Summary
Type III protein secretion systems (T3SS), which have evolved to deliver bacterial proteins into nucleated cells, are found in many species of Gram-negative bacteria that live in close association with eukaryotic hosts. Proteins destined to travel this secretion pathway are targeted to the secretion machine by customized chaperones, with which they form highly ordered complexes. Here, we have identified a mechanism that coordinates the expression of the Salmonella Typhimurium T3SS chaperone SicP and its cognate effector SptP. Translation of the effector is coupled to that of its chaperone, and in the absence of translational coupling, an inhibitory RNA structure prevents translation of sptP. Furthermore, we have found that translational coupling is essential for secretion-competent SicP/SptP complex assembly. The data presented here show how the genomic organization of functionally related proteins can have a significant impact on protein function.
Keywords: bacterial pathogenesis, gene regulation, protein secretion, Salmonella Typhimurium, translational coupling
Introduction
In bacterial genomes, genes encoding proteins that interact or engage in the same functional pathway tend to cluster (Demerec & Hartman, 1959). The evolutionary pressure that drives bacterial gene clustering remains somewhat unclear (Fang et al., 2008, Lawrence & Roth, 1996), but recent evidence shows that prokaryotic mRNAs do not disperse after transcription (Montero Llopis et al., 2010). These data suggest that chromosomal organization determines protein localization immediately upon translation, and this pre-translational localization may facilitate interactions between functional partners. Here, we present an example of extreme coupling: a pair of adjacent, interacting proteins coupled at the transcriptional, translational, and post-translational levels. We believe that this coupling reflects the intimate nature of the interaction between the two proteins, which function in the context of a Salmonella enterica serovar Typhimurium type III secretion system (T3SS) (Galán, 2001).
The T3SS is a bacterial nanomachine used by many Gram-negative bacteria to translocate bacterial “effector” proteins into a eukaryotic host cell (Galan & Wolf-Watz, 2006, Cornelis, 2006). Once delivered, effector proteins can modulate host responses to create an appropriate niche for bacterial survival and replication within the host. The T3SS consists of a structure called the needle complex, which is made of a multi-ring base that spans the bacterial envelope and a needle-like extension that protrudes several nanometers from the bacterial surface (Kubori et al., 1998). A narrow channel, only ~28 Å wide, traverses the entire structure and provides the route by which unfolded protein substrates travel the T3SS pathway (Kubori et al., 1998, Blocker et al., 2001, Marlovits et al., 2004). Evolutionarily related to the flagella, the T3SS is conserved amongst many Gram-negative bacteria that have a symbiotic or pathogenic relationship with a eukaryotic host.
Proteins destined to travel the T3SS pathway are targeted to the secretion machine by multiple signals to ensure specificity. One such signal is encoded within the first 20-30 amino acids of the secreted protein (Sory et al., 1995). The other signal is provided by customized chaperones, which bind significant portions of the amino terminus of the cognate secreted protein (Feldman & Cornelis, 2003). In addition to targeting, chaperones have also been hypothesized to facilitate effector secretion by holding the effector in a partially unfolded conformation, which primes it for insertion into the narrow channel formed by the T3SS apparatus (Stebbins & Galan, 2001). In the absence of their cognate chaperones, effector proteins can be nonspecifically secreted through the evolutionarily related flagellar apparatus (Lee & Galan, 2003) or rapidly degraded (Fu & Galan, 1998).
Since the presence of a chaperone is essential for effector stability and secretion, assembly and disassembly of the chaperone-effector complexes are likely to be tightly regulated to promote appropriate expression and secretion of effector proteins. However, differences amongst T3SS chaperone/effector pairs suggest that tight co-regulation may be more essential for some pairs than for others. T3SS chaperones can be roughly divided into at least two functional groups that have qualitatively different interactions with their respective effector proteins (Page & Parsot, 2002). The first group consists of chaperones that bind and deliver multiple effector proteins. For instance, in the Gram-negative bacterium S. Typhimurium, T3SS chaperone InvB interacts with four effectors (Lee & Galan, 2003, Bronstein et al., 2000, Ehrbar et al., 2004). Crystallographic evidence shows that multi-effector chaperones have limited interactions with their binding partners (Lilic et al., 2006). In contrast, other T3SS chaperones bind only one effector. In S. Typhimurium, chaperones SicP and SigE/PipC fall into this second category (Fu & Galan, 1998, Darwin et al., 2001). Existing data indicate that these uni-effector chaperones have a much more intimate relationship with their cognate effectors than do multi-effector chaperones (Stebbins & Galan, 2001, Lilic et al., 2006). In contrast to multi-effector chaperones, the crystal structures of uni-effector chaperones bound to their effector proteins show that the chaperone-binding domain of the effector protein wraps around the small, globular chaperone in an unfolded conformation to form a very stable complex (Stebbins & Galan, 2001). The elaborate nature of the binding interactions formed between these chaperone/effector pairs, in combination with the fact that these effectors are often degraded in the absence of their chaperones, suggests that regulation may be required to ensure that a chaperone is in position to capture the nascent effector as it is translated. Interestingly, genes encoding uni-effector chaperones are usually encoded adjacent to their cognate effectors, presumably within the same operon. This operonic arrangement of the genes ensures that their expression is co-regulated at the level of transcription.
Previous work suggested that expression of the S. Typhimurium T3SS effector protein SptP may be coordinated with the expression of its uni-effector chaperone SicP at a post-transcriptional level as well. Previous data revealed that in the absence of SicP, SptP is not only rapidly degraded, but also its expression is drastically reduced despite normal levels of transcription (Fu & Galan, 1998). These preliminary experiments suggest a post-transcriptional regulatory mechanism that would ensure the coordinated translation of SicP and SptP so that the complex is assembled immediately after synthesis, both to prevent the premature degradation of SptP and perhaps to topologically position it for secretion. Here, we demonstrate that expression of sicP is translationally coupled to expression of sptP. This coupling is tightly regulated by an mRNA structure, and moreover, translational coupling is required for efficient assembly of a secretion competent SicP/SptP complex. Together, these data describe a finely tuned regulatory relationship between two proteins that function together.
Results
Expression of the chaperone SicP is required for wild type expression of its cognate effector SptP
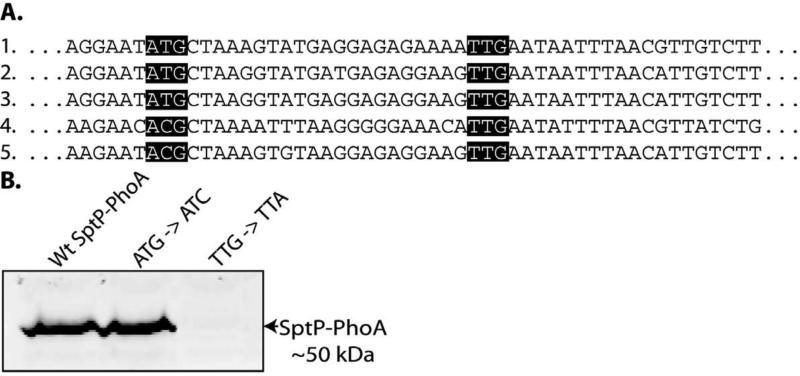
Since previous data suggested a potential post-transcriptional regulatory mechanism affecting the expression of SptP, we examined the effect of SicP on SptP translation. We first sought to experimentally determine the actual translational start site of sptP. An alignment of sptP sequence from S. Typhimurium strain LT2 with sptP sequences from several other Salmonella enterica strains revealed that the annotated ATG start codon is not conserved. In contrast, an alternative TTG start codon located 24 nucleotides downstream is conserved in all of the strains (Figure 1a). To determine which start codon is used, we mutated the ATG to ACG and observed that the reporter was still expressed. However, when the downstream potential start codon was changed from TTG to TTA, reporter expression was abolished (Figure 1b). These results indicate that the downstream start codon is the actual translational start site. Amino-terminal sequence analysis of both intracellular and secreted SptP yielded the peptide MNNLTLSS, which is consistent with the mutagenesis analysis and confirms the TTG codon as the functional start codon.
Fig. 1.
Identification of the sptP translation start codon. (A) Alignment of sptP nucleotide sequence from the following sequenced Salmonella enterica strains: 1) S. Typhimurium LT2; 2) S. Typhi Ty2 and CT18; 3) S. Paratyphi A strain AKU_12601 and ATCC 9150; 4) S. Salamae Sofia; and 5) S. Arizonae serovar 62:z4,z23. (B) Whole cell lysates of strains carrying the indicated changes to the putative sptP start sites in the context of a strain encoding a sptP::phoA translational reporter were analyzed by western immunoblot with an anti-PhoA antibody.
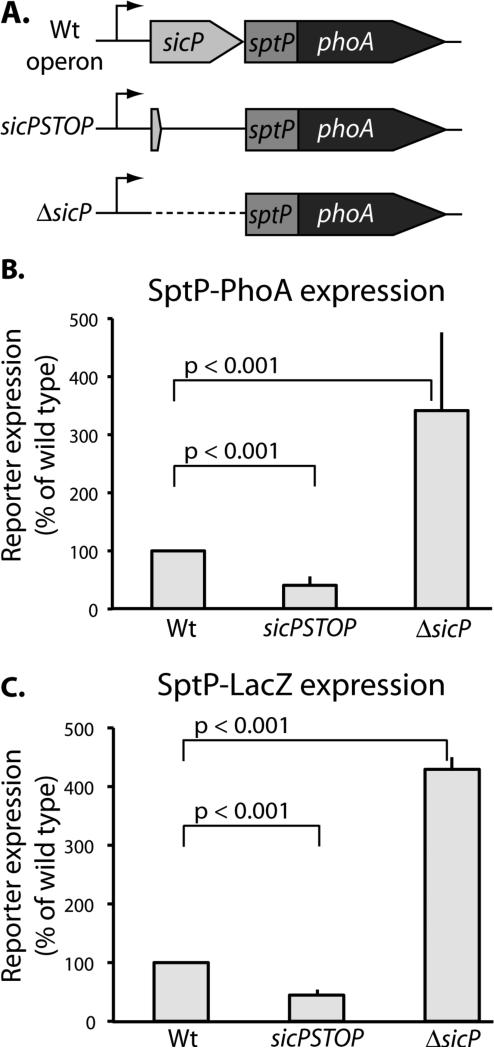
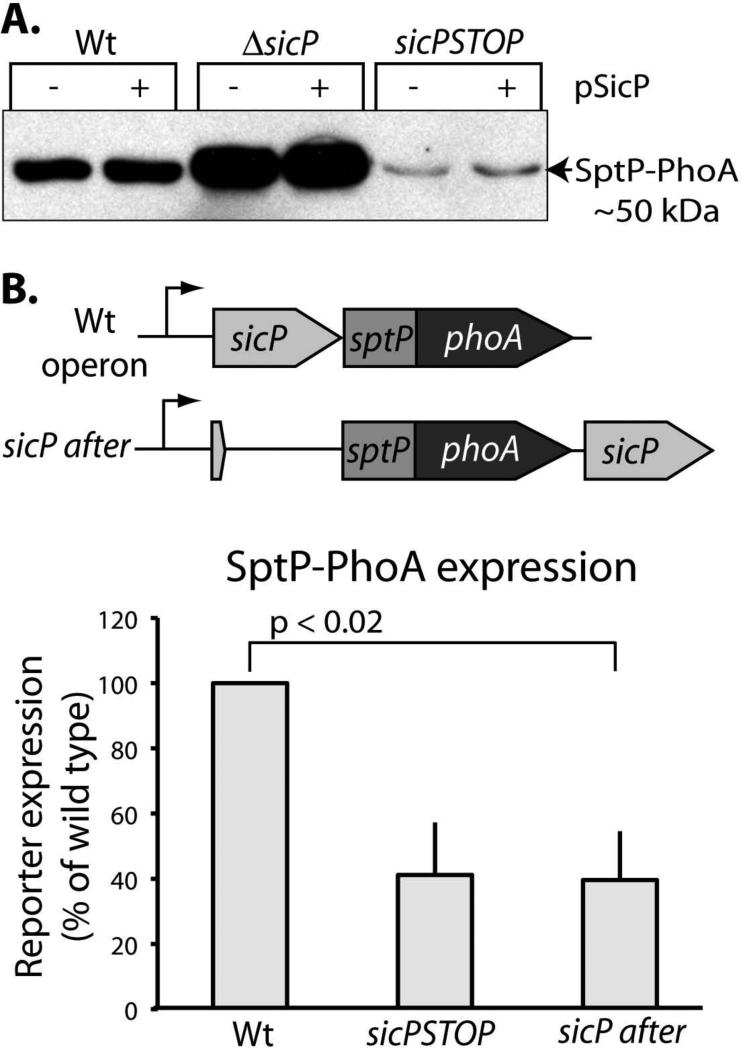
To assess the effect of SicP on SptP translation we constructed a translational fusion reporter gene by linking sptP to phoA. This fusion retains the first 27 amino acids of SptP, which comprise the N-terminal T3SS signal sequence, and replaces the chaperone-binding and catalytic domains with the reporter protein PhoA. Unlike wild type SptP, this reporter is not subject to degradation in the absence of SicP since the construct does not include the chaperone-binding domain of SptP, which de-stabilizes the protein in the absence of SicP. To determine whether SicP regulates sptP translation, we introduced into this reporter background mutations in sicP termed “ΔsicP” and “sicPSTOP” and recombined them into the S. Typhimurium chromosome so that they are expressed in the appropriate context. In the ΔsicP mutant, the sicP coding sequence was eliminated entirely. In the sicPSTOP mutant, a stop codon was introduced into the sicP sequence so that SicP could no longer be produced, but any potential regulatory regions upstream of sptP and within the sicP coding sequence were left intact. We found that in the ΔsicP mutant, the sptP reporter levels increased with respect to wild type. In contrast, reporter levels decreased with respect to wild type in the sicPSTOP construct (Figure 2b). We confirmed these observations with an enzymatic activity assay by making equivalent constructs in which phoA was replaced by lacZ as a reporter gene (Figure 2c). Together, these data suggest that expression of sicP is required for wild type levels of sptP expression.
Fig. 2.
Expression of the chaperone SicP is required for wild type expression of the effector SptP. (A) Schematic representation of the sptP::phoA translational reporter constructs. (B) Whole cell lysates of the indicated strains were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Results represent the average of three independent experiments. Student's t test was used to determine the p values. (C) Strains carrying constructs equivalent to those described in (A) in which sptP::phoA was replaced with sptP::lacZ, were analyzed by Miller assays. LacZ activity was quantified and normalized to wild type. Results represent the average of three independent experiments. Student's t test was used to determine the p values.
Nucleotide sequences upstream of the sptP start codon negatively regulate sptP expression
Since a deletion of sicP resulted in increased reporter expression while a nonsense mutation in sicP resulted in decreased reporter expression, we hypothesized that nucleotide sequences upstream of sptP may repress translation. We therefore constructed a series of deletion mutants in order to define the minimum sequence upstream of sptP required to inhibit sptP translation in the absence of sicP expression (Figure 3a). We found that repression of sptP translation was maintained when at least 49 nucleotides of upstream sequence were present. However, in a larger deletion where only 31 nucleotides of upstream sequence were left, a marked increase in the translation of the sptP::phoA reporter fusion was observed. These data indicate the presence of regulatory elements within the sequences immediately upstream of the start codon of sptP (Figure 3b).
Fig. 3.
A nucleotide region directly upstream of sptP inhibits expression of sptP in the absence of sicP expression. (A) Schematic representation of the sicP deletion mutants. The nucleotides deleted within the sicP coding sequence are indicated. The starting point of all deletions was 14 nucleotides before the sicP start codon (−14) so as to prevent the initiation of sicP translation in these mutants. (B) Whole cell lysates were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Results represent the average of at least three independent experiments. Student's t test was used to determine p values. When compared to sicPSTOP, p<0.01 only for mutants ΔsicPΔ14-372 and ΔsicPΔ14-354 (indicated by *).
sptP translation is repressed by an mRNA structure in the region of its translation start site
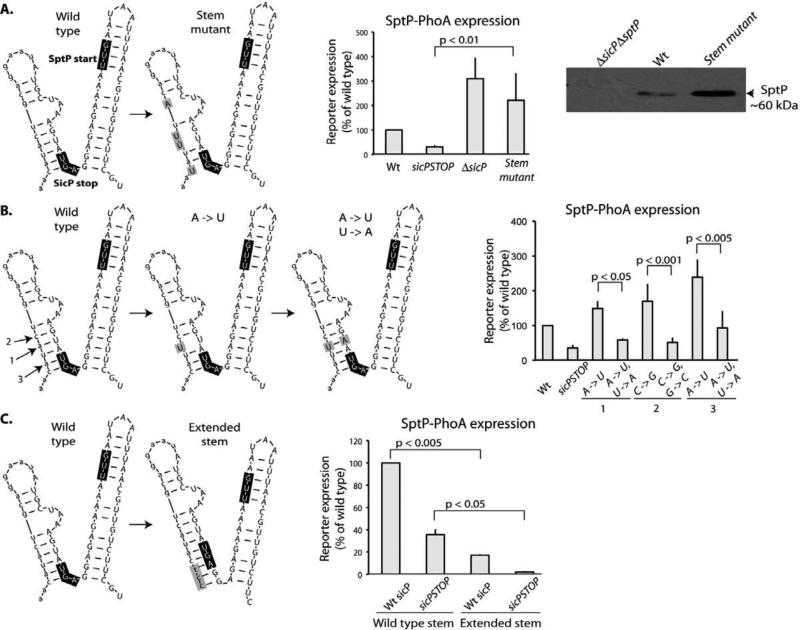
The experiments described above showed that the sequence immediately upstream of sptP is able to inhibit translation of sptP::phoA. We hypothesized that this regulation is due to the effects of an inhibitory RNA secondary structure. The RNA folding program mFOLD (Zuker, 2003) predicted two stem-loop structures in the region around the sptP ribosome-binding site and start codon (Figure 4a). Since the downstream stem-loop identified by mFOLD encompasses the start codon and putative ribosome-binding site of sptP, we hypothesized that this structure could prevent recognition of the translational initiation signal by ribosomes and thereby prevent translation. However, disruption of the predicted structure by the introduction of mutations that should destabilize the predicted stem-loop did not alter sptP::phoA translation (Figure S1). We then tested the potential role of the adjacent, upstream stem-loop in the regulation of sptP translation by introducing nucleotide mutations predicted to disrupt this stem-loop (Figure 4a). In a sicPSTOP background, these mutations resulted in a marked increase in reporter expression (Figure 4a). When the same four-nucleotide changes, which only alter nucleotide sequence and do not alter SicP amino acid sequence, were introduced into a wild type background, we also observed an increase in the SptP levels (Figure 4a). These results confirm that the nucleotide sequences upstream of sptP are important for sptP expression.
Fig. 4.
An mRNA structure inhibits translation of sptP. (A) The mutations indicated in the schematic representation (in grey boxes) of secondary mRNA structure surrounding the sptP start codon (as predicted by mFOLD) were introduced into a strain encoding a sptP::phoA reporter and whole cell lysates were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Additionally, the same mutations were introduced into a wild type strain and whole cell lysates were analyzed by western immunoblot with an anti-SptP antibody. (B) The mutations indicated in the schematic representation (in grey boxes) predicted to disrupt (A -> U) or restore (A -> U/U-> A) base complementarity in the predicted stem surrounding the sptP start codon were introduced into a background strain encoding a sptP::phoA reporter. Whole cell lysates of these strains were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values. (C) The indicated mutations to lengthen the predicted stem surrounding the sptP start codon were introduced into a background strain encoding a sptP::phoA reporter. Whole cell lysates of these strains were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values.
To further probe the RNA secondary structure(s) involved in this regulation, we introduced a pair of mutations designed to verify the existence of the upstream stem-loop predicted by mFOLD (Figure 4b). One nucleotide mutation (A -> U) in the stem-loop was predicted to destabilize the structure by disrupting base pairing in the stem, and the introduction of this single mutation in a sicPSTOP background resulted in an increase in sptP::phoA reporter levels. Furthermore, a mutation (U -> A) predicted to restore base complementarity in the mutated stem was shown to restore inhibition of translation (Figure 4b). We examined two additional sets of complementary base pair mutations and observed similar results (Figure 4b). Therefore, we conclude that the upstream stem-loop identified by mFOLD can inhibit sptP translation in the absence of sicP expression and that translation of sicP may de-stabilize the regulatory stem-loop so that the ribosome can gain access to the translation initiation site.
The data presented above show that the upstream stem-loop depicted in Figure 4 is required for the downregulation of translation of sptP in the absence of sicP expression. However, since we observed a low level of translation even when the stem-loop is intact, we hypothesized that levels of translation would be even lower if the stem were stronger. In order to test this prediction, three unpaired nucleotides upstream of the stem were mutated so that they could base pair with the three nucleotides just downstream of the stem (Figure 4c). As the model predicts, a stronger stem resulted in reduced levels of sptP::phoA translation with respect to a wild type stem in both the wild type sicP and sicPSTOP backgrounds (Figure 4c). Therefore, the strength of the stem correlates inversely with levels of translation of sptP.
The translational regulation of sptP in the absence of sicP expression is independent of other components or regulators of the T3SS
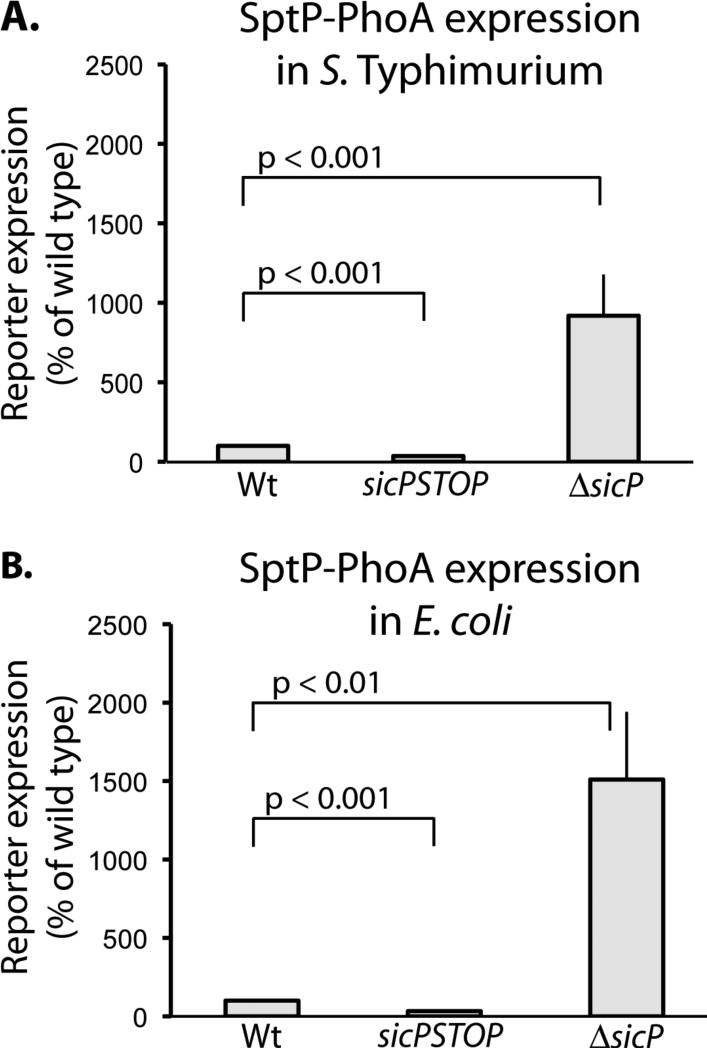
We tested whether other components of the T3SS were required for the translational regulation of sptP expression by introducing into E. coli, which lacks this system, a plasmid encoding sptP::phoA preceded by wild type sicP, sicPSTOP, or ΔsicP. We observed that the translational regulation of sptP::phoA in this background mimicked that observed in S. Typhimurium (Figure 5). Therefore, we conclude that no additional T3SS proteins are required for the translational regulation of sptP.
Fig. 5.
sicP is the only T3SS-specific factor necessary for sicP-expression-dependent regulation of sptP translation. (A) Whole cell lysates of S. Typhimurium bearing plasmids expressing wild type sicP sptP::phoA, sicPSTOP sptP::phoA, and ΔsicP sptP::phoA were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values. (B) Whole cell lysates of E. coli bearing the same plasmids were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values.
Expression of sicP promotes sptP expression only from its native locus
We have identified nucleotide sequences and a putative mRNA structure that are able to repress sptP translation in the absence of sicP expression. We have also shown that sptP translation is dependent upon wild-type sicP expression independently of any other T3SS-specific factors. Next, we examined how expression of sicP from its wild type locus increases sptP translation. In a sicPSTOP background, expressing sicP in trans from a low-copy plasmid did not restore expression of sptP::phoA to wild type levels (Figure 6a). Since levels of SicP expressed from a plasmid with an exogenous promoter differ from wild type levels of SicP, we expressed sicP within the sicPsptP operon on the chromosome downstream from the sptP::phoA reporter gene in an attempt to achieve complementation (Figure 6b). However, in this strain, wild type levels of SptP::phoA reporter expression were not restored (Figure 6c). These results indicate that the SicP protein itself is unlikely to mediate the disruption of the stem-loop structure and promote sptP expression. Therefore, it may not be SicP itself that regulates sptP expression, but rather it may be the translation of sicP just upstream of sptP that is able to promote sptP translation.
Fig. 6.
Providing SicP in trans is not sufficient to promote wild type levels of SptP expression. (A) Whole cell lysates of the indicated strains with or without a plasmid expressing SicP were analyzed by western immunoblot with an anti-PhoA antibody. (B) Whole cell lysates of strains with sicP placed either upstream or downstream of the sptP::phoA reporter (as indicated in the diagram) were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Student's t test was used to determine p values.
Expression of sptP is translationally coupled to the expression of sicP
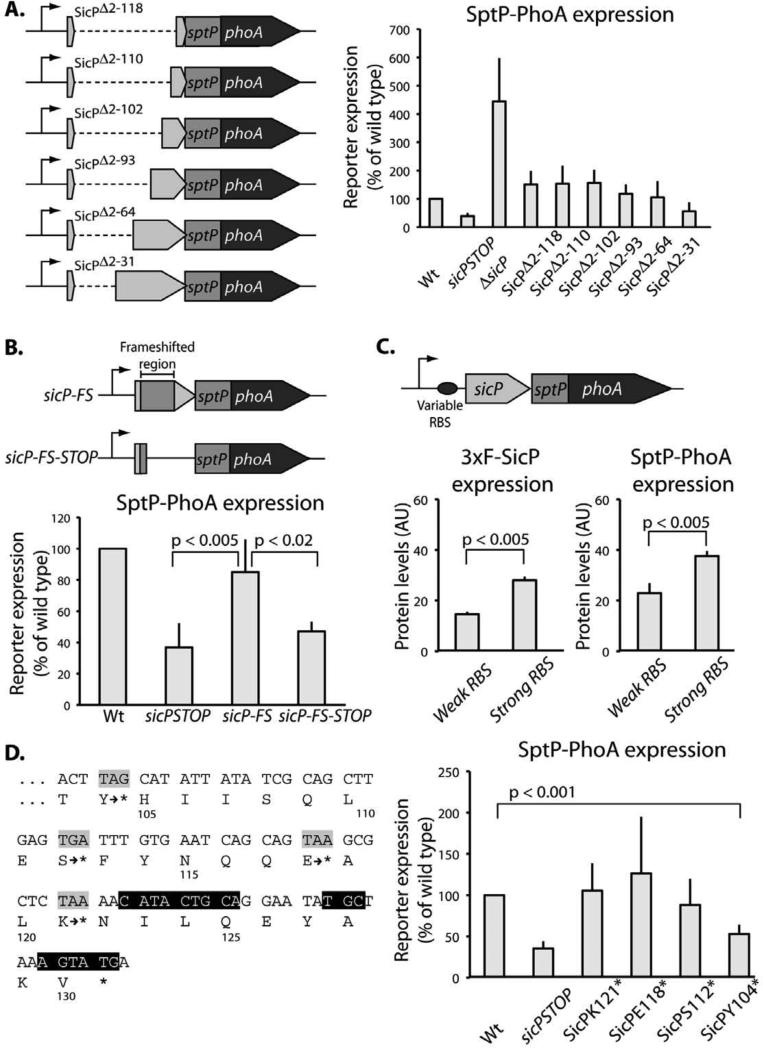
The arrangement of sicP and sptP coding sequences in a single operon with adjacent start and stop codons strongly suggests that translational coupling could account for the positive effect of sicP expression on sptP translation. To test this possibility, mutants containing in-frame deletions of sicP were constructed (Figure 7a). These mutants do not produce a functional chaperone but do maintain translation through the start codon of sptP. We observed that the sptP::phoA reporter fusion was expressed at levels comparable to wild type even when significant portions of upstream sequence were absent (Figure 7a). To minimize disruptions to the native mRNA structure, we designed a frameshifted mutant of sicP. With only a few changes to the nucleotide sequence, this construct does not produce a functional chaperone although it maintains the translation of an unrelated polypeptide through the start codon of sptP, and preserve the distance between the sptP coding sequence and its putative transcription start site (Figure 7b). In this mutant background, wild type levels of SptP::phoA expression were observed. When the translation of the upstream cistron of this mutant was abrogated by the introduction of a stop codon, SptP::phoA levels were significantly reduced (Figure 7b). These results suggest that translation of the upstream open reading frame is required for efficient translation of the downstream open reading frame, which is consistent with translational coupling. To further demonstrate translational coupling between sicP and sptP, we replaced the native ribosome-binding site of sicP with ribosome binding sites of known relative strength (Lovdok et al., 2009) (Figure 7c). We found that increasing the rate of translation of sicP correspondingly increased the rate of translation of sptP::phoA (Figure 7c). Together, these data indicate that sptP is translationally coupled to sicP.
Fig. 7.
Expression of the effector SptP is translationally coupled to expression of the chaperone SicP. (A) Whole cell lysates of strains carrying the indicated chromosomally encoded sicP deletions in the context of the sptP::phoA background were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Results represent the average of at least three independent experiments. The difference in reporter expression between the mutants and wild type was significant (p<0.05, Student's t test) only for the mutants sicPSTOP, ΔsicP, and ΔSicPΔ 2-31. (B) Whole cell lysates of strains carrying the indicated chromosomally-encoded frame-shifted sicP mutants in the context of the sptP::phoA background were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Results represent the average of at least three independent experiments, and p values were determined by the Student's t test. (C) Strains were constructed in which the ribosome binding site (RBS) of sicP was changed to RBSs of defined strength (“weak” and “strong”). Mutations were introduced in the context of a strain that expresses 3xFLAG epitope tagged SicP and carries a sptP::phoA reporter. Whole cell lysates were analyzed by western immunoblot with an anti-FLAG or an anti-PhoA antibody and the signals were quantified. Results represent the average of at least three independent experiments. Student's t test was used to determine p values. (D) Stop codons were introduced at codons 121, 118, 112, or 104 of sicP (corresponding to 2, 11, 29, and 53 nt upstream of the predicted stem) in the context of a strain carrying a sptP::phoA reporter fusion. Black highlighting indicates nucleotides base-paired in the stem. Whole cell lysates were analyzed by western immunoblot with an anti-PhoA antibody. SptP::phoA reporter signal was quantified and normalized to wild type. Results represent the average of at least three independent experiments. Only the mutants sicPSTOP and SicPY104* are significantly different from wild type (Wt). Student's t test was used to determine p values.
These data are consistent with a model in which translation of sicP disrupts the stem in the translation initiation region of sptP so that ribosomes can gain access and translate the downstream open reading frame. If this model is correct, then prematurely halting translation of sicP before the ribosome reaches the stem should result in inefficient translation of sptP::phoA. Accordingly, premature stop codons were introduced into sicP at various points upstream of the mRNA stem-loop structures. A stop codon 53 nucleotides upstream of the two stem-loop structures phenocopies a mutant in which there is no translation of the open reading frame upstream of sptP::phoA, while stop codons within at least 29 nucleotides of the mRNA structures result in wild type levels of SptP::phoA expression (Figure 7d). Since the footprint of a ribosome is ~30 nucleotides (Steitz, 1969), these results are consistent with the proposed model. These data also suggest that there are no other components of the structure involved in regulation upstream of the putative stem defined in Figure 4.
Translational coupling of sicP and sptP is required for optimal secretion of SptP
In order to evaluate the impact of translational coupling on SicP/SptP function, we uncoupled expression of the chaperone from expression of the wild type effector by using the frameshifted sicP mutant described previously (see Fig. 7b). Although this mutant does not produce a functional chaperone, it maintains translation through the start codon of sptP (Figure 7c) resulting in wild type expression of sptP as demonstrated using a reporter fusion (Fig. 7d). The SicP-coding region preceded by its native RBS was then inserted after sptP, therefore in this construct the chaperone is still expressed from the same message as its cognate effector (Figure 8). Binding of SicP to SptP influences two measurable aspects of the effector's SptP fate: its half-life and its secretion. Therefore, to assess the functional importance of translational coupling on SptP's fate we measured both SptP levels in the bacterial cytoplasm and its secretion into culture supernatants. We observed wild type levels of SptP in the bacterial cytoplasm, which indicates that the chaperone is able to bind the effector efficiently in a manner that stabilizes SptP. In contrast, we observed a significant decrease in SptP secretion into culture supernatants when translation of the effector is uncoupled from the translation of its chaperone SicP (Figure 8b). Therefore, these data indicate that despite wild type levels of SptP present in the bacterial cytoplasm, coupled expression of SicP and SptP is required for maximal secretion of the effector.
Fig. 8.
Translational coupling of sicP and sptP is required for optimal secretion of SptP. Whole cell lysates and culture supernatants of the indicated strains were analyzed by Western immunoblot using an antibody against SptP. SptP signal was quantified and normalized to wild type. Results represent the average of at least three independent experiments. Student's t test was used to determine p values.
Discussion
We have shown here that production of the T3SS chaperone SicP and its cognate effector SptP are intimately co-regulated. We have identified an mRNA sequence that folds into an inhibitory structure and prevents translation of sptP in the absence of sicP expression. Furthermore, we have demonstrated that translation of sicP, which is encoded immediately upstream of sptP, is sufficient to relieve the inhibition of sptP translation. Importantly, we have shown that translational coupling of sicP and sptP is required for efficient secretion of the effector.
When sicP is not actively translated, a region of the sicP/sptP mRNA surrounding the sptP start codon and putative ribosome-binding site inhibits the translation of sptP. This segment of mRNA is predicted to fold into two adjacent stem-loop structures, one of which is directly upstream of the sptP start site and another which encompasses the start site and the putative ribosome-binding site. Consistent with the inhibitory role of this mRNA structure, nucleotide mutations predicted to alter base-pairing and disrupt the first stem-loop increased the translation of sptP. Although mRNA folding decreases translation of sptP, translation of sicP (or an unrelated polypeptide expressed in its place) can overcome this inhibition and promote the translation of sptP presumably by melting the mRNA structure and rendering the translational start site of sptP accessible to ribosomes. This process, termed translational coupling, is a strategy used in many prokaryotic operons to coordinate expression of functionally related proteins (reviewed in (McCarthy & Gualerzi, 1990). In the case of SicP and SptP, we have found that translational coupling is required for the efficient formation of a secretion-competent SicP/SptP complex. When SicP was produced from the same transcript as SptP but with their translation decoupled, SptP was not efficiently secreted into culture supernatants.
Efficient translocation of effector proteins into eukaryotic host cells is essential for the survival and replication of bacteria encoding T3SSs. These effector proteins must encounter and bind their cognate chaperone proteins not only to reach the T3SS machine, but also to avoid degradation within the bacterial cytoplasm. Here we present an example of a chaperone/effector pair whose co-expression is assured by a combination of regulatory mechanisms. Since production of the chaperone is a pre-requisite for effector translation, a high degree of efficiency is built into the system. When the effector is translated, a chaperone will already be present to bind the nascent effector, perhaps even during its translation, and thus prevent effector degradation and promote its secretion.
Intriguingly, although we observed a decrease in the secretion of SptP when its translation was uncoupled from SicP, we did not observe a decrease in SptP levels in the bacterial cytoplasm. These results imply that translational coupling of SicP and SptP is not necessarily required for efficient capture of the effector by the chaperone. If the chaperone were unable to bind the effector, the effector would be degraded and cytoplasmic levels of SptP would decline. Given that cytoplasmic levels of SptP are the same with and without translational coupling, a simple increase in SptP degradation cannot explain reduced SptP secretion. Therefore, translational coupling must affect chaperone/effector complex assembly such that complexes resulting from co-translation are more competent for secretion than others.
How could translational coupling make a chaperone/effector complex more competent for secretion? It is possible that a complex formed during coupled expression could be structurally distinct from a complex formed from proteins whose expression is uncoupled. The crystal structure of the SicP/SptP complex reveals a dimer-of-dimers consisting of four SicP and two SptP molecules (Stebbins & Galan, 2001). The chaperone-binding domains of the two SptP molecules each wrap around a dimer of SicP molecules and also make additional contacts on a third SicP molecule such that the two SicP dimers are held in close proximity. It is not known whether the dimer-of-dimers is a physiologically relevant structure or merely an artifact of crystallization, although by gel filtration, purified SicP/SptP complexes can be observed in both a 2:1 and a 4:2 ratio (Stebbins & Galan, 2001). It is possible that both complexes are present within the bacterial cytoplasm. Perhaps the two complexes are not equally efficient at targeting SptP to the T3SS machine for secretion. Translational coupling of the chaperone and effector protein may promote assembly of the complex that most efficiently brings SptP to the T3SS machine. However, it is also possible that the defect in secretion caused by uncoupling chaperone and effector translation could be caused by differences in the kinetics and location of complex assembly. Perhaps translational coupling promotes faster complex assembly by ensuring that sufficient amounts of chaperone protein are synthesized and poised in the right location to capture the nascent effector as it is translated. Uncoupling production of the proteins might slow down complex assembly just enough to handicap SicP/SptP complexes in the “race to the T3SS machine”. The recently discovered T3SS cytoplasmic “sorting platform” that is thought to organize secretion substrates prior to secretion, and thereby imposes a hierarchy on secretion, has been shown to load with different classes of secretion substrates at different stages of T3SS function (Lara-Tejero et al., 2011). In the absence of a class of secretion substrates that would normally be given priority, the platform is loaded instead with substrates belonging to a class that is supposed to be translocated later. Type III secretion-associated chaperones were shown to be essential for platform loading. It is not known whether preference for loading is assigned based on differential affinities of chaperone/substrates complexes for the platform, or whether it is loaded on a “first-come-first-served” basis. If the latter is true, then a small delay in SicP/SptP complex assembly could conceivably result in a large secretion defect as other secretion substrates could enter the platform first and thereby prevent access to “latecomers”. More studies will be required to clarify these issues.
Here, we have explored the regulation coupling expression of T3SS chaperone SicP and its cognate effector SptP. Coupling of their translation is required for maximally efficient secretion of SptP, and we have demonstrated that even a slight perturbation in genomic organization can significantly impact protein secretion. Few examples show at a mechanistic level how genome organization translates into an evolutionary advantage at the level of protein function, even though genomic approaches have shown that clustering of functionally related genes is ubiquitous throughout disparate bacterial genomes. These data highlight the impact of gene location by demonstrating that the location of sicP relative to sptP is critically important for protein secretion.
Experimental Procedures
Bacterial strains, plasmids, chemicals, reagents and culture conditions
All strains used in this study, unless otherwise noted, are derivatives of Salmonella enterica serovar Typhimurium SL1344 (Hoiseth & Stocker, 1981), and strains are listed in Supplementary Table 1. Genetic modifications were introduced into the bacterial chromosome by allelic exchange using an R6K suicide vector as previously described (Kaniga et al., 1994). All plasmids were constructed using standard recombinant DNA techniques (detailed in Supplementary Methods) and are listed in Supplementary Table 2. Primers used to introduce mutations by fusion PCR are listed in Supplementary Table 3. Strains were grown at 37°C in Luria broth supplemented with either 0.3M NaCl to induce expression of the T3SS, 0.1% arabinose (strains carrying pSB3292), or 50 μM isopropyl ß-D-1-thiogalactopyranoside (strains carrying plasmids derived from pWSK29). Strains were grown in the presence of antibiotics at the following concentrations, when appropriate: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; kanamycin, 50 μg ml−1; tetracycline, 12.5 μg ml−1; streptomycin, 100 μg ml−1. All chemicals and reagents, unless otherwise noted, were from Sigma-Aldrich (Saint Louis, MO, USA).
Western immunoblot analysis
Levels of SptP and SptP::phoA reporter fusions in bacterial cell lysates were determined by immunoblot analysis. Samples were grown to late logarithmic phase, normalized by OD600, separated by SDS-PAGE on a 10% gel, and transferred to either PVDF (Pall, Pensacola, FL, USA) or nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). SptP and PhoA were detected using monoclonal antibodies directed to the respective proteins followed by anti-mouse and anti-rabbit infrared fluorescent (emission 800 nm) secondary antibodies, respectively (Pierce, Rockford, IL, USA). Fluorescent signal was detected and quantified using the LI-COR Odyssey Infrared Imaging system (LI-COR Biosciences, Lincoln, NE, USA), which allows for accurate quantification of protein signal over a wide linear range.
Miller assays
Strains were grown to late logarithmic phase. β-galactosidase assays were performed as previously described by J.H. Miller (Miller, 1972).
In vitro secretion assays
Strains were grown to late logarithmic phase in Luria broth supplemented with 0.3M NaCl. Secreted proteins were isolated from 5 ml of culture supernatant as previously described (Kaniga et al., 1995).
Amino Terminal Sequencing of SptP
A S. Typhimurium strain expressing SptP 3xFLAG-epitope-tagged at its C-terminus expressed from its native chromosomal locus was grown to late log phase under T3SS-inducing conditions. SptP-3xFLAG was immunoprecipitated from both whole cell lysates and culture supernatants using ANTI-FLAG M2 Affinity Gel according to the manufacturer's instructions (Sigma, Saint Louis, MO, USA). Immunoprecipitates were subjected to SDS-PAGE and transferred to a PVDF membrane (Pall, Pensacola, FL, USA). The band corresponding to SptP was excised from the membrane and sent to the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (http://medicine.yale.edu/keck/index.aspx) for Edman degradation.
Supplementary Material
Acknowledgements
We thank Ron Breaker and Christine Jacobs-Wagner for useful discussions, and members of the Galán laboratory for critical reading of this manuscript. This work was supported by NIH Grant AI30492 to J. E. G.
References
- Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Molecular microbiology. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- Bronstein PA, Miao EA, Miller SI. InvB is a type III secretion chaperone specific for SspA. Journal of bacteriology. 2000;182:6638–6644. doi: 10.1128/jb.182.23.6638-6644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Robinson LS, Miller VL. SigE is a chaperone for the Salmonella enterica serovar Typhimurium invasion protein SigD. Journal of bacteriology. 2001;183:1452–1454. doi: 10.1128/JB.183.4.1452-1454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M, Hartman PE. Complex loci in microorganisms. Annual Review of Microbiology. 1959;13:377–406. [Google Scholar]
- Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. InvB is required for type III - dependent secretion of SopA in Salmonella enterica serovar Typhimurium. Journal of bacteriology. 2004;186:1215–1219. doi: 10.1128/JB.186.4.1215-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Rocha EP, Danchin A. P ersistence drives gene clustering in bacterial genomes. BMC genomics. 2008;9:4. doi: 10.1186/1471-2164-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Cornelis GR. The multitalented type III chaperones: all you can do with 15 kDa. FEMS microbiology letters. 2003;219:151–158. doi: 10.1016/S0378-1097(03)00042-9. [DOI] [PubMed] [Google Scholar]
- Fu Y, Galan JE. Ident ification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. Journal of bacteriology. 1998;180:3393–3399. doi: 10.1128/jb.180.13.3393-3399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE. Salmonella interaction with host cells: Type III Secretion at Work . Annu. Rev. Cell Dev. Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Bossio JC, Galan JE. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Molecular microbiology. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Tucker S, Trollinger D, Galan JE. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. Journal of bacteriology. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A Sorting Platform Determines the Order of Protein Secretion in Bacterial Type III Systems. Science (New York, N.Y. 2011 doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J, Roth J. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Galan JE. InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE. Journal of bacteriology. 2003;185:7279–7284. doi: 10.1128/JB.185.24.7279-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilic M, Vujanac M, Stebbins CE. A common structural motif in th e binding of virulence factors to bacterial secretion chaperones. Molecular cell. 2006;21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Lovdok L, Bentele K, Vladimirov N, Muller A, Pop FS, Lebiedz D, Kollmann M, Sourjik V. Role of translational coupling in robustness of bacteria l chemotaxis pathway. PLoS biology. 2009;7:e1000171. doi: 10.1371/journal.pbio.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JE, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics . CSH Laboratory Press; Cold Spring Harbor, NY.: 1972. [Google Scholar]
- Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AL, Parsot C. Chaperones of the type III secretion pathway: jacks of all trades. Molecular microbiology. 2002;46:1–11. doi: 10.1046/j.1365-2958.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Steitz JA. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969;224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.