fold increase in loading amount falls into the same magnitude as the SIMAC technical variance. In fact, ANOVA tests (results not shown) show insignificant differences in the relative MS abundances between different loading amounts for all casein phosphopeptides under investigation. In many cases, poor correlations between the amount of loading and the mean relative MS signal of casein phosphopeptides were observed. A similar concern arose in a recent investigation of the quantitative performance of a Fe(III)-IMAC based protocol that was used to enrich phosphopeptides from a whole cell lysate [28]. We also suspect that the poor correlation of MS intensity and the loading amount indicates an effect of the peptide-to-beads ratio on the enrichment performance as shown previously [29-33]. Based on our observation, we suggest that even moderate phosphorylation changes (up to 5 fold) may not be quantifiable if SIMAC enrichment is used with only a small number of technical replicates. However, it should be borne in mind that these conclusions are made based on label-free quantitation. It remains unknown whether metal affinity enrichment approaches such as SIMAC can affect the quantitation of phosphopeptides using labeling methods such as SILAC or iTRAQ. We postulate that by pooling labeled samples before the enrichment of metal-based phosphopeptides, technical variations caused by enrichment techniques will equally affect all samples within a pool, thus provide a possible solution to circumvent the problem.
The linearity test in our study also revealed that different peptide species may have very different linear relationships and linear ranges, as was suggested in the Fe(III)-IMAC study mentioned above [28]. It is unclear whether this is caused by different efficiencies of the SIMAC enrichment or by different electrospray ionization efficiencies between different phosphopeptide species. This outcome means that not all phosphopeptides can be quantified at the same time from a complex sample in a real phosphoproteomics application by LC-MS coupled with enrichment techniques. In a recent application using SILAC-labeled samples mixed in different proportions prior to digestion, phosphopeptide enrichment, and LC-MS analysis [28], Casado and Cutillas determined the linear response and quantitation accuracy of each detected phosphopeptide. This approach allowed the researchers to select phosphopeptides with acceptable linearity and accuracy for reliable quantitation analysis. Similar approaches should be employed in other label or label-free quantitation scenarios to filter out peptides without linear responses. However, it is worth noting that, even in this specific SILAC labeling experiment, as long as the mixed samples were individually enriched by metal-based techniques, the inherent variability of the enrichment step may still have impact on quantitation performance.
Although the SIMAC procedure was designed to effectively enrich both mono-phosphorylated and multiply-phosphorylated peptides [21], we observed significant signal loss of multiply-phosphorylated species such as CP3-2 from casein in Experiment 1 and P8 in Experiment 2. It is known that electrospray ionization is biased against multiply-phosphorylated peptides [7]. However, all multiply-phosphorylated peptides were detectable using Q-TOF if a casein digest or phosphopeptide standards were injected alone without a complex background (data not shown). Thus, we suspect that substantial ion suppression in electrospray was at least in part responsible for the observed signal loss. Furthermore, we hypothesize that this ion suppression is the result of non-phosphorylated peptides carried through the SIMAC workflow when the enrichment began with low phosphopeptide abundance and a highly complex background. Although glycolic acid was used to minimize non-phosphorylated peptide binding [18], non-phosphopeptide species still accounted for the majority of the total ion intensity of the chromatograms. Despite the widespread implementation of such ‘acidic peptide quenching’ agents, studies still report differing results on the optimal enrichment conditions. Recent reports have indicated that the addition of glycolic acid may hamper the selectivity [7; 34] and that DHB can introduce bias against multiply-phosphorylated peptides [7]. In addition, a recent study by Worthington et al. documented another possible source of non-specific binding in the metal affinity-based enrichment technique for phosphopeptides caused by peptides with other types of modifications [35], which may pose additional challenges for achieving quantitative results using SIMAC. Given the inherent semi-quantitative nature of metal-based enrichment methods, as illustrated in this work, researchers should consider the variance introduced by those enrichment methods in large-scale phosphoproteomics studies. Strategic workflow designs that incorporate automated on-line or microfluidic separation and enrichment, or use labeling techniques in prior to enrichment, may help to circumvent the technical drawbacks of phosphopeptide enrichment methods.
Concluding Remarks
Our data suggest that the SIMAC procedure should be considered to be a semi-quantitative method with limited reproducibility regardless of loading conditions and compromised linearity. We also show that the variability generated by the SIMAC procedure cannot be ignored, particularly without sufficient technical repeats. There is still a need for fundamental breakthroughs in IMAC/MOAC enrichment chemistry that achieve substantial improvements of enrichment efficiency and repeatability. Overall, careful evaluation and optimization of phosphopeptide enrichment techniques is recommended if they are to be used in a quantitative phosphoproteomics study.
Supplementary Material
Abbreviations
- ABC
ammonium bicarbonate
- ACN
acetonitrile
- BSA
bovine serum albumin
- DHB
dihydroxybenzoic acid
- DTT
dithiothreitol
- ERLIC
repulsion-hydrophilic interaction chromatography
- ESI-Q-TOF
electrosprary ionization-quadrupole-time-of-flight
- FA
formic acid
- HILIC
hydrophilic interaction liquid chromatography
- IAA
iodoacetamide
- IMAC
immobilized metal affinity chromatography
- MS
mass spectrometry
- MOAC
metal oxide affinity chromatography
- SCX
strong cation exchange
- SDS
sodium dodecyl sulfate
- SIMAC
sequential elution protocol from immobilized metal affinity chromatography
- SPE
solid phase extraction
- TFA
trifluoroacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Muszynska G, Dobrowolska G, Medin A, Ekman P, Porath JO. Model studies on iron(III) ion affinity-chromatography: II Interaction of immobilized iron(III) ions with phosphorylated amino-acids, peptides and proteins. J. Chromatogr. 1992;604:19–28. doi: 10.1016/0021-9673(92)85524-w. [DOI] [PubMed] [Google Scholar]
- [2].Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 1999;71:2883–92. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- [3].Feng S, Ye M, Zhou H, Jiang X, Jiang X, Zou H, Gong B. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol. Cell. Proteomics. 2007;6:1656–65. doi: 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- [4].Yu Z, Han G, Sun S, Jiang X, Chen R, Wang F, Wu R, Ye M, Zou H. Preparation of monodisperse immobilized Ti(4+) affinity chromatography microspheres for specific enrichment of phosphopeptides. Anal. Chim. Acta. 2009;636:34–41. doi: 10.1016/j.aca.2009.01.033. [DOI] [PubMed] [Google Scholar]
- [5].Sano A, Nakamura H. Titania as a chemo-affinity support for the column-switching HPLC analysis of phosphopeptides: application to the characterization of phosphorylation sites in proteins by combination with protease digestion and electrospray ionization mass spectrometry. Anal. Sci. 2004;20:861–4. doi: 10.2116/analsci.20.861. [DOI] [PubMed] [Google Scholar]
- [6].Sano A, Nakamura H. Chemo-affinity of titania for the column-switching HPLC analysis of phosphopeptides. Anal. Sci. 2004;20:565–6. doi: 10.2116/analsci.20.565. [DOI] [PubMed] [Google Scholar]
- [7].Aryal UK, Ross AR. Enrichment and analysis of phosphopeptides under different experimental conditions using titanium dioxide affinity chromatography and mass spectrometry. Rapid Commun. Mass Spectrom. 2010;24:219–31. doi: 10.1002/rcm.4377. [DOI] [PubMed] [Google Scholar]
- [8].Gates MB, Tomer KB, Deterding LJ. Comparison of metal and metal oxide media for phosphopeptide enrichment prior to mass spectrometric analyses. J. Am. Soc. Mass Spectrom. 2010;21:1649–59. doi: 10.1016/j.jasms.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leitner A, Sturm M, Hudecz O, Mazanek M, Smatt JH, Linden M, Lindner W, Mechtler K. Probing the phosphoproteome of HeLa cells using nanocast metal oxide microspheres for phosphopeptide enrichment. Anal. Chem. 2010;82:2726–33. doi: 10.1021/ac902560z. [DOI] [PubMed] [Google Scholar]
- [10].Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- [11].Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- [12].Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–86. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- [13].Wolschin F, Wienkoop S, Weckwerth W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC) Proteomics. 2005;5:4389–97. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- [14].Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–43. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- [15].Ikeguchi Y, Nakamura H. Determination of organic phosphates by column-switching high performance anion-exchange chromatography using on-line preconcentration on titania. Anal. Sci. 1997;13:479–483. [Google Scholar]
- [16].Zhou HJ, Tian RJ, Ye ML, Xu SY, Feng S, Pan CS, Jiang XG, Li X, Zou HF. Highly specific enrichment of phosphopeptides by zirconium dioxide nanoparticles for phosphoproteome analysis. Electrophoresis. 2007;28:2201–2215. doi: 10.1002/elps.200600718. [DOI] [PubMed] [Google Scholar]
- [17].Shin EW, Han JS, Jang M, Min SH, Park JK, Rowell RM. Phosphate adsorption on aluminum-impregnated mesoporous silicates: Surface structure and behavior of adsorbents. Environ. Sci. Technol. 2004;38:912–917. doi: 10.1021/es030488e. [DOI] [PubMed] [Google Scholar]
- [18].Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 2007;21:3635–45. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- [19].Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- [21].Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics. 2008;7:661–71. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- [22].Thingholm TE, Jensen ON, Larsen MR. Enrichment and separation of mono- and multiply phosphorylated peptides using sequential elution from IMAC prior to mass spectrometric analysis. Methods Mol. Biol. 2009;527:67–78. xi. doi: 10.1007/978-1-60327-834-8_6. [DOI] [PubMed] [Google Scholar]
- [23].Carrascal M, Ovelleiro D, Casas V, Gay M, Abian J. Phosphorylation analysis of primary human T lymphocytes using sequential IMAC and titanium oxide enrichment. J. Proteome Res. 2008;7:5167–76. doi: 10.1021/pr800500r. [DOI] [PubMed] [Google Scholar]
- [24].Meyer LJ, Gao J, Xu D, Thelen JJ. Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 2012;159:517–28. doi: 10.1104/pp.111.191700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pan J, Naumann-Busch B, Wang L, Specht M, Scholz M, Trompelt K, Hippler M. Protein phosphorylation is a key event of flagellar disassembly revealed by analysis of flagellar phosphoproteins during flagellar shortening in Chlamydomonas. J. Proteome Res. 2011;10:3830–9. doi: 10.1021/pr200428n. [DOI] [PubMed] [Google Scholar]
- [26].Kange R, Selditz U, Granberg M, Lindberg U, Ekstrand G, Ek B, Gustafsson M. Comparison of different IMAC techniques used for enrichment of phosphorylated peptides. J. Biomol. Tech. 2005;16:91–103. [PMC free article] [PubMed] [Google Scholar]
- [27].Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Casado P, Cutillas PR. A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003079. M110 003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–7. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- [30].Li QR, Ning ZB, Tang JS, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–81. doi: 10.1021/pr900659n. [DOI] [PubMed] [Google Scholar]
- [31].Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem. 2011;83:7635–44. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Montoya A, Beltran L, Casado P, Rodriguez-Prados JC, Cutillas PR. Characterization of a TiO(2) enrichment method for label-free quantitative phosphoproteomics. Methods. 2011;54:370–8. doi: 10.1016/j.ymeth.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal Chem. 2011;83:3758–64. doi: 10.1021/ac200213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6:1103–9. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- [35].Worthington J CP, Timms JF. IMAC/TiO(2) enrich for peptide modifications other than phosphorylation: implications for chromatographic choice and database searching in phosphoproteomics. Proteomics. 2011;11:4583–7. doi: 10.1002/pmic.201100143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.