Abstract
Definitive hematopoiesis requires the master hematopoietic transcription factor Runx1, which is a frequent target of leukemia-related chromosomal translocations. Several of the translocation-generated fusion proteins retain the DNA binding activity of Runx1, but lose subnuclear targeting and associated transactivation potential. Complete loss of these functions in vivo resembles Runx1 ablation, which causes embryonic lethality. We developed a knock-in mouse that expresses full length Runx1 with a mutation in the subnuclear targeting/co-factor interaction domain, Runx1HTY350-352AAA. Mutant mice survive to adulthood, and hematopoietic stem cell emergence appears to be unaltered. However, defects are observed in multiple differentiated hematopoietic lineages at stages where Runx1 is known to play key roles. Thus, a germline mutation in Runx1 reveals uncoupling of its functions during developmental hematopoiesis from subsequent differentiation across multiple hematopoietic lineages in the adult. These findings indicate that subnuclear targeting and co-factor interactions with Runx1 are important in many compartments throughout hematopoietic differentiation.
Keywords: Runx1, hematopoiesis, leukemia, subnuclear targeting
Introduction
Hematopoietic stem cell (HSC) emergence in the developing embryo requires the key Runt-related transcription factor Runx11, which is stringently regulated during differentiation of hematopoietic lineages2,3. Runx1 is a frequent target of point mutations and translocations in human leukemia4, and these mutations cluster in either the N-terminal DNA binding domain or the C-terminal transactivation domains5,6. Many Runx1 mutations cause lethality at embryonic day 12.5 in mouse models: defects in DNA binding ability or critical C-terminal functions result in phenocopies of the complete null1,7,8. This early lethality has made studying the precise roles of Runx1 during hematopoiesis difficult. Genetic restoration of Runx1 expression in Tie2-positive endothelial cells rescued HSC emergence but revealed a secondary genetic bottleneck of Runx1 deficiency related to perinatal respiratory failure9. Similar gene replacement experiments in which the Runx1 N-terminus was fused to the C-terminus of Runx2 or Runx3 rescued HSC emergence, but uncovered additional roles of Runx1 in myeloid and T-lymphoid development10. A conditional null developed to bypass embryonic lethality revealed important roles of Runx1 in most adult hematopoietic lineages11. However, gaps in knowledge remain about the developmental roles of Runx1 beyond stem cell emergence.
Many transcription factors, including Runx1, localize to specific foci within the nucleus and this localization is critically related to biological function12–18. Subnuclear targeting defective mutants of Runx1 have previously been shown to cause cellular differentiation defects17 and altered regulation of target genes, including miRs19. Appropriate subnuclear targeting is associated with Runx1 function as a scaffold within the nucleus, organizing regulatory machinery and mediating combinatorial control of gene expression20. Interestingly, the leukemic fusion protein Runx1-ETO, in which the Runx1 C-terminus is replaced with most of the ETO protein, exhibits aberrant subnuclear targeting12,15,21. In previous studies, we characterized a knock-in mouse with a Runx1 C-terminal truncation that removed domains responsible for subnuclear targeting and co-factor interactions7. This mutation resulted in embryonic lethality at day 12.5, a complete phenocopy of models without Runx1 DNA binding ability, and thus demonstrated that critical biological functions are associated with the Runx1 C-terminus.
To address the precise biological contributions of specific domains of Runx1, we developed a knock-in mouse model, Runx1HTY350-352AAA. This mouse model encodes a triple point mutation in the C-terminal domain required for subnuclear localization22 and interaction of Runx with biologically important co-factors. A key signaling pathway that is perturbed by the mutation is the TGFβ/BMP signaling pathway. We have shown previously that Runx1HTY350-352AAA disrupts interaction with Smads, effectors of TGFβ/BMP signaling13,14,16,22–26. Mice homozygous for Runx1HTY350-352AAA bypass embryonic lethality. However, this point mutation influences several downstream hematopoietic compartments in which there are key roles for wildtype Runx1. Our study establishes that the capability of Runx1 to support interaction with regulatory co-factors and nuclear architecture is required for optimal multi-lineage hematopoietic differentiation. Our findings demonstrate that the biological role of Runx1 during embryonic development can be uncoupled from its roles in differentiation of multiple hematopoietic lineages in the adult.
Materials and Methods
Construction of the Runx1HTY350-352AAA targeting vector
The final exon of the mouse Runx1 locus, exon 8, was targeted by homologous recombination using a 3.97 kb SacII-NotI PCR fragment of intron 7 (left arm) and a 4.0 kb NotI-SalI PCR fragment of intron 7 - exon 8 (right arm). Fragments were generated from mouse AB2.2 genomic DNA by PCR (Primers 5’ to 3’: LAF1 CCGCGGGGCATCTCTCTCCTTCCTCCAGTGTCT; LAR1 GAGGGGATCGAAAAGCTTCCT; LAF2 AGGAAGCTTTTCGATCCCCTC; LAR2 GCGGCCGCGATCACGGAGAGTGCCTCTGACAC; RAF1 GCGGCCGCGTGGGCAGGAGCACTCGCTGT; RAR1 GAGTAGGGAACTAGCGTGGG; RAF2 CCCACGCTAGTTCCCTACTC; RAR2 GACCACCCAGATGCAAACAGG; RAF3 CGCACCTTATCGATTGCAA; RAR3 GTCGACCCGACCAACAGCCAAACCCACCAA). The left arm was created by using primer pairs that produce 1.3 kb and 2.67 kb fragments (LAF1 to LAR1 and LAF2 to LAR2) which were ligated using an internal HindIII site to obtain the entire 3.97 kb fragment. The right arm was created using primer pairs that produce two overlapping fragments of 1.0 kb containing the mutation (RAF1 to RAR1 and RAF2 to RAR2) and 3.0 kb (RAF3 to RAR3) which were ligated using an internal ClaI site to obtain the entire 4.0 kb fragment. The 3.97 kb and 4.0kb fragments were cloned in tandem into the pGEM-5Zf(+) vector (Promega). We then inserted a 2.0 kb NotI-NotI cassette containing a floxed neomycin gene (LoxP site - PGK promoter - Neo cDNA - LoxP site) and a 2.2 kb SalI-SalI cassette with the thymidine kinase gene (PGK promoter - TK cDNA). Vectors containing the Neo and TK cassettes were provided by the Transgenic Animal Modeling Core Facility of the University of Massachusetts Medical School. DNA sequencing confirmed the organization of the final targeting vector and intermediate constructs.
Screening mouse embryonic stem cells for the Runx1HTY350-352AAA allele and generation of the Runx1HTY350-352AAA mice
Targeting vector linearized with AscI was electroporated into PC3 (129S5/SvEvBrd) embryonic stem (ES) cells (Transgenic Animal Modeling Core Facility, University of Massachusetts Medical School) (i.e., 107 ES cells were transfected with 20 µg linearized construct at 230 V and 500 µF). Selection began 24 hours after electroporation by addition of 180 µg/ml of G418 (Invitrogen Life Technologies, Inc., Carlsbad, CA). Thymidine kinase selected against non-homologous recombination. Homologous recombination of the Runx1HTY350-352AAA allele was established by Southern blot using restriction sites and probes external to the targeted region. Hybridization used the PerfectHyb Plus Hybridization kit (Sigma-Aldrich, St. Louis, MO). A single clone with a correctly targeted mutation was identified.
Chimeric mice with a significant ES cell contribution (determined by agouti coat) were mated with wildtype C57BL/6. Germ line transmission was determined by Southern blot of tail DNA from offspring and confirmed by PCR (Primers 5’ to 3’: forward ACTCTGGCAGTCTAGGAAGCC, reverse AGGCGCCGTAGTATAGATGGTA). This reaction amplifies a fragment including the HTY to AAA mutation; CACACCTAC to GCCGCGGCA, which creates a SacII digestion site. After initial screenings by Southern blot analysis, subsequent generations were genotyped by PCR and restriction enzyme digest (Figure 1B). Runx1HTY350-352AAA heterozygotes were crossed to generate homozygous mice confirmed by PCR and Southern blot genotyping.
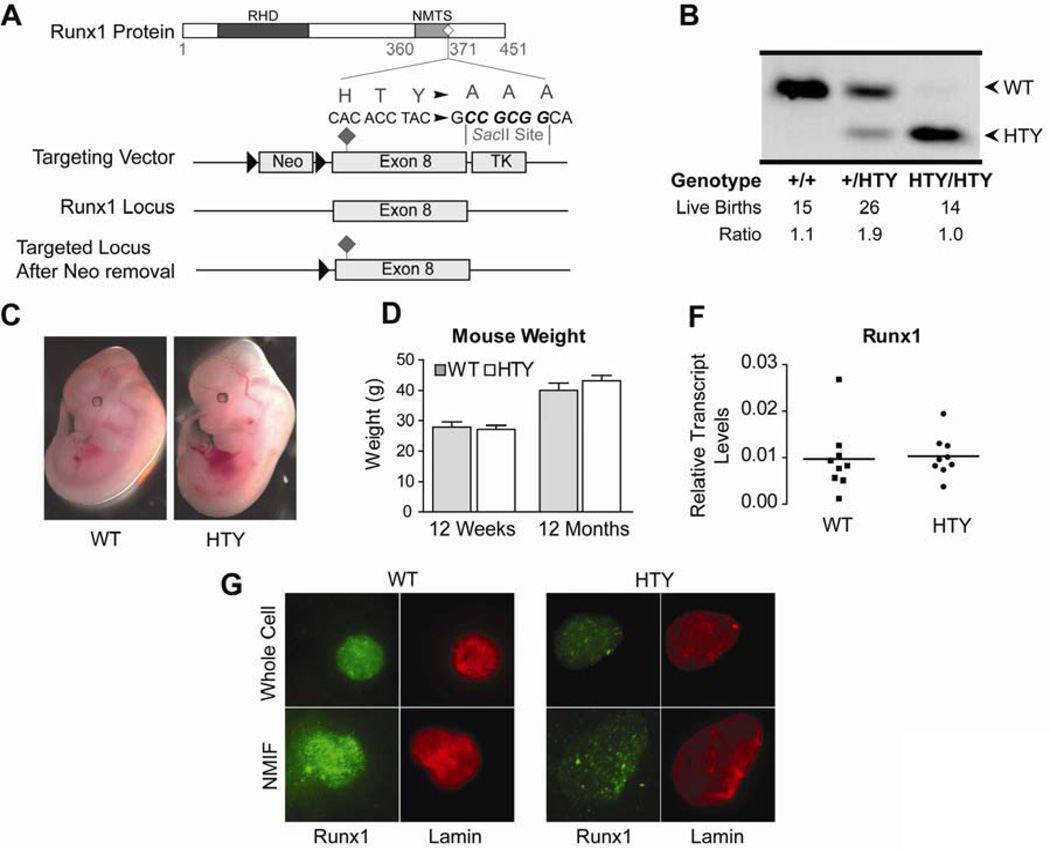
Figure 1. Generation of the Runx1HTY350-352AAA mouse.
(A) Targeting vector for the knock-in mouse contained a floxed Neo cassette as a positive selection marker and a TK cassette for negative selection to ensure homologous recombination in ES cells. (B) Successful targeting to the native Runx1 locus results in a diagnostic SacII site that allows for genotyping by PCR and enzyme digest. From left to right are genotyping results showing the change in digest fragment sizes from a wildtype, heterozygous and homozygous Runx1HTY350-352AAA mouse. (C) Runx1HTY350-352AAA animals were born at Mendelian ratios. (D) Runx1HTY350-352AAA embryos are healthy at embryonic day 12.5 and are comparable to wildtype littermates (3 litters, n=8 WT, 7 HTY). (E) Adult wildtype and Runx1HTY350-352AAA mice at 12 weeks or 12 months are equivalent in weight (n=16 WT, 19 HTY for 12 weeks and 15 WT, 9 HTY for 12 months). (F) qRT-PCR for Runx1 in bone marrow cells shows no difference in expression levels (p=0.8 by Student’s T test). Each point represents the average of technical replicates from one animal, normalized to mCox (n=9 WT, 9 HTY). (G) Whole cell and nuclear matrix-intermediate filament (NMIF) preparations of bone marrow from wildtype and Runx1HTY350-352AAA animals shows that Runx1 is still present within the nuclear matrix (n=3 WT, 3 HTY). All of the images were acquired using same settings of the microscope and the associated MetaMorph Software for accurate comparison. All error bars are SEM.
Immunofluorescence microscopy
Bone marrow cells (100,000 per slide) were spun onto glass slides, formaldehyde (3.7%) fixed, and permeabilized with 0.5% Triton X-100 for whole-cell preparations. Nuclear matrix-intermediate filament (NMIF) preparations were obtained as described27. Runx1 protein was detected by AML1(RHD) antibody (Oncogene Science, Cambridge, MA; 1:200 dilution) followed by Alexa Fluor 488 anti-rabbit secondary antibody (Invitrogen Molecular Probes, Eugene, OR; 1:800 dilution). Lamin A/C was detected with N-18 antibody (Santa Cruz; 1:800 dilution) followed by Alexa Fluor 594 anti-goat (1:800 dilution). Cells were stained with DAPI and mounted in Prolong Gold antifade medium (Invitrogen Molecular Probes). Fluorescence images were captured using a Zeiss Axioplan 2 equipped with a digital charged-coupled device camera (Hamamatsu Photonics, Bridgewater, NJ Cat. No. C4742-95) interfaced with the MetaMorph Imaging System (Universal Imaging Corporation Ltd, Marlow, Buckinghamshire, UK).
Colony forming unit (CFU) assays and ex vivo culture
Two thousand bone marrow cells were plated in duplicate in 35-mm dishes containing Methocult methyl cellulose medium (StemCell Technologies Vancouver, BC, Canada Cat. No. M3434), incubated at 37°C and colonies were counted on day 7. Bone marrow cells were cultured in DMEM with 10% FBS and L-glutamine but without additional cytokines. TGF beta was used at a final concentration of 10 ng per mL.
Flow cytometry
Cells were isolated from the bone marrow, spleen, and/or thymus of age matched mice into RPMI Medium without phenol red (Invitrogen) with 1 mM EDTA, 0.02% sodium azide, and 3% fetal bovine serum. Cells were adjusted to 6×107 cells per mL and incubated in antibody cocktails for 20 min on ice. Antibodies were purchased from BD Bioscience: B220-PE, Gr1-FITC, CD11b-PE, cKit-APC, Sca1-FITC, CD127-PE-Cy7, CD16/32-v450, CD34-alexaFlour700, AA4.1-FITC, IgM-PerCP-Cy5.5, CD43-APC, CD3-PE, CD4-PE-Cy7, CD8-APC, CD41-FITC, CD71-PE, and Ter119-APC. Lineage exclusion used antibodies against Ter119, CD3, CD11b, Gr1 and B220 conjugated to PE. Propidium iodide staining was performed by the University of Massachusetts Flow Cytometry core facility on cells stained with CD41-FITC and fixed in ethanol. Cells were fixed with 1% formaldehyde and processed on an LSRII or FACSCalibur. Analysis of CLP and early B progenitors was performed on live cells (stained as above) using a FACS Aria. Unstained and single stained cells were included as controls (Supplementary Figure 1).
qRT-PCR
RNA was prepared from bone marrow cells using TRIzol following the manufacturer’s protocol (Invitrogen). RNA was treated with DNaseI and 1 µg was subjected to reverse transcription with oligo dT primers. Quantitative RT-PCR was performed on the resulting cDNA using the following primers: Runx1 CCAGCAAGCTGAGGAGCGGCG, TGACGGTGACCAGAGTG; HB alpha ACTTCAAGCTCCTGAGCCACTGC, GCACGGTGCTCACAGAGGCA; HB Beta AACGATGGCCTGAATCACTTG, AGCCTGAAGTTCTCAGGATCCA ; HB gamma TTGGGAAGGCTTCTTGTTGT, AAGCAGAGGACAAGTTCCCA; EPO R CATCTGCGACAGTCGAGTTCTG, CACAACCCATCGTGACATTTTC; GATA1 GGCAAGACGGCACTCTACC, CAAGAACGTGTTGTTGCTCTTC; PU.1 TATCAAACCTTGTCCCCAGC, GCGAATCTTTTTCTTGCTGC.
Histology
Soft tissues were formalin fixed overnight and embedded in paraffin. Bones for marrow sections were paraformaldehyde fixed for 3 days under vacuum and decalcified for 14 days using 0.5 M EDTA prior to embedding. Six micron sections were stained with hematoxylin and eosin by standard procedures. Embryos were dissected from timed pregnancies. Images were captured using an Axioskop 40 (Carl Zeiss, Inc., Maple Grove, MN) with an AxioCam HRc and AxioVision Rel. 4.7 software (Zeiss).
Results
The Runx1HTY350-352AAA homozygous mice bypass embryonic lethality
To investigate the biological importance of Runx1 interactions with regulatory co-factors and the nuclear architecture in vivo, we generated a knock-in mouse with a missense mutation of Runx1 (Runx1HTY350-352AAA; denoted HTY in figures and table) introduced into the Runx1 locus by homologous recombination (Figure 1A). This allele expresses a protein at physiological levels with a triple point mutation in the domain important for subnuclear targeting22 and interaction with several known Runx co-factors, including SMADs13,14,16,22–26 We have extensively characterized the HTY mutant with over-expressed proteins and have shown that the mutant protein exhibits decreased subnuclear targeting and compromised responsiveness to TGFβ signaling22. Gene substitution was confirmed by Southern blot analysis (data not shown) and PCR assays. An engineered SacII site diagnostic for the HTY to AAA mutation was used to discriminate between the wildtype and mutant alleles (Figure 1B). Genotyping demonstrated that Runx1HTY350-352AAA homozygous mice are born at Mendelian ratios, bypassing the embryonic day 12.5 lethality associated with Runx1 ablation or C-terminal deletion (Figure 1C). Runx1HTY350-352AAA homozygous embryos at embryonic day 12.5 were comparable to wildtype littermates (Figure 1D). The weights of adult wildtype and mutant animals were similar at 12 weeks or 12 months of age (Figure 1E). Runx1 RNA levels in the bone marrow of wildtype and mutant animals are equivalent, indicating that the missense mutation does not destabilize the mRNA (Figure 1F). Immunofluorescence microscopy of whole cell and NMIF preparations of bone marrow cells from wildtype and Runx1HTY350-352AAA homozygous mice showed that while both Runx1 proteins are present within the nuclear matrix, association of the mutant Runx1 protein with the matrix may be decreased (Figure 1G). Runx1HTY350-352AAA homozygous embryos and neonates were similar to wildtype with no apparent hematopoietic defects. Hence, the Runx1HTY350-352AAA mutation appears to retain essential functions of Runx1 during HSC emergence and developmental hematopoiesis. Because Runx1 is required throughout hematopoiesis, we postulated that this mutation could have effects in hematopoietic progenitors or at different stages of hematopoietic lineage progression.
The Runx1HTY350-352AAA mutation compromises growth control in hematopoietic progenitors
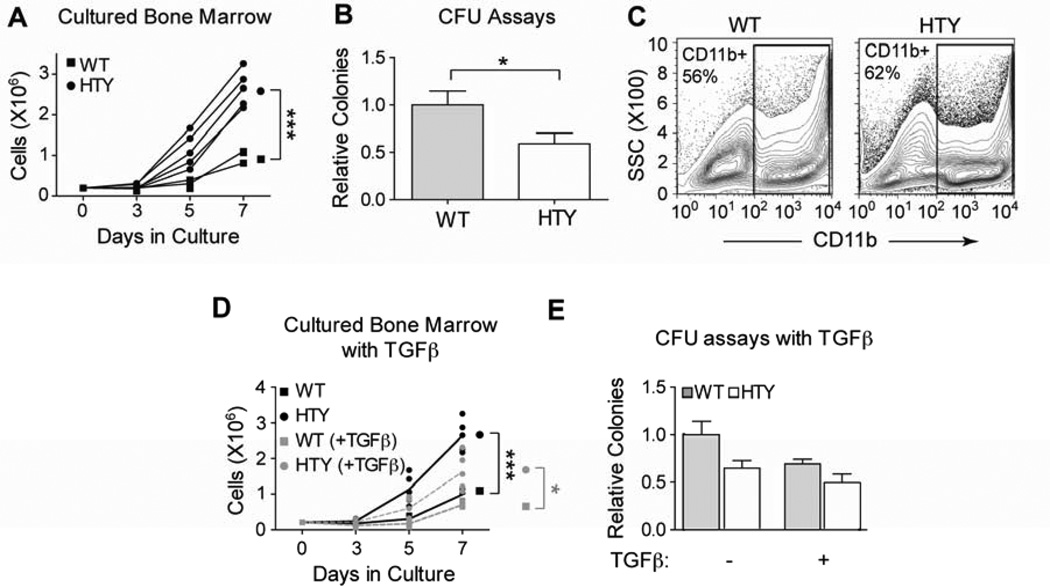
Runx1 has been proposed to preserve long-term hematopoietic repopulating capacity by maintaining HSC quiescence and appropriate apoptosis28. Expression of a naturally occurring truncated form, Runx1a, in HSC enhanced proliferation and increased short-term engraftment29,30. To examine hematopoietic progenitor properties in the Runx1HTY350-352AAA mice, bone marrow cells from wildtype and homozygous mutant animals were cultured ex vivo. After 7 days, there were significantly more cells in cultures from the knock-in animals (Figure 2A, p<0.001). In contrast, when the same cells were instead plated in methylcellulose for colony forming unit (CFU) assays, they formed fewer colonies than wildtype (Figure 2B, p=0.027), but colonies were of similar size (data not shown). These results indicate that the population has diminished colony-forming potential, perhaps because of a decreased representation of progenitors. The majority of cells taken from day 7 ex vivo cultures from both wild-type and Runx1HTY350-352AAA mice express the myeloid marker CD11b (Figure 2C). Given the known roles of Runx1 in myeloid development, these results warranted further investigation of committed progenitor populations in the Runx1HTY350-352AAA mice.
Figure 2. Increased ex vivo growth and diminished colony forming ability of Runx1HTY350-352AAA marrow.
(A) Ex vivo cultures of bone marrow cells from wildtype or Runx1HTY350-352AAA animals (each point represents the mean cell number of technical replicates from one animal; n=3 WT, 5 HTY). (B) Myeloid colony forming unit assays performed with wildtype or Runx1HTY350-352AAA bone marrow cells (n=28 WT, 34 HTY). Because there was no discernable difference in colony size, only total colony number was counted. (C) Cells from the experiment reported in panel A were harvested at day 7 and the percentage of cells expressing the myeloid antigen CD11b was determined using flow cytometry (n=3 WT, 3 HTY). (D) Ex vivo cultures as in panel A, but with the addition of TGF beta to a final concentration of 10ng per ml (each point represents technical replicates from one animal; n=3 WT, 5 HTY). (E) Colony forming unit assays were performed as in panel B, but with the addition of TGF beta to some cultures to a final concentration of 10ng per ml (n=3 WT, 5 HTY). * p<0.05, *** p<0.001, calculated by Student’s T test. All error bars are SEM.
TGF beta is involved in maintaining HSC quiescence; however, the exact mechanism is poorly understood31,32. SMAD proteins mediate TGF beta signaling and Runx1 interacts with SMAD proteins during hematopoietic development. We have previously shown that the Runx1HTY350-352AAA disrupts interaction between Runx1 and Smad proteins in hematopoietic cells, both biochemically and in situ22. We expanded our in vitro and ex vivo studies to investigate whether TGFβ signaling is perturbed by the Runx1HTY350-352AAA mutation in the genetically engineered mice. Both ex vivo growth and CFU assays were repeated with the addition of TGF beta in the medium. Growth was suppressed by TGF beta in both wildtype and Runx1HTY350-352AAA ex vivo cultures, but the Runx1HTY350-352AAA cytokine-treated cells still proliferated more rapidly than wild type treated cells (Figure 2D, p=0.024). Similarly, adding TGF beta to the CFU assays suppressed colony-forming capability for both wild type and mutant bone marrow cells; there was no statistically significant difference in the ratio of WT to HTY for the control and TGFβ treated marrow (Figure 2E). These data indicate that TGF beta-mediated growth suppression may proceed through Smad-independent mechanisms, as the TGFβ effect remains intact in the bone marrow cells of Runx1HTY350-352AAA homozygous mice.
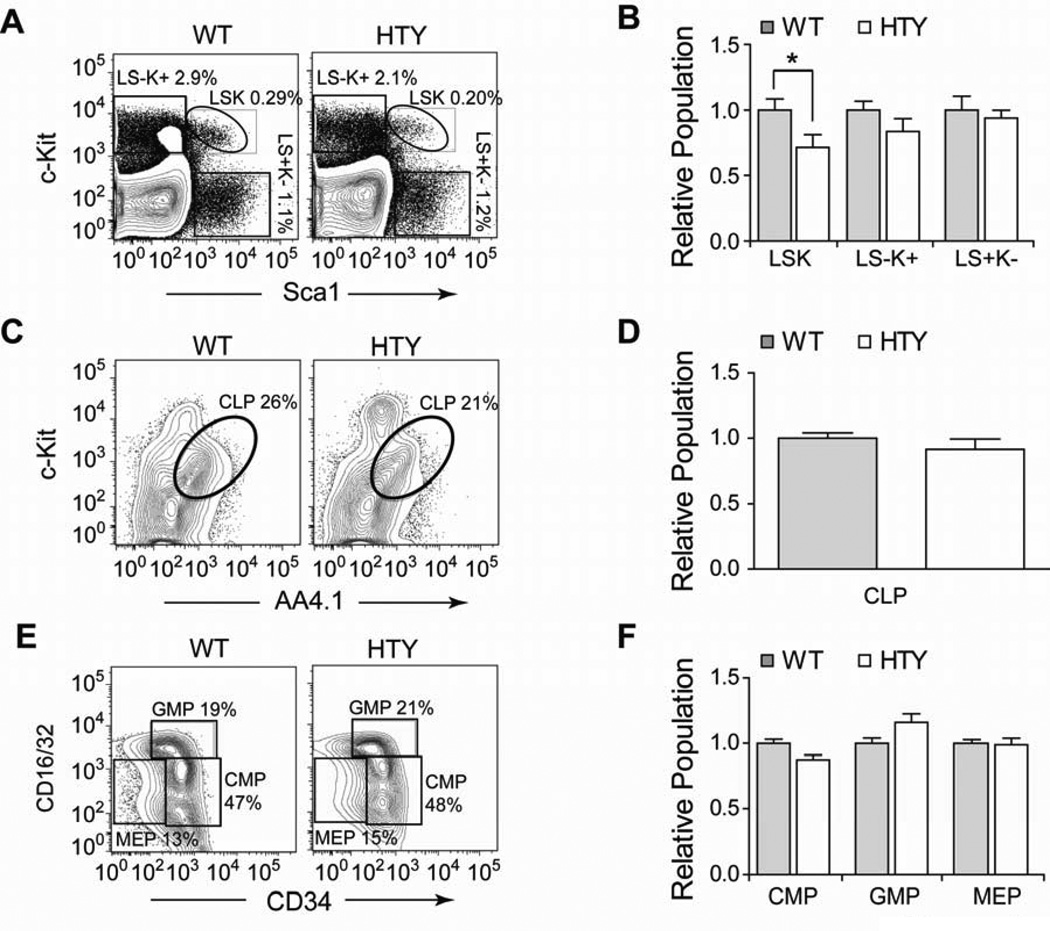
To assess alterations in the earliest progenitor populations, bone marrow from Runx1HTY350-352AAA mice was analyzed by flow cytometry. There was a small yet statistically significant decrease in the lineage negative, Sca1 positive, c-Kit positive population (LSK) which contains HSC, with no significant reductions in the lineage negative, Sca1 negative, c-Kit positive (LS-K+) myeloid precursors or the lineage negative, Sca1 positive, c-Kit negative (LS+K−) lymphoid precursors33 (Figures 3A and B). The frequency of common lymphoid progenitor (CLP) cells in the Runx1HTY350-352AAA mice was similar to wildtype (Figures 3C and D). The LS-K+ compartment contains common myeloid progenitors (CMP), which further differentiate into either granulocyte-monocyte progenitors (GMP) or megakaryocyte erythroid progenitors (MEP). We determined that these discrete populations were not significantly different in Runx1HTY350-352AAA bone marrow when compared to wildtype mice (Figures 3E and F).
Figure 3. Progenitors in Runx1HTY350-352AAA bone marrow.
(A) Bone marrow cells from wildtype or Runx1HTY350-352AAA animals were stained using antibodies to c-Kit, Sca1, CD16/32, CD34, IL7Ralpha, AA4.1 and Lineage markers (CD3, CD11b, B220, Ter119, Gr1) and analyzed by flow cytometry. Lineage negative cells were gated and are plotted in panel A to highlight the Lin-Sca1+Kit+ (LSK), myeloid potential L-S-K+ and lymphoid potential L-S+K− fractions. (B) Quantification of multiple experiments performed as depicted in A, normalizing to the wildtype average of each experiment (n=13 WT, 13 HTY). (C) Lineage negative and IL7R alpha positive bone marrow cells were gated and c-Kit versus AA4.1 was plotted to measure CLPs. (D) Quantification of multiple experiments in C (n=6 WT, 6 HTY). (E) Lineage negative, c-Kit positive, Sca1 negative (L-S-K+) cells from panel A were plotted as CD16/32 versus CD34 to measure the CMP, GMP and MEP progenitor populations. (F) Quantification of multiple experiments as in E, normalizing to the wildtype average of each experiment (n=10 WT, 10 HTY). * p<0.05, calculated by Student’s T test. All error bars are SEM.
Deregulation of B and T lymphoid cells with Runx1HTY350-352AAA
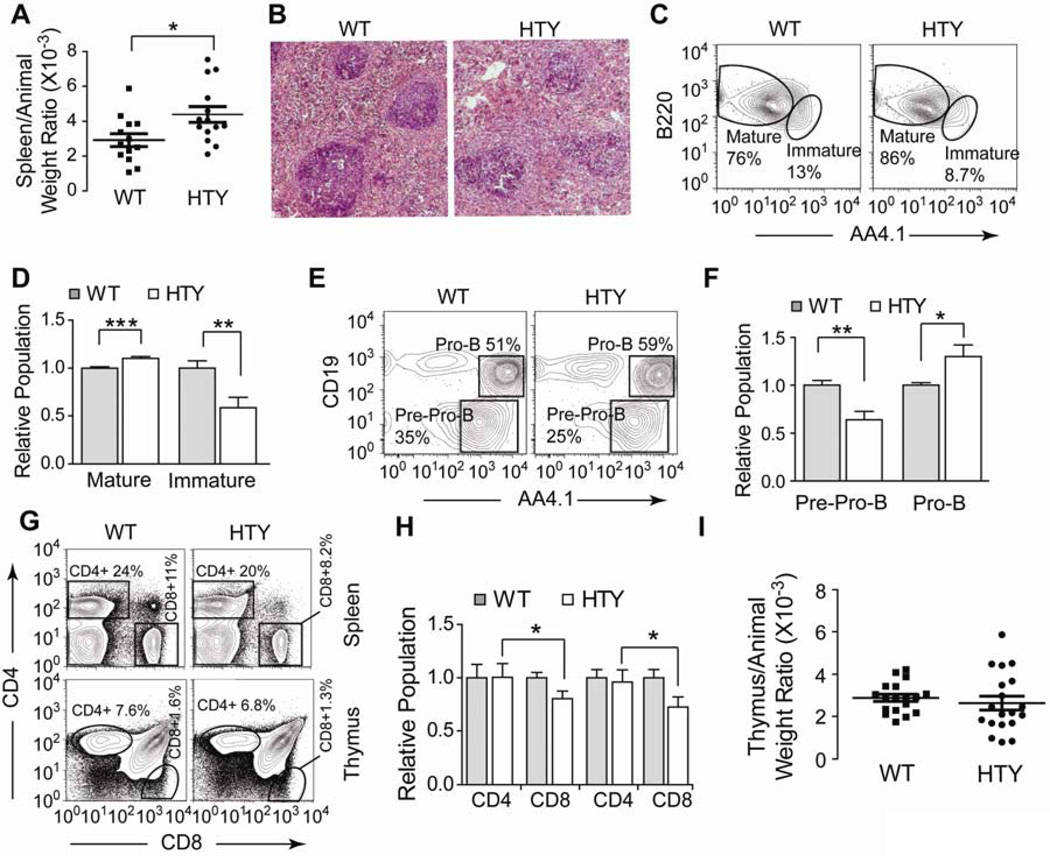
We further examined B and T lymphopoiesis in the Runx1HTY350-352AAA mice. Spleens of the Runx1HTY350-352AAA homozygous animals were significantly enlarged (Figure 4A, p=0.019), consistent with the splenomegaly that occurs following conditional Runx1 ablation34. Histological examination of Runx1HTY350-352AAA spleens showed deregulation of the white pulp, with decreased delineation of follicles and lower cell density (Figure 4B). Bone marrow of Runx1HTY350-352AAA mice exhibited slightly increased mature and decreased immature B cell populations (Figures 4C and D, p<0.0001 and p<0.001), consistent with a moderate impairment of B-lymphopoiesis. Furthermore, there was a decrease in the Pre-Pro-B and increase in the Pro-B populations (Figures 4E and F, p=0.005 and p=0.04). Taken together, these findings indicate that defective Runx1 function from the HTY350-352AAA mutation perturbs B-lymphopoiesis at multiple stages.
Figure 4. B and T lymphopoiesis in Runx1HTY350-352AAA animals.
(A) At sacrifice, animals were weighed then the spleen and/or thymus was removed and weighed. Displayed is the spleen weight normalized to total animal weight. Each point represents one animal (n=14 WT, 15 HTY). (B) Spleens were formalin fixed, sectioned and then stained by H & E. Representative fields are shown (n=3 WT, 3 HTY). (C) Bone marrow cells from wildtype or Runx1HTY350-352AAA animals were stained for B220, IgM, AA4.1 and CD43. B220 positive and IgM positive cells were gated to show expression of AA4.1 to separate Immature and Mature B cells. (D) Quantification of multiple experiments performed as in C (n=12 WT, 12 HTY). (E) Bone marrow cells from wildtype or Runx1HTY350-352AAA animals were stained for B220, IgM, AA4.1, Ly6C, CD49b, CD19 and CD43. Cells positive for Ly6C, CD49b and IgM were excluded by gating, and the CD43+ B220+ subset is shown to measure AA4.1+CD19− pre-Pro-B cells and AA4.1+ CD19+ pro-B cells. (F) Quantification of multiple experiments as in E (n=6 WT, 6 HTY). (G) Spleen or thymus cells were isolated and stained with CD4 and CD8. Spleen cells were gated and plotted as CD4 versus CD8 to measure CD4+ and CD8+ T cell populations. (H) Quantification of multiple experiments as shown in G (n=12 WT, 14 HTY; age-matched animals were used). (I) Displayed is the thymus weight normalized to total animal weight (n=18 WT, 19 HTY). * p<0.05, ** p<0.01, *** p<0.001, calculated by Student’s T test. All error bars are SEM.
We observed fewer CD8 single positive cells in both the spleen and thymus of Runx1HTY350-352AAA animals (Figures 4G and H, p=0.038 and p=0.038), with no significant alterations in the CD4 single positive or CD4/CD8 double positive populations (data not shown). The discrete populations of CD4/CD8 double negative T cell progenitors within the thymus were not altered (data not shown), nor was thymus size of Runx1HTY350-352AAA animals (Figure 4I). Our findings indicate that the Runx1 mutation does not interfere with early T-lymphoid development, but likely influences CD4 versus CD8 fate decisions.
Runx1HTY350-352AAA causes expansion of both myeloid and megakaryocytic lineage cells
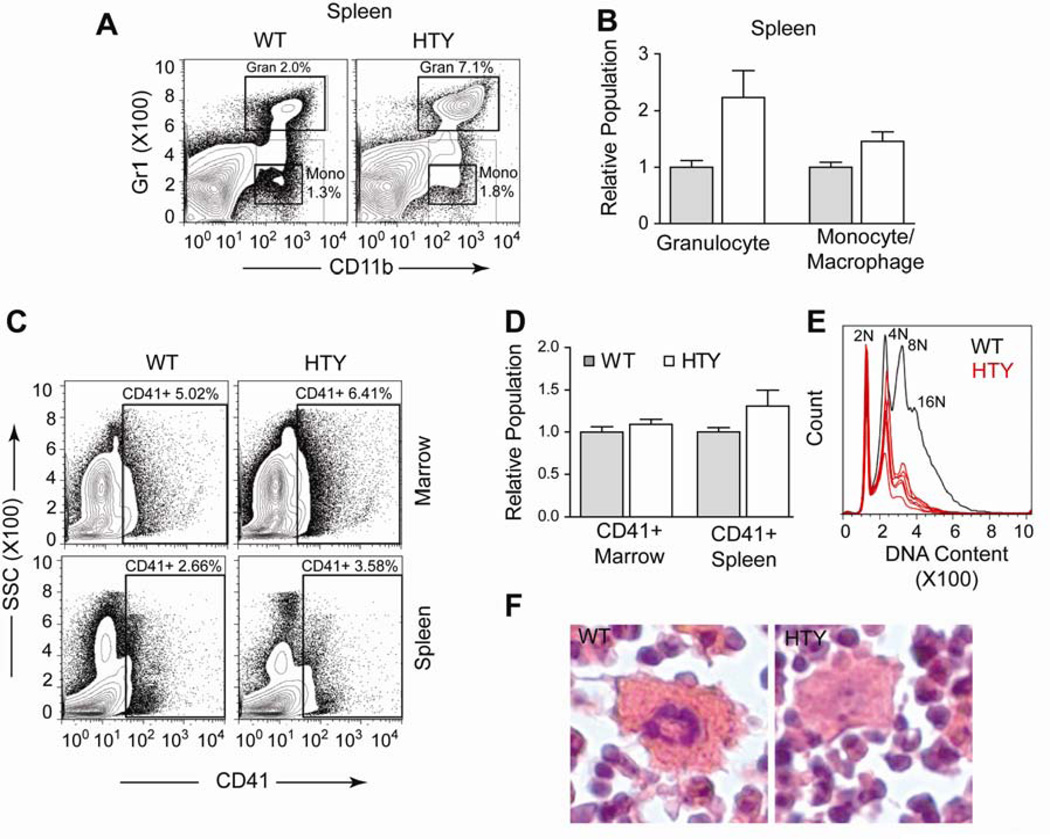
Runx1 mutations are frequently implicated in myeloid leukemia and Runx1 point mutations are associated with familial platelet disorders5,35,36. Conditional Runx1 ablation causes defects in myeloid and megakaryocytic lineages35. Therefore, we anticipated the most striking differences in our knock-in mouse would be observed there. We noted an increase in both the mature granulocyte (CD11b positive, Gr1 positive; p<0.0001) and monocyte/macrophage (CD11b positive, Gr1 intermediate; p<0.0001) populations in the spleens of Runx1HTY350-352AAA mice (Figures 5A and B). However, in the marrow these populations were not significantly different (data not shown). The splenic expansion is consistent with the growth factor independent proliferation observed in murine myeloid cell lines harboring similar mutations17. This expansion also suggests that the observed splenomegaly (Figure 4A and B) may be a consequence of myeloid expansion or infiltration, similar to that observed upon ablation in Runx1 conditional mice35.
Figure 5. Expansion of splenic myeloid compartment and delay in megakaryocytic maturation in Runx1HTY350-352AAA animals.
(A) Spleen cells were stained with antibodies to Gr1 and CD11b to measure the Gr1 high, CD11b positive granulocyte and the Gr1 intermediate CD11b positive monocyte / macrophage populations. (B) Quantification of multiple experiments performed as shown in A (n=7 WT, 9 HTY) (C) Bone marrow or spleen cells were stained for CD41 to measure the megakaryocyte population. (D) Quantification of multiple experiments shown in C (n=12 WT, 14 HTY). (E) Bone marrow cells were stained for CD41, fixed with ethanol and then stained with propidium iodide to detect DNA content. Displayed is the CD41 positive fraction, with normal hyperploidy of a representative wildtype sample plotted in black, with several Runx1HTY350-352AAA samples plotted in red to show the reduction in 8N and 16N cells. (F) Histological sections of bone marrow were H & E stained. Megakaryocytes that appear smaller and less hyperploid in the Runx1HTY350-352AAA animals are shown. *** p<0.001, calculated by Student’s T test. All error bars are SEM.
Megakaryocytic maturation is known to be particularly sensitive to Runx111. We observed that the CD41+ compartments of both bone marrow and spleen were comparable in Runx1HTY350-352AAA and wildtype mice (Figures 5C and D). This finding was unexpected because conditional Runx1 ablation is known to cause thrombocytopenia34,36. Therefore, we examined the maturation status of the CD41+ compartment, using propidium iodide staining to measure DNA content. This analysis showed a marked decrease in 8N and 16N cells compared with the wildtype reference, suggesting delayed megakaryocyte maturation (Figure 5E). We confirmed this finding with histological analysis. While some megakaryocytes in the marrow appeared normal, we observed many smaller megakaryocytes with less nuclear hematoxylin staining compared with wildtype marrow (Figure 5F). These results indicate that Runx1HTY350-352AAA may cause a delay in megakaryocyte maturation.
Red blood cell maturation is perturbed in Runx1HTY350-352AAA homozygous mice
As part of the characterization of the Runx1HTY350-352AAA mice, blood was collected at 8 weeks of age for complete blood counts (Table 1). Screening of a large number of animals (n=33 wildtype, 26 mutant) revealed subtle, yet consistent and statistically significant alterations. We observed an increase in platelet number and size, prompting the investigation into megakaryocyte development described in Figure 5. We also observed a subtle, sub-clinical anemia. Absolute red blood cell count was increased, but the cell volume was reduced significantly, lowering the hematocrit compared to wildtype (Table 1, p=0.025). Because the role of Runx1 in erythropoiesis is poorly understood, these results prompted additional examination.
Table I.
Complete Blood Counts from WT and HTY Mice
| Complete Blood Counts at 8 weeks |
WT | n=33 | HTY | n=26 | p value |
|---|---|---|---|---|---|
| White Blood Cells | 6.38 | ± 2.33 | 5.59 | ± 1.64 | 0.13 |
| Lymphocytes | 5.22 | ± 2.03 | 4.52 | ± 1.48 | 0.12 |
| Granulocytes | 0.71 | ± 0.42 | 0.64 | ± 0.39 | 0.53 |
| Monocytes | 0.45 | ± 0.29 | 0.43 | ± 0.18 | 0.79 |
| Hematocrit | 50.84 | ± 5.56 | 47.74 | ± 5.16 | 0.025 |
| Mean Corpuscle Volume | 51.05 | ± 3.87 | 45.20 | ± 1.34 | <0.00001 |
| Red Blood Cells | 9.96 | ± 0.83 | 10.58 | ± 1.26 | 0.035 |
| Hemoglobin | 16.62 | ± 1.00 | 17.34 | ± 1.59 | 0.050 |
| Mean Cell Hemoglobin | 16.72 | ± 0.64 | 16.45 | ± 0.73 | 0.15 |
| Hemoglobin Concentration | 32.92 | ± 2.37 | 36.42 | ± 0.73 | <0.00001 |
| Red Cell variability | 17.61 | ± 1.77 | 20.69 | ± 1.34 | <0.00001 |
| Platelet Volume | 6.01 | ± 0.21 | 6.21 | ± 0.20 | <0.001 |
| Platelet Count | 698.09 | ± 136.48 | 795.56 | ± 149.25 | 0.012 |
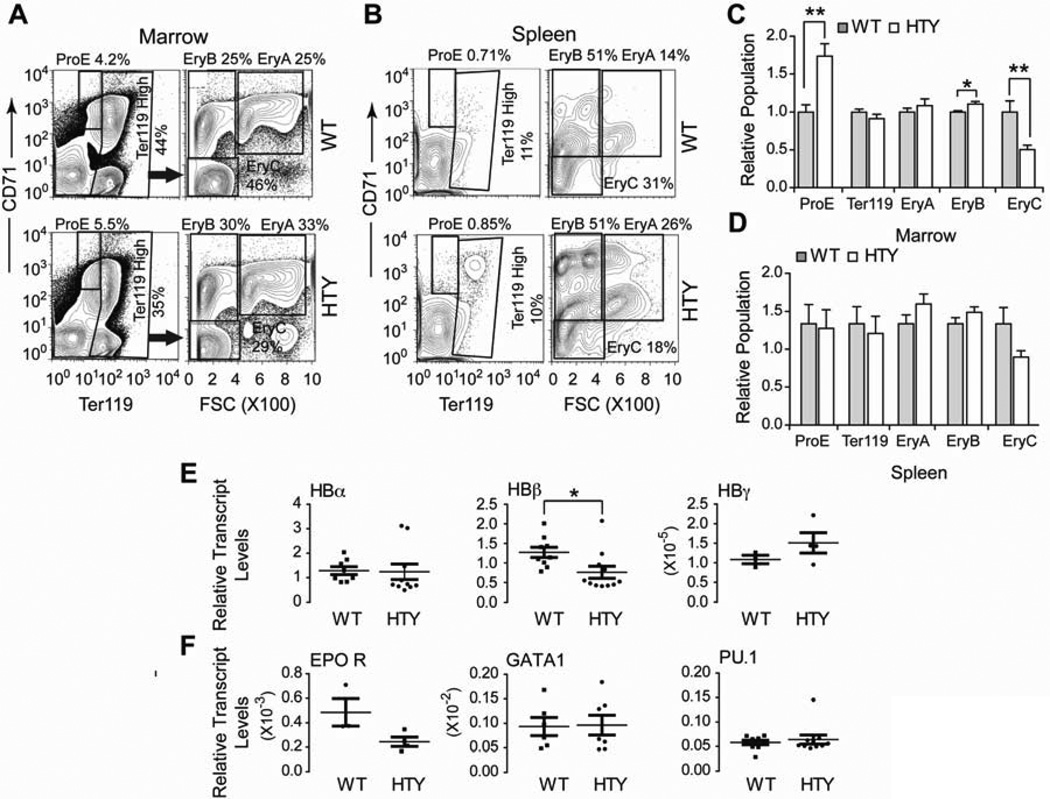
We characterized erythroid progenitor populations using a validated method of flow cytometry analysis with Ter119 and CD7137,38. Examination of bone marrow revealed an increase in early erythroid progenitor populations (ProE, EryA and EryB) and fewer mature red cell progenitors (EryC) (Figures 6A and C). The increase in ProE and EryB progenitors and the decrease in more mature EryC cells were all statistically significant (p<0.001, p=0.012 and p<0.01, respectively; n=12 WT, 14 HTY). In the spleens of Runx1HTY350-352AAA mice, ProE cell populations were similar to wildtype, and the increase of EryA and EryB with a subsequent decrease in EryC was still observed (Figures 6B and D). However, none of the differences in the spleen cells reached statistical significance (p=0.14, p= 0.18 and p=0.054 for EryA, EryB and EryC). Gene expression analysis of bone marrow cells showed a decrease of hemoglobin beta transcript levels (p=0.025), without significant alteration of hemoglobin alpha (Figure 6E, p=0.91). Fetal hemoglobin gamma was not significantly increased (p=0.25), inconsistent with a delay in globin switching in the mutant mice39 (Figure 6E). Alteration of the alpha and beta hemoglobin balance may explain the smaller size of the red blood cells (Table 1). Neither the levels of the master erythroid transcription factor GATA1, nor those of PU.1, a known Runx1 target and direct inhibitor of GATA1, were significantly altered (Figure 6F). However, EPO receptor expression showed a trend of decreased expression in the Runx1HTY350-352AAA marrow (Figure 6F, p=0.068, n=3 WT, 4 HTY), perhaps accounting, in part, for a defect in erythroid progenitors. Thus, the Runx1HTY350-352AAA mutation provides a new dimension to understanding contributions of Runx1 in erythroid development.
Figure 6. Delayed red blood cell maturation in Runx1HTY350-352AAA animals.
(A) Bone marrow cells were stained for CD71 and Ter119 and plotted to measure the CD71 positive, Ter119 intermediate ProE population. Ter119 high cells were plotted CD71 versus forward scatter (FSC) to measure the EryA, EryB and EryC progenitor populations. (B) Spleen cells were stained and analyzed as in A. (C) Quantification of several experiments shown in A (n=12 WT, 14 HTY). (D) Quantification of several experiments shown in B (n=12 WT, 14 HTY). (E) qRT-PCR of bone marrow cells for hemoglobin genes (F) qRT-PCR for factors important for erythroid maturation and an important antagonist of erythropoiesis. Each point represents the average of technical replicates from one animal (n=3 to 11). * p<0.05, ** p<0.01, calculated by Student’s T test. All error bars are SEM.
Discussion
Mutations in specific domains of Runx1 that disrupt subnuclear targeting and co-factor interactions alter target gene regulation, block differentiation and confer a pre-leukemic phenotype17,19. It was therefore of interest to investigate the functional activities of these domains in vivo. For this study, we developed and characterized Runx1HTY350-352AAA knock-in mice that bypass the embryonic lethality observed with Runx1 ablation or C-terminal truncation, but have defects in multiple hematopoietic lineages. Thus, the Runx1HTY350-352AAA mutation discriminates specific roles for Runx1 during embryogenesis and adult hematopoiesis1,7.
Regulation of bone marrow cell growth is perturbed in the Runx1HTY350-352AAA homozygous animals. We observed decreased colony numbers in ex vivo methylcellulose assays, but colony size was unchanged. These differences may represent a reduction in progenitor number but not in their individual capacity to expand. In contrast, bone marrow cells from mutant animals showed increased growth in culture. This apparent inconsistency may reflect distinct cell populations being measured by the two assays.. These observations are combined with expansion or contraction of progenitor compartments and a reduction of LSK cells in vivo (Figures 2B, 3A–F, 4C–F, 6A–C). Studies by others have described that conditional ablation of Runx1 in adult mice causes an increase in LSK cells, which are less competitive over time34,35. However recent reports indicate a more subtle impact on long term repopulation of the Runx1 deficient hematopoietic stem cells28,. The Runx1HTY350-352AAA mutation is germ line and therefore deregulates progenitor growth control very early during development. These cells are rapidly expanded at the onset of definitive hematopoiesis and by adulthood it is possible only less competitive cells remain, potentially explaining the smaller LSK population with reduced progenitors. These data suggest that Runx1HTY350-352AAA mice are compromised in the normal roles of Runx1 that maintain control of progenitor growth and proliferation40. However, long term repopulation capability of the HSC from this mutant remains to be seen.
The Runx1HTY350-352AAA homozygous mice have splenomegaly, abnormalities in the white pulp of the spleen and deregulated B cell maturation. Several cell compartments of the B lineage are affected. Pre-Pro-B and Pro-B progenitors are altered in addition to the balance of immature and mature circulating B cells. Conditional Runx1 ablation34,35 or expression of leukemic fusion proteins that inhibit Runx1 function41 inhibit B-cell development in its early stages. Further, Runx1 directly regulates the key B lineage gene, Ebf1, and selective deletion of Runx1 in early B-cell progenitors causes a block in early B-lymphopoiesis42. Runx1HTY350-352AAA in contrast alters the balance of multiple compartments, implying that this domain of Runx1 has functional roles at multiple stages.
Down-regulation of Runx1 is required during T-cell maturation for silencing of CD4 in CD4/CD8 double positive cells43. A knock-in mouse model that removed the C-terminal VWRPY motif decreased the CD8 positive T cell population in the spleen44. Similarly, Runx1HTY350-352AAA homozygous mice have fewer CD8 single positive T-cells in spleen and thymus. Thymus size and early T-cell progenitors were not significantly altered in Runx1HTY350-352AAA homozygous mice, in contrast to the smaller thymus and differentiation block observed after Runx1 excision in adults34,35. These results define contributions of precise domains with the known roles of Runx1 in control of both B and T lymphopoiesis41,45–50. While the exact mechanism is not fully understood, impaired co-factor interactions likely interfere with lymphoid target gene regulation. These data suggest that the HTY350-352 mutation compromises normal physiological functions of Runx1 and consequently deregulates B and T-lymphopoiesis.
The spleen harbors a major reserve of myeloid cells in healthy mice51 and these cells undergo expansion with Runx1 loss35. We observed significant expansion of both the monocyte and granulocyte compartments in Runx1HTY350-352AAA homozygous spleens, suggesting that the splenomegaly is predominantly caused by aberrant expansion of resident myeloid cells. These data are consistent with known contributions of Runx1 mutants to myeloid proliferative disease6,12.
Thrombocytopenia is one of the most common findings in patients with Runx1 mutations, and in animal models employing conditional Runx1 ablation35,36. The Runx1HTY350-352AAA homozygous mice are defective in megakaryocyte maturation and have a slight expansion of CD41 positive cells. Despite perturbed maturation, and unlike conditional Runx1 ablation34,35, Runx1HTY350-352AAA homozygous mice are still able to maintain platelet counts within normal ranges. Thus Runx1 functions associated with HTY350-352 are required for megakaryocytic maturation but do not block platelet production.
Abnormalities in primitive erythrocytes from Runx1 null mice and perturbation of erythropoiesis by the leukemic fusion Runx1-ETO implicate Runx1 in erythropoiesis52,53. Subtle changes in the characteristics of red blood cells were observed with Runx1 haploinsufficiency or conditional ablation, but were not explored further34,35. We find that Runx1HTY350-352AAA homozygous mice have a lower hematocrit, resulting from smaller red blood cells, that may be explained by decreased beta hemoglobin expression and the consequential presence of alpha rich hemoglobin. In patients with beta thalassemia, alpha rich hemoglobin is less stable and resultant red blood cells have difficulty retaining shape54. Red cell maturation was also perturbed in the Runx1HTY350-352AAA homozygous mice, causing an increase in the earliest erythroid progenitors, with a subsequent decrease in the more mature cells. Thus, Runx1HTY350-352AAA causes a defect in erythroid maturation. At the onset of erythroid maturation, Runx1 levels decrease. It is possible that the HTY350-352AAA mutation interferes with this endogenous down-regulation, causing the maturation delay.
The phenotype of Runx1HTY350-352AAA homozygous mice is contrasted to Runx1 haploinsufficiency, as seen by IRES-GFP knock-in to the Runx1 locus or heterozygous knock-out55,56. Runx1-IRES-GFP knock-in mice have decreased CD4 and increased CD8 single positive T cells in the spleen55. Heterozygous Runx1 knock-out mice had no splenomegaly and increased progenitor ability56. Thus the phenotypes observed in the Runx1HTY350-352AAA homozygous mice are not due to lower amounts of Runx1, but to altered biological activity.
Runx factors organize and scaffold regulatory machinery, including factors that support chromatin structure and nucleosome organization at strategic sites of target gene promoters and in focal nuclear microenvironments20. Subtle alterations in the scaffolding function of Runx1 could have profound regulatory effects, as were noted in the in vitro characterization of subnuclear targeting mutations of both Runx1 and Runx217,19,22,57. As the point mutation is within the domain important for subnuclear localization, alterations in subnuclear organization and/or perturbed co-regulatory protein interactions are likely responsible for the hypomorphic hematopoietic phenotypes of the Runx1HTY350-352AAA knock-in mouse. Because the C-terminal domain of Runx1 interacts with effectors of many signaling pathways 13,14,16,22–26, it will be interesting to explore whether the HTY mutation results in perturbation of certain Runx1 functions while enhancing others.
Our findings reinforce the pivotal role of Runx1 as a master regulator of hematopoiesis. Runx1HTY350-352AAA represents a germline Runx1 mutation that bypasses embryonic lethality but alters multiple differentiated lineages in the adult. Runx1HTY350-352AAA homozygosity results in defective growth control of hematopoietic progenitors, deregulation of B-lymphoid and myeloid lineages, as well as maturation delays in megakaryocytic and erythroid development. Our study establishes a novel dimension in Runx1 mediated regulatory control that separates its roles in embryogenesis, definitive hematopoiesis and differentiation across hematopoietic lineages.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Stephen Baker of the University of Massachusetts Medical School, Department of Information Services, for assistance with statistical analysis and Dr. Merav Socolovsky (Department of Cancer Biology, University of Massachusetts Medical School) for invaluable guidance in erythroid flow cytometry technique. The authors also wish to thank Judy Rask for help in preparation of the manuscript.
This work was supported by grants from the National Institutes of Health (P01 CA082834) (G.S.S.). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-Interest Disclosure: The authors declare no competing financial interests.
References
- 1.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. Prepublished on 1996/01/26 as DOI. [DOI] [PubMed] [Google Scholar]
- 2.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22(2):158–168. doi: 10.1634/stemcells.22-2-158. Prepublished on 2004/03/03 as DOI 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- 3.Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100(13):7731–7736. doi: 10.1073/pnas.1232420100. Prepublished on 2003/06/11 as DOI 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059–1064. doi: 10.1126/science.278.5340.1059. Prepublished on 1997/11/14 as DOI. [DOI] [PubMed] [Google Scholar]
- 5.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103(6):2316–2324. doi: 10.1182/blood-2003-09-3074. Prepublished on 2003/11/15 as DOI 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 6.Kuo MC, Liang DC, Huang CF, et al. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23(8):1426–1431. doi: 10.1038/leu.2009.48. Prepublished on 2009/03/14 as DOI 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- 7.Dowdy CR, Xie R, Frederick D, et al. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum Mol Genet. 2010;19(6):1048–1057. doi: 10.1093/hmg/ddp568. Prepublished on 2009/12/26 as DOI 10.1093/hmg/ddp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.North T, Gu TL, Stacy T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126(11):2563–2575. doi: 10.1242/dev.126.11.2563. Prepublished on 1999/05/05 as DOI. [DOI] [PubMed] [Google Scholar]
- 9.Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice DP, van 't Hof RJ, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010;340(2):539–546. doi: 10.1016/j.ydbio.2010.02.005. Prepublished on 2010/02/16 as DOI 10.1016/j.ydbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Fukushima-Nakase Y, Naoe Y, Taniuchi I, Hosoi H, Sugimoto T, Okuda T. Shared and distinct roles mediated through C-terminal subdomains of acute myeloid leukemia/Runt-related transcription factor molecules in murine development. Blood. 2005;105(11):4298–4307. doi: 10.1182/blood-2004-08-3372. Prepublished on 2005/02/17 as DOI 10.1182/blood-2004-08-3372. [DOI] [PubMed] [Google Scholar]
- 11.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. Prepublished on 2004/02/18 as DOI 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 12.Barseguian K, Lutterbach B, Hiebert SW, et al. Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc Natl Acad Sci U S A. 2002;99(24):15434–15439. doi: 10.1073/pnas.242588499. Prepublished on 2002/11/13 as DOI 10.1073/pnas.242588499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol. 2003;23(2):607–619. doi: 10.1128/MCB.23.2.607-619.2003. Prepublished on 2003/01/02 as DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17(11):2994–3004. doi: 10.1093/emboj/17.11.2994. Prepublished on 1998/06/26 as DOI 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNeil S, Zeng C, Harrington KS, et al. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc Natl Acad Sci U S A. 1999;96(26):14882–14887. doi: 10.1073/pnas.96.26.14882. Prepublished on 1999/12/28 as DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25(42):5777–5786. doi: 10.1038/sj.onc.1209591. Prepublished on 2006/05/03 as DOI 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- 17.Vradii D, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proc Natl Acad Sci U S A. 2005;102(20):7174–7179. doi: 10.1073/pnas.0502130102. Prepublished on 2005/05/05 as DOI 10.1073/pnas.0502130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng C, van Wijnen AJ, Stein JL, et al. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-alpha transcription factors. Proc Natl Acad Sci U S A. 1997;94(13):6746–6751. doi: 10.1073/pnas.94.13.6746. Prepublished on 1997/06/24 as DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaidi SK, Dowdy CR, van Wijnen AJ, et al. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69(21):8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. Prepublished on 2009/10/15 as DOI 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein GS, Lian JB, Stein JL, et al. Combinatorial organization of the transcriptional regulatory machinery in biological control and cancer. Adv Enzyme Regul. 2005;45:136–154. doi: 10.1016/j.advenzreg.2005.02.009. Prepublished on 2005/09/02 as DOI 10.1016/j.advenzreg.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Bakshi R, Zaidi SK, Pande S, et al. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J Cell Sci. 2008;121(Pt 23):3981–3990. doi: 10.1242/jcs.033431. Prepublished on 2008/11/13 as DOI 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Vradii D, Gutierrez S, et al. Subnuclear targeting of Runx1 is required for synergistic activation of the myeloid specific M-CSF receptor promoter by PU.1. J Cell Biochem. 2005;96(4):795–809. doi: 10.1002/jcb.20548. Prepublished on 2005/09/09 as DOI 10.1002/jcb.20548. [DOI] [PubMed] [Google Scholar]
- 23.Javed A, Afzal F, Bae JS, et al. Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs. 2009;189(1–4):133–137. doi: 10.1159/000151719. Prepublished on 2008/08/30 as DOI 10.1159/000151719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javed A, Bae JS, Afzal F, et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283(13):8412–8422. doi: 10.1074/jbc.M705578200. Prepublished on 2008/01/22 as DOI 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leboy P, Grasso-Knight G, D'Angelo M, et al. Smad-Runx interactions during chondrocyte maturation. J Bone Joint Surg Am. 2001;83-A(Suppl 1)(Pt 1):S15–S22. Prepublished on 2001/03/27 as DOI. [PubMed] [Google Scholar]
- 26.Afzal F, Pratap J, Ito K, et al. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204(1):63–72. doi: 10.1002/jcp.20258. Prepublished on 2004/12/02 as DOI 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 27.Nickerson JA, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci U S A. 1997;94(9):4446–4450. doi: 10.1073/pnas.94.9.4446. Prepublished on 1997/04/29 as DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai X, Gaudet JJ, Mangan JK, et al. Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS One. 2011;6(12):e28430. doi: 10.1371/journal.pone.0028430. Prepublished on 2011/12/07 as DOI 10.1371/journal.pone.0028430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto M, Enver T. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med. 2007;4(5):e172. doi: 10.1371/journal.pmed.0040172. Prepublished on 2007/05/17 as DOI 10.1371/journal.pmed.0040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran D, Shia WJ, Lo MC, et al. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013;121(15):2882–2890. doi: 10.1182/blood-2012-08-451641. Prepublished on 2013/02/02 as DOI 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortunel NO, Hatzfeld A, Hatzfeld JA. Transforming growth factor-beta: pleiotropic role in the regulation of hematopoiesis. Blood. 2000;96(6):2022–2036. Prepublished on 2000/09/09 as DOI. [PubMed] [Google Scholar]
- 32.Ruscetti FW, Akel S, Bartelmez SH. Autocrine transforming growth factor-beta regulation of hematopoiesis: many outcomes that depend on the context. Oncogene. 2005;24(37):5751–5763. doi: 10.1038/sj.onc.1208921. Prepublished on 2005/08/27 as DOI 10.1038/sj.onc.1208921. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, Fossati V, Israel M, Snoeck HW. Lin-Sca1+kit− bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol. 2008;181(11):7507–7513. doi: 10.4049/jimmunol.181.11.7507. Prepublished on 2008/11/20 as DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25(6):929–939. doi: 10.1038/sj.onc.1209136. Prepublished on 2005/10/26 as DOI 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- 35.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. Prepublished on 2005/03/24 as DOI 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166–175. doi: 10.1038/13793. Prepublished on 1999/10/03 as DOI 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Pop R, Sadegh C, Brugnara C, Haase VH, Socolovsky M. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 2006;108(1):123–133. doi: 10.1182/blood-2005-11-4458. Prepublished on 2006/03/11 as DOI 10.1182/blood-2005-11-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koulnis M, Porpiglia E, Porpiglia PA, et al. Contrasting dynamic responses in vivo of the Bcl-xL and Bim erythropoietic survival pathways. Blood. 2012;119(5):1228–1239. doi: 10.1182/blood-2011-07-365346. Prepublished on 2011/11/17 as DOI 10.1182/blood-2011-07-365346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lal A, Vichinsky E. The role of fetal hemoglobin-enhancing agents in thalassemia. Semin Hematol. 2004;41(Suppl 6)(4):17–22. doi: 10.1053/j.seminhematol.2004.08.004. Prepublished on 2004/11/10 as DOI. [DOI] [PubMed] [Google Scholar]
- 40.Ichikawa M, Goyama S, Asai T, et al. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J Immunol. 2008;180(7):4402–4408. doi: 10.4049/jimmunol.180.7.4402. Prepublished on 2008/03/21 as DOI. [DOI] [PubMed] [Google Scholar]
- 41.Kuo YH, Gerstein RM, Castilla LH. Cbfbeta-SMMHC impairs differentiation of common lymphoid progenitors and reveals an essential role for RUNX in early B-cell development. Blood. 2008;111(3):1543–1551. doi: 10.1182/blood-2007-07-104422. Prepublished on 2007/10/18 as DOI 10.1182/blood-2007-07-104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfbeta facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209(7):1255–1262. doi: 10.1084/jem.20112745. Prepublished on 2012/06/06 as DOI 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong WF, Nakazato M, Watanabe T, et al. Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double-negative to double-positive stages. Immunology. 2010;130(2):243–253. doi: 10.1111/j.1365-2567.2009.03230.x. Prepublished on 2010/01/28 as DOI 10.1111/j.1365-2567.2009.03230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura M, Fukushima-Nakase Y, Fujita Y, et al. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103(2):562–570. doi: 10.1182/blood-2003-06-2109. Prepublished on 2003/09/25 as DOI 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- 45.Blyth K, Slater N, Hanlon L, et al. Runx1 promotes B-cell survival and lymphoma development. Blood Cells Mol Dis. 2009;43(1):12–19. doi: 10.1016/j.bcmd.2009.01.013. Prepublished on 2009/03/10 as DOI 10.1016/j.bcmd.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Egawa T. Runx and ThPOK: a balancing act to regulate thymocyte lineage commitment. J Cell Biochem. 2009;107(6):1037–1045. doi: 10.1002/jcb.22212. Prepublished on 2009/05/30 as DOI 10.1002/jcb.22212. [DOI] [PubMed] [Google Scholar]
- 47.Grossmann V, Kern W, Harbich S, et al. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica. 2011;96(12):1874–1877. doi: 10.3324/haematol.2011.043919. Prepublished on 2011/08/11 as DOI 10.3324/haematol.2011.043919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashidate T, Murakami N, Nakagawa M, et al. AML1 enhances the expression of leukotriene B4 type-1 receptor in leukocytes. FASEB J. 2010;24(9):3500–3510. doi: 10.1096/fj.10-156844. Prepublished on 2010/04/17 as DOI 10.1096/fj.10-156844. [DOI] [PubMed] [Google Scholar]
- 49.Lukin K, Fields S, Lopez D, et al. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc Natl Acad Sci U S A. 2010;107(17):7869–7874. doi: 10.1073/pnas.1003525107. Prepublished on 2010/04/14 as DOI 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene. 2005;24(11):1873–1881. doi: 10.1038/sj.onc.1208404. Prepublished on 2005/02/03 as DOI 10.1038/sj.onc.1208404. [DOI] [PubMed] [Google Scholar]
- 51.Swirski FK, Nahrendorf M, Etzrodt M, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325(5940):612–616. doi: 10.1126/science.1175202. Prepublished on 2009/08/01 as DOI 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokomizo T, Hasegawa K, Ishitobi H, et al. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111(8):4075–4080. doi: 10.1182/blood-2007-05-091637. Prepublished on 2008/02/06 as DOI 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- 53.Choi Y, Elagib KE, Delehanty LL, Goldfarb AN. Erythroid inhibition by the leukemic fusion AML1-ETO is associated with impaired acetylation of the major erythroid transcription factor GATA-1. Cancer Res. 2006;66(6):2990–2996. doi: 10.1158/0008-5472.CAN-05-2944. Prepublished on 2006/03/17 as DOI 10.1158/0008-5472.CAN-05-2944. [DOI] [PubMed] [Google Scholar]
- 54.Old JM. Screening and genetic diagnosis of haemoglobin disorders. Blood Rev. 2003;17(1):43–53. doi: 10.1016/s0268-960x(02)00061-9. Prepublished on 2002/12/20 as DOI. [DOI] [PubMed] [Google Scholar]
- 55.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103(7):2522–2529. doi: 10.1182/blood-2003-07-2439. Prepublished on 2003/11/25 as DOI 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 56.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104(12):3565–3572. doi: 10.1182/blood-2003-12-4349. Prepublished on 2004/08/07 as DOI 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 57.Zaidi SK, Javed A, Pratap J, et al. Alterations in intranuclear localization of Runx2 affect biological activity. J Cell Physiol. 2006;209(3):935–942. doi: 10.1002/jcp.20791. Prepublished on 2006/09/15 as DOI 10.1002/jcp.20791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.