Abstract
The minicircle (MC), composed of eukaryotic sequences only, is an interesting approach to increase the safety and efficiency of plasmid-based vectors for gene therapy. In this paper, we investigate micro-MC (miMC) vectors encoding small regulatory RNA. We use a construct encoding a splice-correcting U7 small nuclear RNA, which results in a vector of 650 base pairs (bp), as compared to a conventional 3600 bp plasmid carrying the same expression cassette. Furthermore, we construct miMCs of varying sizes carrying different number of these cassettes. This allows us to evaluate how size influences production, super-coiling, stability and efficiency of the vector. We characterize coiling morphology by atomic force microscopy and measure the resistance to shearing forces caused by an injector device, the Biojector. We compare the behavior of miMCs and plasmids in vitro using lipofection and electroporation, as well as in vivo in mice. We here show that when the size of the miMC is reduced, the formation of dimers and trimers increases. There seems to be a lower size limit for efficient expression. We demonstrate that miMCs are more robust than plasmids when exposed to shearing forces, and that they show extended expression in vivo.
Keywords: nano, RNA therapeutics, RNA editing, pre-mRNA
Introduction
In gene therapy, nonviral plasmid-based vectors are considered safer than the more commonly used viral vectors. However, they are less effective in terms of cellular uptake, intracellular transport and long-term expression. There are many factors to consider when designing and optimizing plasmid-based vectors, such as which functional elements to include, how to eliminate immunogenic sequences and how to formulate the vector for delivery.1,2 Regulatory agencies in both the United States and Europe recommend avoiding antibiotics resistance genes—commonly used as selection markers—in DNA vectors used for clinical trials, see e.g., FDA Guidance for Industry: Guidance for Human Somatic Cell Therapy and Gene Therapy from 1998, and a number of different strategies to avoid antibiotics resistance genes markers have been developed.3 Furthermore, the integrity of plasmid-based DNA vector is considered an important property by regulatory authorities. FDA recommends a super-coiled plasmid fraction above 80% in their Guidance for Industry: Considerations for Plasmid DNA Vaccines for Infectious Disease Indications from 2007. Producing smaller vectors by excising the bacterial sequences of the plasmid, e.g., the origin of replication and selection markers, is a way of addressing these problems. The resulting vector would only contain the expression cassette. The minicircle (MC) is the most common vector of this type. There are a number of published systems for MC production.4,5,6,7,8 In principle, a parental plasmid is constructed consisting of the eukaryotic expression cassette flanked by recombination sites. Outside these sites are all the sequences needed for plasmid propagation in bacteria, e.g., origin of replication and antibiotics resistance genes. Induction of recombination produces an MC devoid of bacterial sequences, containing only the gene of interest with suitable regulatory sequences.
Nonviral gene transfer is not only of interest for transfection of protein-encoding plasmids. There also exist a number of regulatory RNAs, which are highly interesting in a gene therapy setting. Some of these, e.g., shRNA and U7-encoded splice-switching RNAs, are produced from very small expression cassettes. We here introduce the term micro-MC (miMC) for MC vectors encoding short regulatory RNAs. MCs encoding shRNA have been studied by Zhao et al.9 in lymphoma cells and Liu et al.10 for inhibition of Hepatitis B virus replication. To our knowledge, the MC has not previously been used for splice correction. Vectors of these sizes approach the minimal limit for MC formation and numerous aspects are affected by this. The flexibility of a linear polymer, such as DNA, can be described by the persistence length, i.e., the maximum length where the directions of the ends of the molecule are correlated; the molecule is modeled as a flexible rod. At two times the persistence length, the correlation between the positions of the ends is lost. For DNA, the persistence length is 50 nm which corresponds to roughly 150 base pairs (bp). Approaching these short lengths, the bending of DNA into a circle meets with increased resistance. While it has recently been shown11 that engineered linear DNA as short as 67 bp can form circles, DNA in nature is rarely, if ever, relaxed to the extent of linear DNA. Nearly all the DNA in cells exists as negatively super-coiled structures. Coiling of plasmid DNA is achieved through gyrases in an ATP-dependent reaction. As discussed by Bates and Maxwell,12 there must be a lower limit for how small a DNA circle can be while still being super-coiled. Fogg et al.13 investigated coiling in MCs, and observed that at lengths below 300 bp the rigidity of a DNA helix prevents it from readily recombining. However, while previous studies on small plasmid-based constructs have focused either on the conformation of the construct or the therapeutic effect, little attention has been devoted to how the size of the construct affects the expression. The coiling of a plasmid is known to affect gene expression,14 and negative super-coiling leads to reduced binding between the two strands, which facilitates interaction of transcription factors and promoter regions. The degree of superhelicity is also a determining factor in the assembly of transcriptionally active chromatin.15 The strain on the double-stranded DNA in these small circular constructs could thus influence the ability of the transcription machinery to bind to the promoter region.
Given all this, we set out to study the effect and behavior of an MC when the minimal size limit is approached. To do so, the 570 bp U7-based expression cassette for splice-correcting small nuclear RNA (snRNA) was cloned into an MC-producing plasmid. The splice-correction model based on HeLa cells stably expressing a luciferase gene with a part of the mutated intron 2 from human β-globin gene was originally described by Kang et al.16 Expression of the gene with the mutated intron results in a nonfunctional luciferase protein. Masking the mutated site, however, redirects the splicing machinery that concomitantly gives rise to productive splicing of the luciferase pre-mRNA. This may be mediated by a synthetic splice-correcting antisense oligonucleotide transfected into the cell, but the antisense oligonucleotide can also be expressed from a viral17 or, as here, from a plasmid-based vector.18 In this study, we have compared the expression as well as the size-effect on coiling and stability of differently sized miMCs, with the smallest construct approaching the known size limit for successful recombination. We have visualized the miMCs with atomic force microscopy (AFM) to examine the degree of super-coiling and compared vector size to ability to withstand shearing, followed the expression levels of snRNAs and assessed the biological effect in vitro and in vivo.
Results
Production of miMC
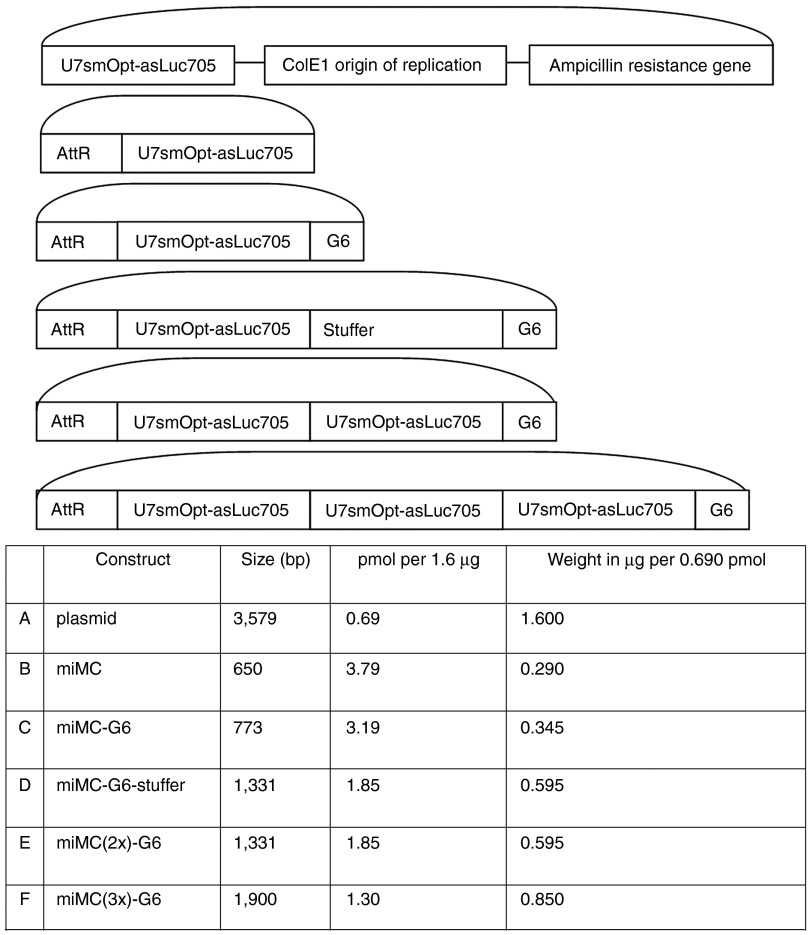
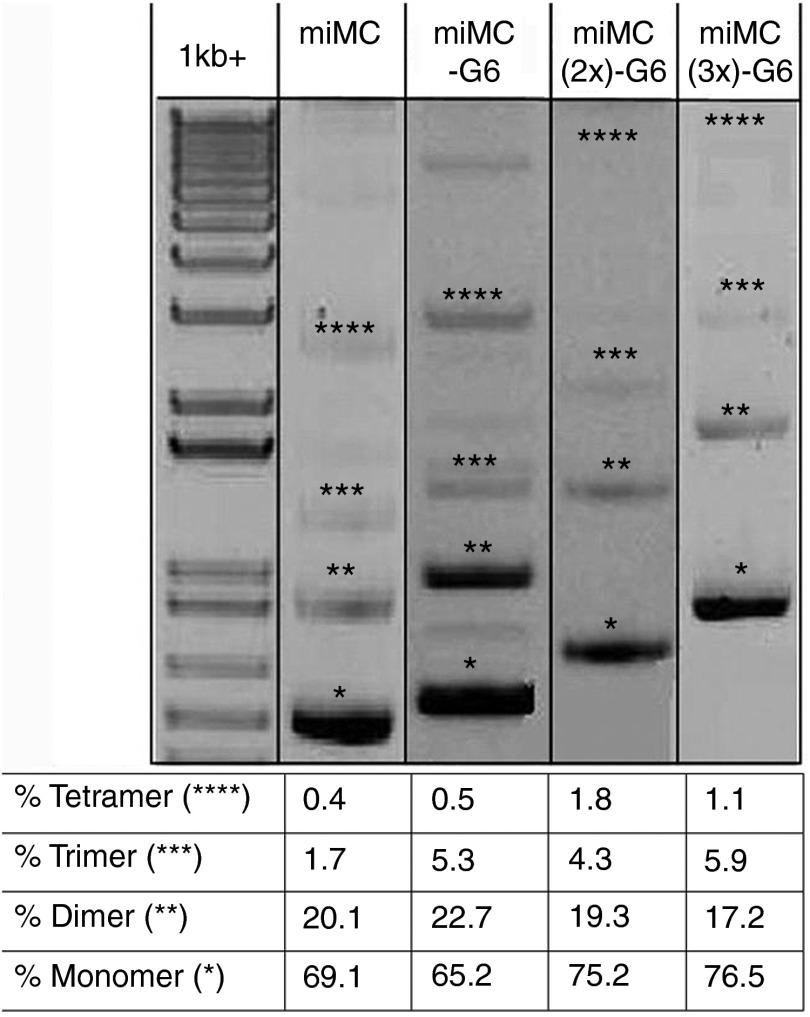
To study the behavior of MC constructs approaching the minimal size for plasmid-based vectors, we created a miMC construct containing a relatively small expression cassette encoding a splice-correction sequence in fusion with a U7 snRNA, expressed from the U7 promoter, as well as larger constructs containing several cassettes (annotated miMC(2x) and miMC(3x)) or nonexpressing sequences (annotated G6 and stuffer). See Figure 1 for construct maps and terminology. The smallest miMC is only 650 bp and the largest 1.9 kb (kilo base pairs). We use gel electrophoresis to look at the fermentation products, see Figure 2. During the production of miMCs, formation of di- and trimers frequently occurs. The majority of the recombination events results in monomeric miMCs; the molar percentage is 70% monomer, 8% dimer, and 1.6% trimer. It is noteworthy that the amount of concatamers diminishes with increasing size of the miMC; the smaller the MC construct, the greater the difficulty to form monomeric construct.
Figure 1.
Construct maps and comparison of sizes, weights, and molar equivalents of plasmid and micro-minicircles (miMCs). (A) plasmid, (B) miMC (C) miMC-G6 (D) miMC-G6-Stuffer (E) miMC(2x)-G6, and (F) miMC(3x)-G6. U7smOpt-asLuc705: Expression cassette for the U7 splice-correction snRNA targeting the 705 luciferase mutant. AttR: The product of attB/AttP recombination by ΦC31 integrase. G6 and Stuffer: Stuffer sequences devoid of regulatory sequences.
Figure 2.
Gel analysis of different micro-minicircle (miMC) constructs and quantification of the different amounts of concatamers found in QIAGEN purified material as quantified by Quantity One Software (BioRad, Hercules, CA). 1 kb+ lane contains size marker. *Monomers, **dimers, ***trimers, and ****tetramers.
Coiling of miMCs
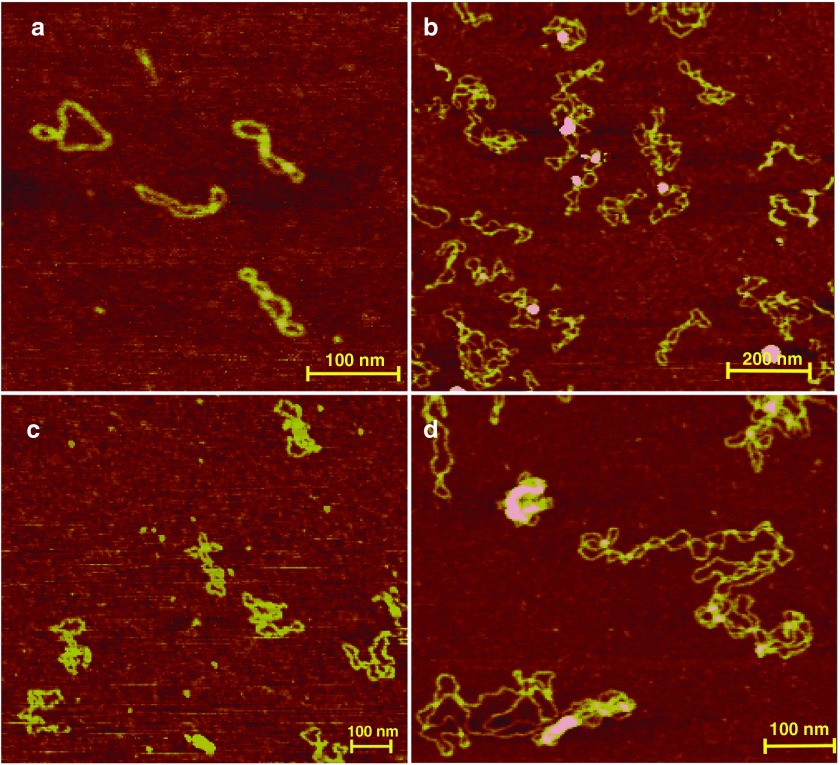
The coiling of a plasmid is one of many factors known to affect gene expression.14 Therefore, we analyzed the structure of the miMCs by AFM to discern differences in degrees of coiling between the different constructs. Despite the small size of the monomeric miMC, it forms distinct coiling patterns. Several isoforms are visible by AFM, with one to three crossings as well as open circular molecules (examples are depicted in Figure 3a). The predominant forms have two or three turns. It is evident that the miMC coiling differs from the plasmid seen in Figure 3d. Already, the two times larger miMC(2x)-G6 construct shows a more plasmid-like coiling pattern (see Figure 3b), making it hard to distinguish the number of turns, but variants with four or five turns are common. In the images of miMC(3x)-G6 (Figure 3c), many molecules with five or six turns are visible.
Figure 3.
Atomic force microscopy (AFM) image of micro-minicircle (miMC) and plasmid constructs (a) miMC, scale bar = 100 nm; (b) miMC(2x)-G6, scale bar = 200 nm; (c) miMC(3x)-G6, scale bar = 100 nm; and (d) plasmid, scale bar = 100 nm.
Shearing resistance of miMCs
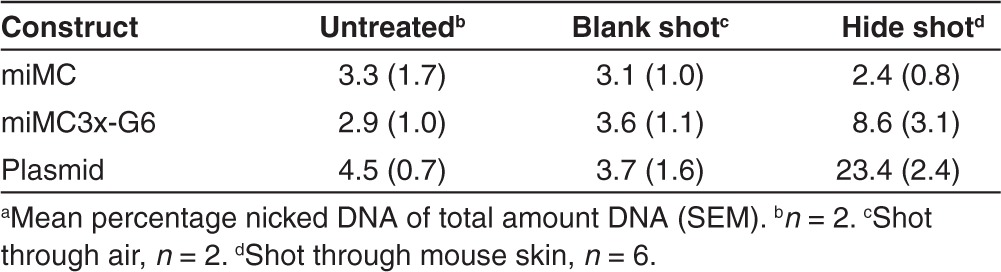
It occurred to us that constructs with many turns possibly could be more susceptible to shearing forces. In order to study this, we tested their behavior with a commonly used tool to deliver DNA in vivo, namely Biojector. This is a pneumatic delivery method which utilizes a high-pressure stream of the DNA solution to penetrate the skin. Pneumatic delivery methods are known to be associated with nicking of plasmid DNA.19 The ability of miMCs and plasmid to resist shearing forces caused by Biojector mediated injection through mouse skin was evaluated and compared to the plasmid construct. Table 1 provides the percentage of nicked miMC or plasmid construct after gel analysis and quantification using Quantity One Software. The smallest miMC is hardly affected at all by biojection through mouse skin, with only 2.4% nicking, while the plasmid is almost ten times more susceptible to shearing, resulting in 23.4% nicking. The larger miMC(3x)-G6 shows some nicking, 8.6%, which corroborates with the data comparing expression from miMCs with different sizes, as seen below.
Table 1. Comparing integrity of miMC and plasmid after delivery by Biojector through mouse skina.
Dose titration of splice-correcting constructs
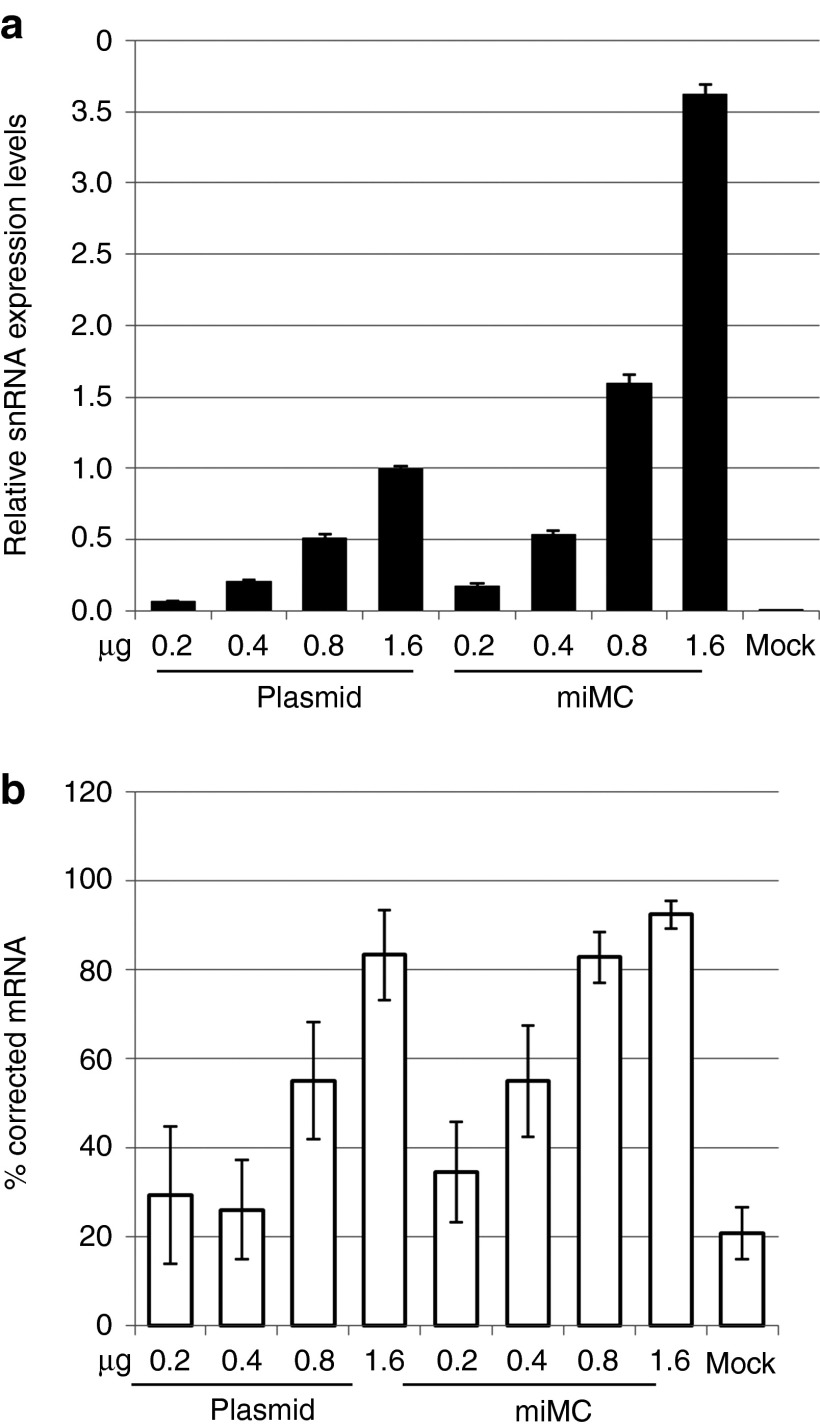
Although our aim is to use the miMCs in vivo, we first sought to determine some parameters using cell lines, since this is normally a first step in the analysis of a new vector system. A dose-titration experiment was performed in vitro in HeLaLuc705 cells to evaluate which construct dose range to use for the comparison between miMC and plasmid constructs. Both the expression from splice switching constructs themselves and the resulting corrected splicing in the luciferse mRNA was measured with RT-PCR. As seen in Figure 4a, both miMC and plasmids show a dose:response correlation between the expression of snRNA and the amount of DNA in the transfection in the range between 0.2 and 1.6 µg. However, as seen in Figure 4b, correction observed with the lower doses are just above the background while maximal correction is reached at the highest doses. This range is rather narrow; the molar equivalent of 1.6 µg plasmid is 0.29 µg for the miMC. Using lower doses would make it difficult to detect the correction, and higher doses reach saturation, as seen by the high correction levels in Figure 4b. As a consequence of this narrow range, the RT-PCR luciferase mRNA assay is not always sensitive enough to reflect the differences in snRNA levels.
Figure 4.
Dose titration of splice-correcting plasmid and micro-minicircle (miMC) in vitro in HeLaLuc705 cells 24 hours after transfection. (a) Expression of splice-correcting 705 snRNA, as determined by qPCR, equilibrated against the endogenous control RNU24 and normalized to expression from 1.6 µg plasmid (data shown as mean + 95% confidence interval, n = 2). (b) Splice-corrected luciferase mRNA assayed by two step RT-PCR (data shown as mean + SEM, n = 2).
Comparing splice-correcting miMCs and plasmid vector
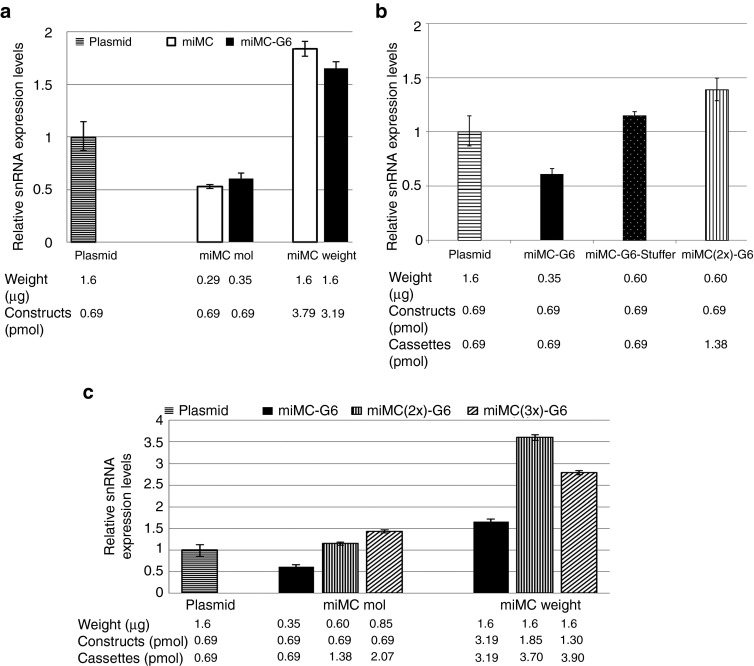
Next, we evaluated the expression and splice-correction effect of the miMC construct compared to the full plasmid vector in vitro in HeLaLuc705 cells. miMC constructs of two sizes, miMC of 650 bp and miMC-G6 of 773 bp, both containing a single cassette encoding the splice-correcting U7-RNA, were compared with the plasmid of 3,579 bp containing the same expression cassette. In all experiments, comparisons of the effects were made on molar and weight basis relative to 1.6 µg plasmid (=0.69 pmol). As seen in Figure 5a, the smaller constructs have a lower expression level than the plasmid; this is evident in the comparison on molar level. On weight basis, the number of miMC cassettes is more than three times higher and the increase of expression is proportional to the dose increase. Despite the differences in snRNA expression, this is barely detectable at the corrected luciferase mRNA level (Supplementary Figure S1); the lower amount of snRNA expressed by the miMC construct under our assay conditions is still enough to induce the same level of correction as from the larger plasmid construct.
Figure 5.
Expression efficiencies of micro-minicircles (miMCs) depending on size and number of cassettes in vitro in HeLaLuc705 cells, 24 hours after transfection. (a) Comparison between miMC and plasmid in vitro. (b) Assay of how size and number of casettes affects of expression from miMC using 0.69 pmol of each construct plasmid, miMC-G6, and miMC-G6-Stuffer which contains stuffer sequences making it twice the size of miMC-G6 and miMC(2x)-G6 which contains two expression casettes. (c) Expression from miMCs containing one, two, or three casettes. The expression of splice-correcting 705 snRNA as determined by qPCR and equilibrated against the endogenous control RNU24 in HeLaLuc705 cells and normalized to the expression from 1.6 µg plasmid. Transfections were made on equimolar basis and on weight basis referring to amounts of splice-correcting vector. Data shown as mean + 95% confidence interval, n = 3.
Comparing expression efficiencies for miMCs depending on size and number of cassettes
As seen in Figure 2, miMCs naturally form a mixture of mono-, di-, and trimers and even tetramers during propagation in E. coli, with monomers as the major form, on average 70% of molar basis. This prompted us to assay how multimerization influences the expression and splice correction in vitro in HeLaLuc705 cells. It is important to be able to discriminate between the influence of concatamerization and size effects. To study the effect of size, a comparison between constructs of different sizes containing one single expression cassette was performed. To study the effect of concatamerization on expression and splice-correcting effect, we compare miMCs containing one or two copies of the expression cassette but with the same total miMC size. For the size evaluation, we compared the plasmid (3579 bp) to two miMC constructs, where one is almost twice the size of the other due to insertion of stuffer sequences (miMC-G6 of 773 bp and miMC-G6-stuffer of 1331 bp). In Figure 5b, a notable difference in expression between the smaller miMC construct and the miMC of twice the size but still only one U7-cassette is evident; the larger construct even surpasses expression from the plasmid. To analyze the effect of concatamerization, we compared the stuffer-enlarged miMC (miMC-G6-stuffer) to the dimeric construct (miMC(2x)-G6), both containing 1,331 bp. These comparisons were made on molar basis counting the number of constructs. Figure 5b shows that the concatameric construct were not proportionally better. While it does give an elevated expression as compared to the construct with only a single cassette but having the same size, it does not reach two times the expression as would be expected since it contains twice as many expression cassettes.
Next, we investigated miMC vectors containing one (miMC-G6), two (miMC(2x)-G6) or three expression cassettes (miMC(3x)-G6) and compared them to the plasmid on molar as well as on weight basis. Molarity was calculated on the number of vectors, which results in double amount of cassettes for the dimer and three times the amount of cassettes for the trimer. For the weight-based comparison, the number of cassettes is nearly equal. As seen in Figure 5c, with miMCs of different size due to the concatamerization, the increase of snRNA expression in cells transfected with dimer and trimer constructs is almost proportional to the increase of the number of cassettes. When compared on a weight basis, a disadvantage of multimeric constructs is evident for the trimer.
These comparisons indicate that our smallest miMC construct is of suboptimal size and that multimerization makes the construct more susceptible to nicking, which negatively influences the expression efficiency. However, the differences are not large enough for the mRNA splice-correction assay to be sufficiently sensitive to readily measure these effects, as is evident from Supplementary Figure S2.
Studying different transfection methods
To elucidate how the miMC performs in vitro using different transfection methods, the smallest and the largest of the miMC constructs (miMC and miMC3x-G6) were compared to the plasmid on equimolar level using Lipofectamine 2000 and Neon-mediated electroporation of HeLaLuc705 cells. When using Lipofectamine 2000, the total amount of DNA could affect the process of formation of lipoplex. Therefore, the smallest vector was assayed with and without extra DNA, i.e., pFiller. The amount of vector cassettes in the cell was quantified as well as the levels of transcribed snRNA and the effect on luciferase mRNA correction.
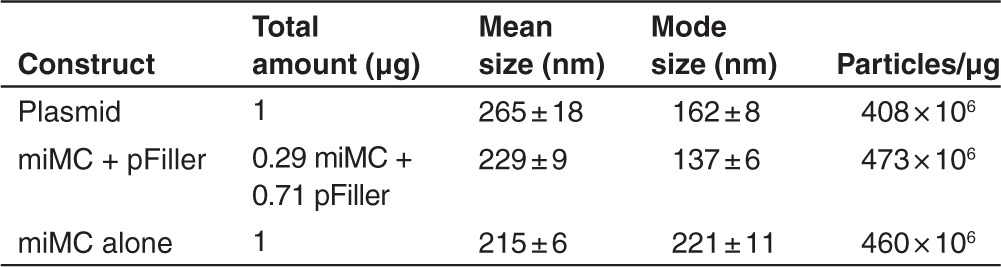
In the study of lipofection, it is notable that the total amount of DNA used in the lipoplex-forming reaction influences both uptake and the expression levels (Figure 6a,b). This is most evident in the experiments with the smallest construct, where there is a substantial difference in expression levels with and without pFiller. Using lipofection, the miMC transfected with pFiller behaves as in previous experiments, but without the filler DNA the expression is considerably lower (compare Figures 5 and 6). We hypothesize that one possible reason for the difference in transfection efficacy observed between miMC and plasmids may be due to differences in the size of the DNA:Lipofectamine-complexes. We therefore analyzed the size distribution of lipoplexed particles containing miMC or plasmids. As seen in Table 2, the formed complexes differ in size and numbers depending on whether miMCs or plasmids were used. These differences could potentially account for some of the observed differences in transfection efficiencies. When using electroporation, however, more DNA does not enhance transfection efficiency or expression. Figure 6 shows that when comparing delivery of equimolar amounts of miMC with or without pFiller, the delivery and especially expression is hampered by the higher total DNA dose. The increased total amount of DNA also impairs the miMC weight group where the dose increase is proportional only to the low expression from the pFiller molar group (Figure 6b).
Figure 6.
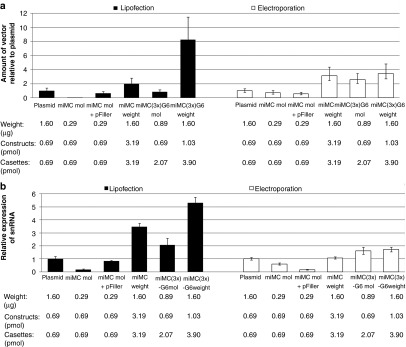
Studying the effect of different transfection methods on splice-correction effect from the micro-minicircle (miMC) construct and from miMC(3x)-G6 to plasmid in vitro in HeLaLuc705 cells, assayed after 24 hours (n = 3): Lipofection by Lipofectamine 2000 and electroporation using the Neon Transfection System. (a) Amount of vector DNA measured by qPCR for the expression casette, as compared to transfections with plasmid and equilibrated to genomic copy numbers by quantification of the Tfrc gene. Data shown as mean + SEM. (b) Expression of splice-correcting 705 snRNA from miMCs as determined by qPCR and equilibrated against the endogenous control RNU24 in HeLaLuc705 cells and normalized to the expression from 1.6 µg plasmid. Labels: weight (µg), vector (pmol), and casettes (pmol) refers to values for splice-correcting vector also when filler DNA is added. Data shown as mean + 95% confidence interval.
Table 2. Lipofectamine 2000 complex size measurements.
Comparing lipofection and electroporation as transfection methods for miMC, we see that electroporation is favorable for lower DNA amounts whereas Lipofectamine 2000 is more effective when using larger doses. On the reporter mRNA level, we see that the system begins to reach saturation (Supplementary Figure S3).
Effect of miMCs in vivo
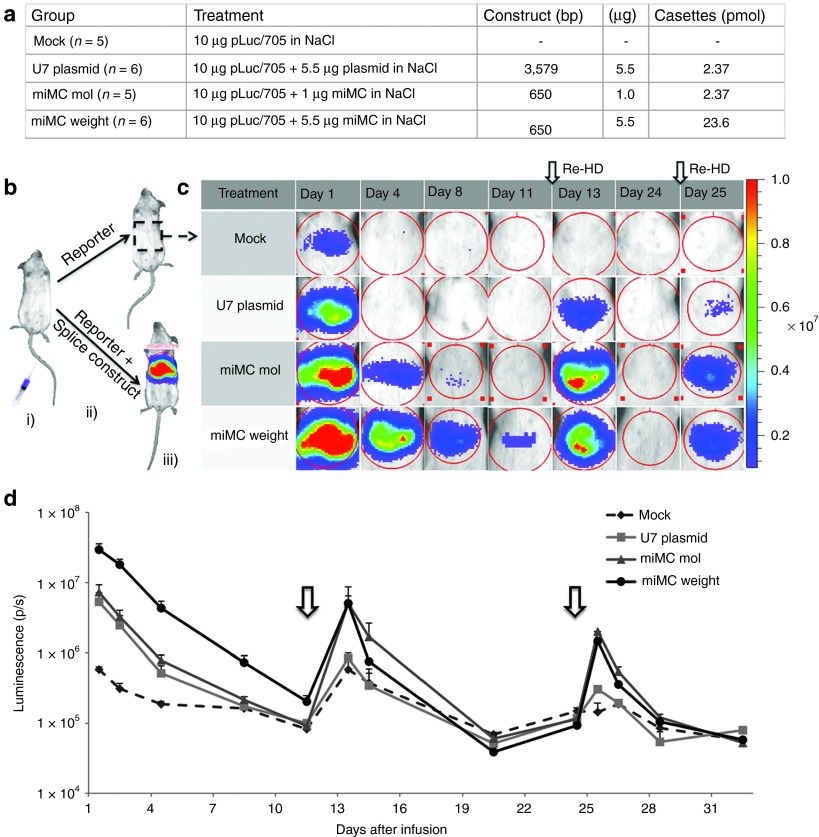
To compare the long-term in vivo effect of splice-correcting miMCs and plasmids, hydrodynamic coinfusion into mouse liver was performed with splicing construct and reporter plasmid, see schematics in Figure 7b. Hydrodynamic infusion is a well-documented gene transfer technique, known to give a high transfection efficiency of the liver in mice.20 Initial experiments were performed with the original pLuc705 plasmid where the reporter gene is driven by the CMV-promoter which, especially in the liver, is known to be silenced over time. To ensure enough expression of the reporter plasmid, reinfusions of reporter plasmid pCMVLuc/705 alone using hydrodynamic infusion were performed at days 12 and 24. The resulting luminescence levels were markedly higher than before reinfusion (see Figure 7c,d). Reinfusion using the empty pFiller did not affect the luciferase levels (data not shown). Following reinfusion, the splice-correcting plasmid group had similar luciferase levels as the control group (pCMVLuc/705 alone), suggesting that this construct behaved in a similar way as the reporter plasmid. The miMC groups on the other hand had a six times higher signal, showing activity from the miMC splice-correcting vector at day 13. Interestingly, the activity after reinjection was similar in both miMC groups despite the fact that the weight group was given 5.5 times as much splice-correcting miMC as the molar group, indicating possibly saturated levels. Accordingly, this group was not included in follow-up experiments. The second reinfusion of pCMVLuc/705 at day 24 was conducted in all groups, and again resulted in high activity in the miMC groups. At day 25, both miMC groups showed a six times higher level of luciferase activity compared to the plasmid, thus displaying a high and sustainable expression from the splice-correcting miMC.
Figure 7.
Splice correction from micro-minicircles (miMCs) and plasmid in vivo using the pCMVLuc/705 reporter construct. (a) Experimental setup for the different treatment groups. (b) Schematic presentation of the in vivo procedure with (i) hydrodynamic infusion to the liver of either (ii) pCMVLuc/705 + U7-splicing construct (miMC or plasmid) or pCMVLuc/705 + “mock.” When the luciferase pre-mRNA is correctly processed, the luciferase protein together with its substrate (luciferin) will emit light and the luminescence can be (iii) detected and measured in the IVIS. (c) IVIS-images of luminescence of the mouse liver over time for the four different groups. Each row depicts the same representative mouse for each group measured at the indicated time-points. The red color indicates a stronger signal and blue indicates a weaker. At day 0, all animals received a coinfusion (hydrodynamic infusion to the liver) with pCMVLuc/705 + splicing agent (miMC or plasmid) or mock. At day 12 and 24, reinfusion was made with pCMVLuc/705 to determine the residual activity of the splicing agent (MC or pU7). (d) Graph presenting the mean values of luminescence at log scale for each group of mice over time. Reinfusions of pCMVLuc/705 are indicated by arrows. Data shown as mean + SEM.
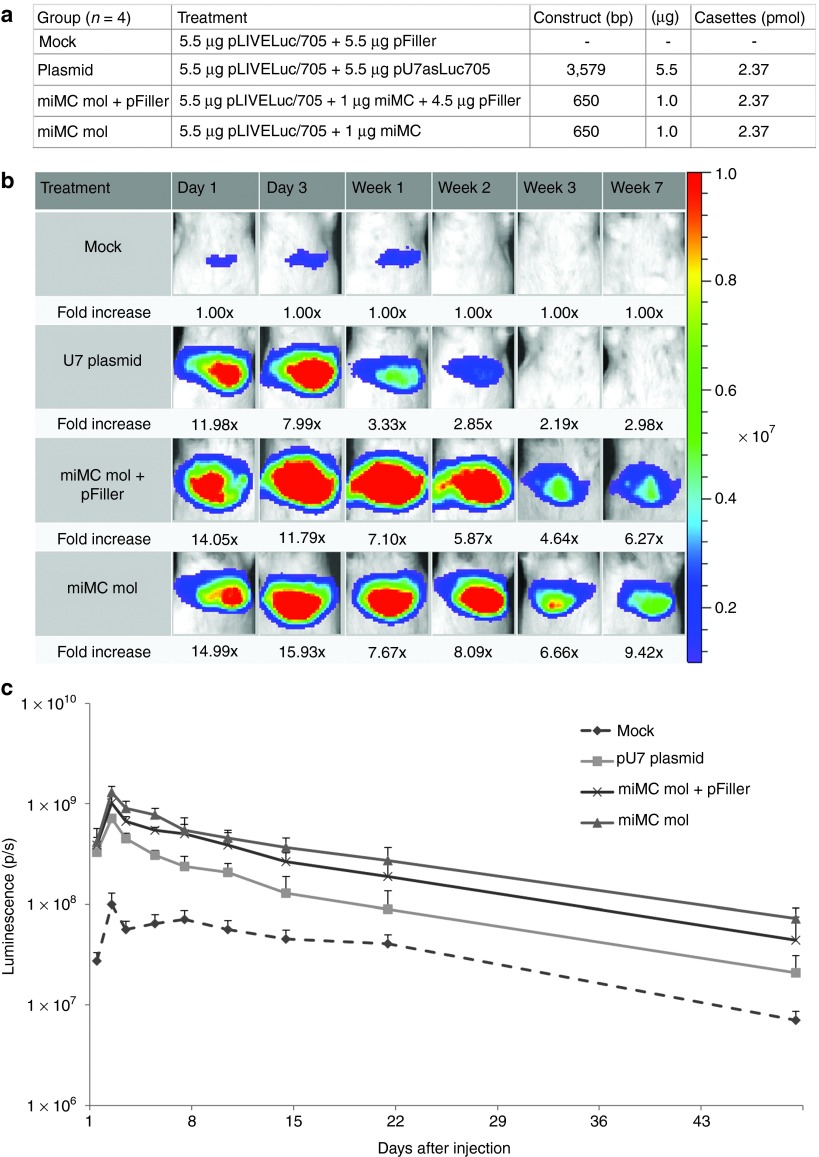
A reporter construct with more long-lived expression, where the reporter gene driven by a liver-specific promoter, pLIVE/705 (see Materials and Methods) was subsequently constructed and the experiment was repeated without the need for supplying additional reporter plasmid (Figure 8). At day 0, all animals were given a hydrodynamic coinfusion of the reporter plasmid pLIVELuc/705 either with pFiller alone or with splicing-plasmid or miMCs in equimolar amounts (with or without pFiller). Throughout this in vivo experiment, all mice treated with splice-correcting constructs had a higher expression than the control mice, which only received the reporter plasmid with pFiller. One day after infusion, both miMC groups (with and without pFiller) had about 20% higher luminescent signal as compared to the mice treated with splice-correcting plasmid. Over time, the signal from the plasmid declined more rapidly compared to the miMC construct and at the final time point (7 weeks) the miMC groups had over 200% (with pFiller) and 300% (without pFiller) higher signal than the plasmid group (Figure 8b,c).
Figure 8.
Splice correction using of micro-minicircles (miMCs) in vivo using the pLIVE.Luc/705 reporter construct. (a) Experimental setup for the different treatment groups. (b) IVIS-images of luminescence of the mouse liver over time for the four different groups. Each row depicts the same representative mouse for each group measured at the indicated time-points. The red color indicates a stronger signal and blue indicates a weaker. At day 0, all animals received a coinfusion (hydrodynamic infusion to the liver) with pLIVE.Luc/705 + splicing agent (plasmid or miMC ± pFiller) or mock. Numbers indicate luciferase signal normalized to pLIVE.Luc/705 alone at each given time point. (c) Graph presenting the mean values of luminescence at log scale for each group of mice over time. Data shown as mean + SEM.
Following the final luciferase measurements, mouse livers were harvested and analyzed for levels of plasmid or miMC vector, snRNA expression, and mRNA correction, see Supplementary Figure S3a–c. The amounts of vector remaining in the liver were similar between the different treatment groups. The levels of snRNA as detected by northern blot showed that the miMC gave nearly two times higher expression as compared to the plasmid when delivered without pFiller. Also, the mRNA data shows an increased correction using miMC, where the same molar amount of vector gives nearly four times higher correction as compared to the plasmid.
Discussion
In this study, we aimed to investigate the effect of approaching the minimal size limit for MC construction extending a previous study on regular MCs.21. Therefore, we created an MC containing the cassette for expression of a splice-correcting oligonucleotide from the U7 promoter. The resulting miMC was merely 650 bp. We have studied how the coiling pattern and stability is influenced by size, and correlate these findings to our investigation of how such a small construct performs in vitro and in vivo.
When approaching the minimal size limit for MC formation, several parameters are affected. This became evident already in the fermentation process, since the bacteria did not only form monomeric miMCs, see Figure 2, but also di-, tri-, and tetramers. The smaller the size of the cassette, the higher the quantity of multimers. Bates et al.12 report that it is thermodynamically possible for DNA gyrase to induce coiling in DNA circles as small as 174 bp, and as discussed above, Vafabakhsh et al.11 show that circularization of linear DNA is possible at even shorter lengths. Yet, in the production of our miMCs we observed that the amount of monomeric miMC decreased with decreasing size of the cassette, while concurrently an increasing amount of multimers is formed. Fogg et al. also report an inverse relationship between the DNA length and the efficiency of the intermolecular recombination.13 This could be due to a sterical limitation, i.e., the length of the miMC DNA being too short to allow the required recombination to take place efficiently. It would simply be more favorable for the integrase to use recombination sites from two different plasmids than bending just one. To our knowledge, there have been no previous reports on the effect of concatamerization of MCs intended as gene therapy vectors. Artificial multimeric plasmid vectors have been studied22 and in 2003, Voss et al.23 investigated the production of multimeric plasmids as vectors for gene therapy. They state that there are of particular interest for pharmaceutical purpose because they contain multiple copies of a therapeutic gene and can therefore be more efficient vectors. These constructs do, however, contain multiple copies of not only the gene of interest but also of the bacterial backbone, whereas the multimeric MC does not. Voss et al. report that for the larger multimers, the plasmid becomes unstable and disintegrates into monomers again. In the miMC case, the multimers were produced naturally by the recombinase in the fermentation process. In the experiments where we compared expression and effect from multimeric miMCs, multimerization did not seem to affect the functionality. We saw a correlation between the expression from the larger constructs and the increase in cassette-dose. However, a factor that must be considered when using multimeric constructs is that loss of one vector leads to loss of several cassettes. This could explain why the expression from the trimeric constructs did not show the same increase as from the dimers (Figure 5c). In a study published by Catanese et al.,24 MCs showed an increased resistance to shearing forces associated with sonication, and we were able to confirm this observation in our constructs (data not shown). When plasmid-based vectors are used for DNA vaccination, a promising route of administration for gene transfer through the skin uses pneumatics.25 However, pneumatics is also associated with shearing of plasmid DNA.19 The Biojector is a needle-free system for drug delivery forcing liquid medication through a tiny orifice that is held against the skin. Our studies demonstrate that the miMC could resist the shearing forces caused by the Biojector to a much greater extent than the full-length plasmid, as seen in Table 1. The larger miMC construct showed some nicking, which corroborates with the in vitro expression data. It is noteworthy that the plasmid DNA that reaches the cell after biojection is partially destroyed and no longer meets the FDA requirements of 80% super-coiled fraction, whereas the nicked miMC fraction is ten times lower. These findings have implications for using miMC as a vector for immunization by pneumatics as well as for the use of other transfection methods known to cause shearing of pDNA. To our knowledge, this is the first time the fate of a normal plasmid has been compared to that of an MC after pneumatic delivery.
When analyzing mRNA correction in cell culture, the plasmid and miMC constructs performed equally well. However, using RT-qPCR to measure the expression of the small U7-RNA, we noted significant differences between the constructs. When examining constructs of different size with the same number of cassettes (Figure 5b), it is quite clear that the size of the constructs affects the amount of snRNA produced. As seen in Figure 3, the AFM analysis of coiling patterns displayed a difference between our smallest miMC and our larger constructs. In our experiments using the same fermentation protocol, since the strain and protocol influences apology26,27, the smallest construct exhibited patterns that Fogg et al.13 associate with high writhe and increasingly negative super-coiling. Thus a lack of negative super-coil is probably not the explanation for the lowered expression. It is however possible that the tight coiling of these small constructs hampers transcription efficiency. Often, bending and twisting of the DNA occur at lengths that are short compared to the persistence length, and especially so in sites for transcriptional regulation events. It is known that the curvature of the DNA affects the accessibility to cis-DNA elements by inducing different chromatin formations28 and that bending of the DNA changes the affinity of promoter binding proteins.29
Another possible explanation for the observed differences in expression is that the vector size affects transfection efficiency. Comparing different transfection methods, i.e., lipoplexion using Lipofectamine 2000 and electroporation using the Neon transfection system, we noticed that the total amount of DNA is important for transfection efficiency and expression, but the effect differs considerably between the two methods. Using more DNA in electroporation may not always be advantageous. The impairing effect of an increased DNA amount was not only evident when comparing the molar groups with or without pFiller but also when looking at the weight groups; neither the delivery nor the expression increased proportionally to the total DNA dose (Figure 6a). This indicates that when using electroporation, the miMC allows high expression from a lower dose of total DNA. Also, regarding lipofection, it was clear that the amount of DNA played a role in transfection efficiency, but here more total DNA markedly increased the delivery and expression of the miMC (Figure 6b). Our hypothesis is that the lipoplex formation requires a certain amount of DNA to form stable particles, and using smaller amounts of DNA results in lower yields of these particles, subsequently leading to fewer cells being transfected. The nanoparticle analysis of lipoplex particle size strengthens this hypothesis; the plasmid and the miMCs form about the same amount of particles per µg DNA. Our findings are in line with findings reported from transfection with polyethyleneimine, where adding filler DNA increased the number of transfected cells between 1.5–3 times as compared to the same amount of plasmid without filler DNA.30 When comparing the smallest miMC transfected together with the filler plasmid to the miMC containing one cassette but also stuffer DNA, making it twice the size, they performed equally well. This suggests that lowered transfection efficiency for lipofection is contributing to the lowered transcription from the smaller constructs.
In order to study the long-term effect of miMC in vivo, we used hydrodynamic tail-vein injections to administer reporter plasmid together with the different splice-correcting constructs to mouse liver. While the miMCs initially only have a slightly higher effect compared to the plasmid, it is evident that the splice-correcting effect of the plasmid diminishes faster than that of the miMC construct (Figures 7 and 8). This data set corroborates the studies by Chen et al. made on protein expression from MC constructs in liver. Their studies have shown that bacterial sequences covalently linked to the expression cassette, as in plasmid vectors, hamper the expression and contributes to the silencing of the vector.6,31 A possible mechanism for this is that bacterial sequences adopt a heterochromatin structure that spreads into the expression cassette.32 However, Schüttrumpf et al.33 recently showed that the predominant reason for transient expression after hydrodynamic infusion in mouse liver was loss of the vector, regardless of whether it was a plasmid or MC construct. Nevertheless, they observed a higher expression from the remaining MCs, which is in line with our findings; we observe equal amounts of remaining vector in the miMC and plasmid groups but higher expression and effect from the miMC. In the Schüttrumpf-study, this was attributed to a small, but significant, difference in the methylation pattern between the two constructs. In the light of our findings on shearing, the increased stability of the miMC is also likely to contribute to the prolonged activity. This is in accordance with Zhao et al.9 who reported that super-coiled MC DNA has a higher serum stability.
In summary, we show that production of miMC in the size range that approaches the sterical limit for recombination causes concatamerization during the recombination process and that the expression is hampered as compared to larger constructs. However, we also show that the smaller the construct, the higher the resistance towards shearing. This raises the question of which is the optimal size for a gene therapy vector. Our data suggest that when working with very small constructs, it is favorable to clone cassettes containing two copies of the gene, to increase the size slightly and thereby reducing the number of concatamers in production and increasing the expression efficiency while still benefiting from the stability of these small constructs.
Materials and methods
Constructs and cloning. The plasmid encoding a short splice-correcting RNA, pU7asLuc705, designed by Kang et al.16 was used. From this construct, the expression cassette was cloned into the pMC-plasmid developed by Kay et al.,8 resulting in pmiMC-U7asLuc705. This miMC producing plasmid was further modified by adding one or two cassettes and/or stuffer DNA to produce different sizes of the final miMC construct, as seen in Figure 1. The G6 site used to enlarge the miMC is a binding site for anchor molecules which could later be used to enhance transport.34,35 The miMC construct miMC-G6-Stuffer contains a part of the enhanced Green Fluorescent Protein gene, lacking regulatory sequences and start codon, to make the construct even larger. A filler plasmid, pFiller, devoid of eukaryotic genes, to be used as noncoding DNA stuffer was constructed by removing the promoter and luciferase-coding sequence from the CpG-free plasmid construct CpG 61 lucS (InVivogen, San Diego, CA). For in vivo experiments, the pCMVLuc/705 plasmid16 encoding the gene for firefly luciferase with the mutated 705 human β-globin mini-intron sequence inserted was used as reporter in the splice-correcting systems. The mutated luciferase gene was also cloned into the pLIVE vector (Mirus Bio LLC, Madison, WI). The pLIVE is designed for high level, prolonged expression of transgenes in the mouse liver. Gene expression is driven from a chimeric promoter composed of the mouse alpha fetoprotein enhancer II and the minimal mouse albumin promoter and two introns flanking the transgene have been engineered into the vector.
miMC production. Production of the miMC vectors was done according to the protocol by Kay et al.8 Briefly, E. coli strain ZYCY10P3S2T transformed with miMC producing parental plasmids was grown on Luria Broth Agar plates with 50 µg/ml kanamycin (Km) and 1% glucose over night. Colonies were expanded in 5 ml Terrific Broth supplemented with 50 µg/ml Km and 1% glucose at 37 °C for 6 hours and was subsequently expanded by inoculation of 400 ml Terrific Broth with 400 µl culture followed by incubation at 225 RPM 37 °C over night in a 1 l flask. The following morning, the miMC formation was induced by adding 400 ml Luria Broth containing 0.02% L-arabinose (Sigma-Aldrich, St Louis, MO) and 40 mmol/l NaOH. The culture was then incubated in a 2 l flask at 32 °C and shaking at 225 RPM for 5 hours. Samples taken during the fermentation were analyzed to verify that the induction was successful. The miMCs were purified using either the Plasmid MEGA or Plasmid Plus MEGA kits (QIAGEN, Hilden DE) according to standard protocol with the following modification: Due to the small size of the miMC, each QIAGEN tip was used for a larger bacterial culture than recommended in the standard protocol. The amounts of resuspension, lysis, and neutralization buffers were adjusted to the weight of the bacterial pellet, using 10 ml of each buffer per gram wet pellet and loaded onto two columns/400 ml over night culture. The monomer fraction of the miMCs was gel-purified on a 1% agarose gel while carefully avoiding nicking of the DNA by UV light or intercalating dyes. The monomer miMCs were extracted from the gel by QIAquick Gel Extraction Kit (QIAGEN, Hilden, DE) according to the manufacturer's protocol and further purified by phenol:chloroform extraction.
Atomic force microscopy. The substrate (AP-Mica) was prepared in the following way: Two glass caps were dried at 120 °C for 15 minutes and placed into a 3 l desiccator. Mica was cut in 8 × 8 mm2 slices and cleaved to get a fresh surface. The cleaved Mica was immediately placed into the desiccator which then was purged with N2 for 2 minutes. Forty µl of APTES (3-aminopropyltriethoxy silane) was added to one glass cap and 14 µl of N,N—diisopropylethylamine into the other. The desiccator was again cleaned with N2 for 2 minutes. The Mica was incubated with the chemicals in the desiccator for 2 hours, whereafter the caps were taken out and the desiccator was purged. The activated Mica was incubated in the desiccator for 1–2 days before sample preparation. The miMC or plasmid DNA was diluted to 0.5 µg/ml in a 50 mmol/l NaCl solution. The activated Mica was incubated with 30 µl DNA for 5 minutes and then rinsed with 2–3 ml distilled water. The samples were dried with N2 and placed in a metallic holder for AFM imaging using a NanoScope V Atomic Force Microscopy (Veeco, Santa Barbara, CA) with J piezoelectric scanner. Measurements were made in air in soft tapping mode with a scan rate of 1 Hz. A diamond-like carbon tip cantilever with a resonant frequency of 115–190 kHz was used (NT-MDT, Moscow, Russia).
Stability testing. For testing the resistance towards shearing forces, a needle-free device from Biojector (Bioject Medical Technologies, OR) was used. Three microgram of miMC, miMC(3x)-G6 or plasmid in 100 µl NaCl was loaded into the device, and ejected either through mouse hide or, as blank shot control, through air. For untreated and sample biojected into empty air samples, n = 2. For biojection through mouse hide, n = 6. The mouse hide was prepared from BalbC mice, three biojections through female hide and three through male hide for each construct. After biojection and recovery of DNA, the samples were analyzed on agarose gels. The gels were subsequently stained with ethidium bromide and visualized with Fluor-S Multilmager documentation equipment with a cooled CCD camera. The portion of nicked and super-coiled DNA was quantified by Quantity One Software (BioRad, Hercules, CA).
Cells and transfections. HeLa cells stably expressing the gene for luciferase with the mutated 705 human β-globin mini-intron sequence inserted16 (HeLaLuc705) were used to monitor transcription, measured as splice correction of the luciferase mRNA. The cells were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum (both from Invitrogen, Carlsbad, CA). For transfections of HeLa cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), 2.5 µl/µg DNA was formulated in 200 µl Optimem (Invitrogen) per well according to the manufacturer's protocol. The day before transfection, cells were plated in six-well plates (Costar, Corning Incorporated, Corning, NY), at a density of 400,000 cells/well, to reach a confluency of 80% at transfection. To identify the optimal dose for the experiments, a titration with 0.2, 0.4, 0.8, or 1.6 µg of plasmid or miMC per well in six-well plates was performed. To study how the size of the constructs affects the transcription, and thus the function of the miMCs, 1.6 µg plasmid or miMC, or the molar equivalent amount of miMC constructs (see Figure 1) was cotransfected with 1.6 µg plasmid encoding β-Galactosidase (β-Gal). β-Gal was used in order to be able to detect differences in transfection efficiency (data not shown). To have the same total amount of DNA also in the samples with the molar equivalent amount of constructs, pFiller was added up to 1.6 µg. For electroporation, 1.6 µg plasmid, the same weight-amount or the molar equivalent of the constructs referred to as miMC and miMC(3x)-G6, was transfected into HeLaLuc705 in 100 µl-tips using the Neon Transfection System (Invitrogen) according to manufacturer's protocol. The cells were harvested by trypsination using TrypLE Express (Invitrogen) 24 hours after transfection. The cells in each well were divided and analyzed for RNA and protein expression. Three experiments with duplicate wells for each transfection were assayed, unless otherwise stated.
Quantification of Lipoplex particle size. The NS500 nanoparticle analyzer (NanoSight, Amesbury, UK) was used to measure the size distribution of particles of plasmid or miMC DNA after complexation with Lipofectamine 2000 (Invitrogen). The complexation was done using the same ratios as for transfection and according to the manufacturer's protocol. The complexed mixtures were loaded into the NS500 and the number of particles and their movement were recorded for 3 × 60 seconds and analyzed using the NS500 software.
RNA extraction and analysis. Total RNA was extracted from in vitro samples using miRNeasy Kit (QIAGEN) according to the manufacturer's protocol without DNase treatment. RNA quality was assayed using absorbance (A260/230) and by analysis on 1% agarose gel. For in vivo samples, the mouse liver was grinded in liquid nitrogen, 100 mg was lysed with a Tissue Lyser II (QIAGEN) in Trisol (Invitrogen) and the total RNA subsequently purified with phenol:chloroform extraction.
snRNA analysis. Amount of splice-correcting antisense oligonucleotide expressed from the constructs in vitro was analyzed on StepOne Plus real time PCR system (Applied Biosystems, Foster City, CA) using a Custom TaqMan Small RNA Assay from Applied Biosystems. This analysis targets the U7asLuc705 snRNA, and was normalized to the human small nucleolar RNA, C/D box 24 (RNU24). The cDNA was produced from 47 ng total RNA using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) according to protocol. qPCR was performed with the TaqMan Fast Universal PCR Master Mix No AmpEraseUNG (Applied Biosystems) according to protocol. Calculation and statistics were done in the StepOne Plus Software (Applied Biosystems) using the ΔΔCt method after efficiency verification using a standard curve. Amounts of snRNA were compared to that expressed from 1.6 µg plasmid.
For in vivo samples, the qPCR method could not be used to detect the snRNAs, perhaps due to inhibitors being copurified with the RNA from the liver tissue. Therefore, the expression of U7asLuc705 snRNA was measured using northern blotting. The following DNA/LNA probe was used for detection of the U7asLuc705: taTgtAacTgaGgtAagAggTtaGatCt (uppercase LNA, lowercase DNA). As endogenous control, a probe targeting U1 snRNA. A probe targeting U1 snRNA was used as endogenous control, it has been described by Verbeeren et al.36 as were the blotting and hybridization conditions. The U7 probe was hybridized the same way except that the final wash was carried out at 65 °C.
mRNA analysis. For the dose-titration assay, the amount of corrected luciferase mRNA was assayed by two step RT-PCR using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche Diagnostics, Mannheim, Germany) and HotStart Taq Plus DNA polymerase kit (QIAGEN), with the following primers: Fwd5′-TTGATATGTGGATTTCGAGTCGTC and Rev5′-TGTCAATCAGAGTGCTTTTGGCG. For all other experiments, the OneStep RT-PCR Kit (QIAGEN) was used. The product was analyzed on 2% agarose gel containing SYBR Safe (Invitrogen). The bands were visualized with the Fluor-S MultiImager and quantified by Quantity One Software (BioRad), and the percentage of corrected band as compared to uncorrected was calculated. As endogenous control in the multiplex RT-PCR for analysis of in vitro samples, hypoxanthine-guanine phosphoribosyl transferase mRNA was amplified by the following primers: Fwd-5′GACTTTGCTTTCCTTGGTCAG and Rev-5′GGCTTATATCCAACACTTCGTGGG. For the in vivo material, primers for mouse glyceraldehyde 3-phosphate dehydrogenase mRNA with the following sequenced was used as endogenous control: Fwd-5′ATGGGTGTGAACCACGAGAA and Rev-5′AGTTGCTGTTGAAGTCGCAG.
Vector quantification. Cells were lysed using DirectPCR Lysis Reagent (cell) (Viagen Biotech, Los Angeles, CA) according to manufacturer's protocol. In vivo material was disrupted using the Tissue Lyser II system (QIAGEN) prior to incubation with the DirectPCR Lysis Reagent. Amount of plasmid or miMC vector in the cell lysate was analyzed with a StepOne Plus real time PCR system (Applied Biosystems) using a Custom TaqMan Assay from Applied Biosystems. This analysis targets the U7asLuc705-encoding cassette and the amounts of vector were normalized to amount of cells using TaqMan copy number reference assay RNaseP for in vitro samples and TertC for in vivo samples. qPCR was performed with the TaqMan Fast Universal PCR Master Mix No AmpEraseUNG (Applied Biosystems) according to protocol. Calculation and statistics were done in the StepOne Plus Software (Applied Biosystems) using the ΔΔCt method after efficiency verification using a standard curve. Amounts of vector were compared to what was achieved after transfection with 1.6 µg plasmid.
In vivo experiments. Female 20–25 g NMRI-mice were treated with a 2.0 ml hydrodynamic infusion through the tail vein while anesthetized.20 In the pCMVLuc/705-reporter plasmid experiment, mice were divided into groups (n = 5–6) that received a coinfusion at day 0 of 10 μg reporter plasmid pCMVLuc/705 together with either pU7asLuc705, splicing miMC in different ratios or saline as control, see Figure 7a. At day 12, half of the mice received a second hydrodynamic infusion with 10 μg pCMVLuc/705 and the other group received pFiller as control. This procedure was then repeated at day 24.
In the pLIVELuc/705-reporter plasmid experiment, mice were divided into groups (n = 4) receiving a coinfusion at day 0 of 5.5 μg reporter plasmid pLIVELuc/705 combined with 5.5 μg splicing plasmid (pU7asLuc705), 1.5 μg splicing miMC (±4.5 μg pFiller), or 5.5 μg pFiller only as control, see Figure 8a. In both experiments, gene expression was assessed by sequential imaging of the reporter protein, i.e., firefly luciferase activity. Anesthetized mice were injected intraperitoneally with 150 mg/kg of D-Luciferin (Caliper, Hopkinton, MA) and imaged for 1–5 minutes to analyze luminescence, using a high sensitivity CCD camera, IVIS Spectrum (PerklinElmer, Waltham, MA). The maximum photon/second of acquisition was determined within a region of interest, to give the most consistent measure for comparative analysis. Livers were harvested postmortem in the pLIVELuc/705-experiment, for RNA and DNA analysis, after the last imaging time point (7 weeks after infusion). The animal experiments were approved by The Swedish Local Board for Laboratory Animals. The experiments were performed in accordance with the ethical permission and designed to minimize the suffering and pain of the animals.
SUPPLEMENTARY MATERIAL Figure S1. Correction on luciferase mRNA level assayed by one step RT-PCR and quantified as compared to uncorrected mRNA in HeLaLuc705 cells. Figure S2. Studying the effect of different transfection methods in vitro on miMC performance mRNA correction and protein levels as well as quantification of amount of vector-DNA: Lipofection using Lipofectamine 2000 or electroporation using Neon Transfection System of HeLaLuc705 cells. Figure S3. Vector amounts, expression of snRNA, and correction of Luciferase mRNA in vivo after hydrodynamic delivery of miMC or plasmid to mouse liver (n=4).
Acknowledgments
We thank Zhi-Ying Chen and Mark A. Kay (Departments of Pediatrics and Genetics, Stanford School of Medicine) for providing the MC-producing system. We are grateful to Richard Stout, BioJect, Tualatin, OR, who provided the Biojector for mouse skin experiments. This work was supported by Stockholm County Council, Swedish Association of Local Authorities and Regions, the Swedish Research Council, the Swedish Society for Medical Research (SSMF), the Sigrid Jusélius Foundation and the EU FP7 (CHAARM grant no. 242135). The authors declared no conflict of interest.
Supplementary material
References
- Viola JR, El-Andaloussi S, Oprea II, Smith CI. Non-viral nanovectors for gene delivery: factors that govern successful therapeutics. Expert Opin Drug Deliv. 2010;7:721–735. doi: 10.1517/17425241003716810. [DOI] [PubMed] [Google Scholar]
- Gill DR, Pringle IA, Hyde SC. Progress and prospects: the design and production of plasmid vectors. Gene Ther. 2009;16:165–171. doi: 10.1038/gt.2008.183. [DOI] [PubMed] [Google Scholar]
- Vandermeulen G, Marie C, Scherman D, Préat V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol Ther. 2011;19:1942–1949. doi: 10.1038/mt.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Bigger BW, Tolmachov O, Collombet JM, Fragkos M, Palaszewski I, Coutelle C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J Biol Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Mayrhofer P, Blaesen M, Schleef M, Jechlinger W. Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. J Gene Med. 2008;10:1253–1269. doi: 10.1002/jgm.1243. [DOI] [PubMed] [Google Scholar]
- Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nat Biotechnol. 2010;28:1287–1289. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Fogg JM, Zechiedrich L, Zu Y. Transfection of shRNA-encoding Minivector DNA of a few hundred base pairs to regulate gene expression in lymphoma cells. Gene Ther. 2011;18:220–224. doi: 10.1038/gt.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang Z, Feng T. [A minicircle DNA vector-mediated siRNA to stably suppress hepatitis B virus replication and expression]. Wei Sheng Wu Xue Bao. 2012;52:191–197. [PubMed] [Google Scholar]
- Vafabakhsh R, Ha T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science. 2012;337:1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates AD, Maxwell A. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 1989;8:1861–1866. doi: 10.1002/j.1460-2075.1989.tb03582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg JM, Kolmakova N, Rees I, Magonov S, Hansma H, Perona JJ, et al. Exploring writhe in supercoiled minicircle DNA. J Phys Condens Matter. 2006;18:S145–S159. doi: 10.1088/0953-8984/18/14/S01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y, Miyashita T, Nakamura H, Kuroki K, Nagata A, Imamoto F. In vivo correlation between DNA supercoiling and transcription. Gene. 1981;13:173–184. doi: 10.1016/0378-1119(81)90006-8. [DOI] [PubMed] [Google Scholar]
- Sekiguchi JM, Kmiec EB. DNA superhelicity enhances the assembly of transcriptionally active chromatin in vitro. Mol Gen Genet. 1989;220:73–80. doi: 10.1007/BF00260859. [DOI] [PubMed] [Google Scholar]
- Kang SH, Cho MJ, Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37:6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Gorman L, Suter D, Emerick V, Schümperli D, Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahlsing HL, Yankauckas MA, Sawdey M, Gromkowski SH, Manthorpe M. Immunization with plasmid DNA using a pneumatic gun. J Immunol Methods. 1994;175:11–22. doi: 10.1016/0022-1759(94)90327-1. [DOI] [PubMed] [Google Scholar]
- Suda T, Liu D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- Stenler S, Andersson A, Simonson OE, Lundin KE, Chen ZY, Kay MA, et al. Gene transfer to mouse heart and skeletal muscles using a minicircle expressing human vascular endothelial growth factor. J Cardiovasc Pharmacol. 2009;53:18–23. doi: 10.1097/FJC.0b013e318194234e. [DOI] [PubMed] [Google Scholar]
- Leahy P, Carmichael GG, Rossomando EF. Transcription from plasmid expression vectors is increased up to 14-fold when plasmids are transfected as concatemers. Nucleic Acids Res. 1997;25:449–450. doi: 10.1093/nar/25.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C, Schmidt T, Schleef M, Friehs K, Flaschel E. Production of supercoiled multimeric plasmid DNA for biopharmaceutical application. J Biotechnol. 2003;105:205–213. doi: 10.1016/j.jbiotec.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Catanese DJ, Jr, Fogg JM, Schrock DE, 2nd, Gilbert BE, Zechiedrich L. Supercoiled Minivector DNA resists shear forces associated with gene therapy delivery. Gene Ther. 2012;19:94–100. doi: 10.1038/gt.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Epstein J, Baraceros FM, Gorak EJ, Charoenvit Y, Carucci DJ, et al. Induction of CD4(+) T cell-dependent CD8(+) type 1 responses in humans by a malaria DNA vaccine. Proc Natl Acad Sci USA. 2001;98:10817–10822. doi: 10.1073/pnas.181123498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoep JL, van der Weijden CC, Andersen HW, Westerhoff HV, Jensen PR. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur J Biochem. 2002;269:1662–1669. doi: 10.1046/j.1432-1327.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Reyes-Domínguez Y, Contreras-Ferrat G, Ramírez-Santos J, Membrillo-Hernández J, Gómez-Eichelmann MC. Plasmid DNA supercoiling and gyrase activity in Escherichia coli wild-type and rpoS stationary-phase cells. J Bacteriol. 2003;185:1097–1100. doi: 10.1128/JB.185.3.1097-1100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa J, Amano M, Fukue Y, Tanaka S, Kishi H, Hirota Y, et al. Left-handedly curved DNA regulates accessibility to cis-DNA elements in chromatin. Nucleic Acids Res. 2003;31:6651–6662. doi: 10.1093/nar/gkg854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin JD, McCormick RJ, Sharp PA, Fisher DE. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995;373:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- Rajendra Y, Kiseljak D, Manoli S, Baldi L, Hacker DL, Wurm FM. Role of non-specific DNA in reducing coding DNA requirement for transient gene expression with CHO and HEK-293E cells. Biotechnol Bioeng. 2012;109:2271–2278. doi: 10.1002/bit.24494. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Riu E, He CY, Xu H, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Riu E, Chen ZY, Xu H, He CY, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- Schüttrumpf J, Milanov P, Abriss D, Roth S, Tonn T, Seifried E. Transgene loss and changes in the promoter methylation status as determinants for expression duration in nonviral gene transfer for factor IX. Hum Gene Ther. 2011;22:101–106. doi: 10.1089/hum.2009.212. [DOI] [PubMed] [Google Scholar]
- Lundin KE, Hasan M, Moreno PM, Törnquist E, Oprea I, Svahn MG, et al. Increased stability and specificity through combined hybridization of peptide nucleic acid (PNA) and locked nucleic acid (LNA) to supercoiled plasmids for PNA-anchored “Bioplex” formation. Biomol Eng. 2005;22:185–192. doi: 10.1016/j.bioeng.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Svahn MG, Hasan M, Sigot V, Valle-Delgado JJ, Rutland MW, Lundin KE, et al. Self-assembling supramolecular complexes by single-stranded extension from plasmid DNA. Oligonucleotides. 2007;17:80–94. doi: 10.1089/oli.2006.0045. [DOI] [PubMed] [Google Scholar]
- Verbeeren J, Niemelä EH, Turunen JJ, Will CL, Ravantti JJ, Lührmann R, et al. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell. 2010;37:821–833. doi: 10.1016/j.molcel.2010.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.