Abstract
Background
Little is known for certain about the genetics of schizophrenia. The advent of genomewide association has been widely anticipated as holding promise as a means to identify reproducible DNA sequence variation associated with this important and debilitating disorder.
Methods
738 cases with DSM-IV schizophrenia (all participants in the CATIE study) and 733 group-matched controls were genotyped for 492,900 single nucleotide polymorphisms (SNPs) using the Affymetrix 500K two chip genotyping platform plus a custom 164K fill-in chip. Following multiple quality control steps for both subjects and SNPs, logistic regression analyses were used to assess the evidence for association of all SNPs with schizophrenia.
Results
We identified a number of promising SNPs for follow-up studies, although no SNP or multi-marker combination of SNPs achieved genomewide statistical significance. Although a few signals coincided with genomic regions previously implicated in schizophrenia, chance could not be excluded.
Conclusions
These data do not provide evidence for the involvement of any genomic region with schizophrenia detectable with moderate sample size. However, planned GWAS for response phenotypes and inclusion of individual phenotype and genotype data from this study in meta-analyses holds promise for the eventual identification of susceptibility and protective variants.
Keywords: schizophrenia, genome-wide association, CATIE
Introduction
In the past several decades, it has become generally accepted that schizophrenia (SCZ) is a complex trait with a substantial genetic component. The accumulated evidence suggests that SCZ is a relatively common disorder with lifetime morbid risk of 0.72% (1, 2). Genetic factors have been strongly and consistently implicated via family, adoption, and twin studies (3, 4). A meta-analytic estimate of the broad sense heritability (5) of SCZ was 81% (95% CI, 73%–90%) (4) and the recurrence risk to siblings (λsibs) (6) was 8.55 (95% CI 7.86–9.57) in a population-based Swedish national sample of over 7.7 million individuals (2), consistent with prior estimates (3).
These results have been used in support of the application of an array of linkage and association methods (7) attempting to identify genomic regions that confer risk for or protection against the development of SCZ. These efforts have been considerable: we have identified 31 independent samples in which genomewide linkage has been applied (3,108 multiplex SCZ pedigrees and over 8.3 million genotypes) † and >1,100 association studies of 525 candidate genes for SCZ have been published ‡.
Despite these efforts, little is known for certain about the genetics of SCZ. The genomewide linkage studies have not converged to identify a set of high priority genomic regions (8) ; indeed, no genomic region has been implicated in more than four of 31 genomewide linkage studies and a perhaps implausibly large 58% of the genome has been implicated at least once. The accumulated evidence for specific candidate genes such as NRG1 (9, 10), DTNBP1 (11, 12), or DISC1 (13) does not constitute unequivocal and consistent replication (14). These findings stand in contrast to other results in human complex trait genetics – for example, genetic variation in intron 1 FTO was associated with body mass index in 13 cohorts and 38,759 individuals with remarkable consistency (15). The possibility that all members of a consensus set of the best candidate genes for SCZ are false positives has not been convincingly excluded.
In the past two years, the widely anticipated method of genomewide association study (GWAS) has become technically and economically feasible. These studies entail individual genotyping of considerable numbers of cases and controls for 100,000 or more genetic markers (single nucleotide polymorphisms, SNPs). Evident successes in identifying highly compelling candidate genes for age-related macular degeneration (16), body mass index (15), inflammatory bowel disease (17, 18), type 1 diabetes mellitus (18), and type 2 diabetes mellitus (19–22) support the utility of GWAS. To date, two GWAS that employed individual genotyping have been published for psychiatric disorders – a small study of SCZ (23) and the Wellcome Trust Case-Control Consortium study of bipolar disorder (18). Multiple GWAS are known to be in progress for attention-deficit hyperactivity disorder, autism, bipolar disorder, and major depressive disorder as well as for SCZ.
In 2006, academic investigators from the NIMH-funded Clinical Antipsychotic Trials of Intervention Effectiveness project (CATIE) (24, 25) entered into a scientific collaboration with Eli Lilly and Company to conduct individual GWAS genotyping and joint analyses on the CATIE samples. The collaboration contract required that genotype and phenotype data be made available to the scientific community with intellectual property rights consistent with NIH policies intended to maximize the public benefit resulting from the research. The genotype and phenotype data reported here were deposited with the controlled-access repository of the NIMH in 6/2007 †.
We report here the primary analyses aimed at identifying SNPs associated with susceptibility to SCZ using 492,900 SNPs that were genotyped in 738 participants with SCZ and 733 group-matched controls from a United States population based sample.
Methods
We have attempted to follow published guidelines for GWAS (26, Box 1).
Subjects
All cases were participants in the CATIE project (NIMH contract NO1 MH90001) which was conducted between 1/2001-12/2004. CATIE was a multi-phase randomized controlled trial of antipsychotic medications involving 1,460 persons with SCZ followed for up to 18 months (24, 27). The philosophy of the trial was to assess controlled treatment with antipsychotic drugs in a broad range of patients with SCZ under “real world” conditions. To maximize representativeness, subjects were ascertained from an array of clinical settings across the US. 1,894 subjects were evaluated and 1,460 (77.0%) entered into CATIE. No subject was known to be related to any other subject. The optional genetic sub-study began about a year after the trial began and 51% of CATIE participants donated a DNA sample.
Preliminary diagnoses were established by referring psychiatrists and final study diagnoses of DSM-IV SCZ (28) were independently established by CATIE personnel using the Structured Clinical Interview for DSM-IV (SCID) (29) including review of all available information (including psychiatric and general medical records) along with one or more subject interviews. Interviewers were experienced Master’s-level clinicians who were trained to criterion via a standard protocol (29). Any diagnostic uncertainties were resolved via discussion with one of the CATIE senior clinicians. Study inclusion criteria were: definite diagnosis of SCZ (28, 29), age 18–67 years, clinical decision that oral medication was appropriate, adequate decisional capacity, and provision of written informed consent. Exclusion criteria are detailed elsewhere (24). Briefly, patients were excluded if they had: received a diagnosis of schizoaffective disorder, mental retardation, or another cognitive disorder; a history of serious adverse reactions to the proposed treatments; only one psychotic episode; a history of treatment resistance (defined by the persistence of severe symptoms despite adequate trials of one of the proposed treatments or prior treatment with clozapine); pregnant or breastfeeding; or a serious and unstable medical condition. Individuals with psychoactive drug use disorders were included but only when there was positive evidence that SCZ was an independent diagnosis.
Controls were ascertained from a US national sampling frame as part of the NIMH Genetics repository (MH059571, PI Dr. Pablo Gejman, release v4.0, 6/2006). Controls were collected by Knowledge Networks (KN), a survey and market research company whose panel contains approximately 60,000 households (>120,000 unrelated adults). Households were selected via random digit dialing and proportionally from 25 major US population areas and financial incentives were provided for participation. The KN panel is generally representative of the US population but with a slight bias toward higher income and education. To be eligible for matching to cases, we required that potential controls deny any history of SCZ, schizoaffective disorder, bipolar disorder, auditory hallucinations, and delusional beliefs. These controls are being used for multiple additional GWAS and meta-analysts need to use caution when combining results or data across studies that use these common controls.
Case-control matching
There were 3,487 individuals in v4.0 of the control dataset and 2,645 controls were eligible for matching to CATIE cases – 842 individuals were removed from consideration due to at least one of the following: a self-reported possible history of a psychotic disorder (SCZ, schizoaffective disorder, bipolar disorder, delusional disorder, or auditory hallucinations), age <18 or age >67, lack of documentation of informed consent, or missing data for a matching variable (age, sex, or self-reported race). Cases were group-matched to controls by five-year age band, sex, and self-reported race. Most cases (91.4%) were successfully matched to controls. The exception was for 66 cases (8.6%) that could not be matched due to a deficiency of African-American males aged 18–38 years in v4.0 of the control cohort. Rather than eliminate cases, African-American females in this age band were selected as controls.
DNA Sampling & Cell Line Establishment
Peripheral venous blood samples were sent to the Rutgers University Cell and DNA Repository (RUCDR) where cell lines were established via EBV transformation. RUCDR employs stringent quality control procedures and the success rate for immortalization exceeds 99%. Sample DNA concentrations were quantified and normalized via the use of Picogreen dsDNA Quantitation Kits (Molecular Probes, Eugene, OR).
Ethical Issues
The CATIE study was approved by the institutional review board at each site, and written informed consent was obtained from the subjects or their legal guardians (including an additional consent for genetic studies). Written informed consent was obtained from all controls. All biological samples, phenotypes, and genotypes were de-identified before deposit into the NIMH/RUDCR repositories. Although the analyses described here are generally considered not to be “human subjects research” as defined by the relevant US statutes (45 CFR part 46), this research was reviewed and approved by the IRB at UNC-Chapel Hill.
GWAS Genotyping
Individual genotyping was conducted by Perlegen Sciences (Mountain View, CA, USA) using three genotyping chips – Affymetrix 500K “A” chipset (Nsp I and Sty I chips), Santa Clara, CA, USA) (30) and a custom 164K chip created by Perlegen (31) to provide additional genome coverage. Genotype calls were generated with a proprietary Perlegen algorithm (32) applied to the .cel files from cases and controls together. The Perlegen calling algorithm shows excellent agreement with genotype calls from BRLMM (33) in a subset of the controls used in this study (see below) as well as in genotyping of 270 HapMap samples as part of GAIN (34) †. Supplemental Methods Part 1 provides additional details.
Genotyping Quality Control
There were 500,568 SNPs on the Affymetrix 500K chips and 164,871 on the Perlegen custom chip (665,439 SNPs in total). Of these, 157,048 SNPs failed Perlegen’s quality control (20.2% of the Affymetrix 500K and 33.9% of the Perlegen 164K SNPs). The QC process is detailed below (35).
First, the Perlegen genotyping algorithm yielded a quality score for each individual genotype; after inspection of the quality score distributions, a more stringent quality score cutoff (≥7) than that originally applied by Perlegen was used.
Second, duplicate agreement was investigated. DNA samples from cases and controls were on 18 × 96 well plates. Two samples from each plate were chosen at random and included a second time on that plate (from the same DNA aliquot) for a total of 36 pairs of samples that were genotyped twice, 18 pairs for cases and 18 pairs for controls. The proportion of SNPs with non-missing genotype calls that disagreed in these duplicated samples was 0.00291 for cases and 0.00339 for controls.
Third, in order to investigate the impact of the genotype calling algorithm and genotyping facility, Perlegen genotype calls were directly compared to BRLMM (33) genotype calls. Of the controls used here, 277 individuals were independently genotyped using the Affymetrix 500K “A” chips at the MIT/Harvard Broad Institute by Dr. Pamela Sklar ‡. There were 370,704 pairs of SNPs (168,299,616 genotypes) that passed quality control at both genotyping centers. The proportion of genotypes that were called at both centers was 0.98407 and the proportion of disagreements among called genotypes was 0.00731. These results suggest that the impact of calling algorithm and genotyping facility was not substantial.
Fourth, the genotyping data were used to select the final set of subjects for analysis. The identity-by-state matrix for all autosomal genotypes for all pairwise combinations of subjects was generated using PLINK (36). Four pairs of cases were cryptic duplicates and one member of each pair was removed. The numbers of heterozygous and homozygous genotypes on chrX (excluding the pseudo-autosomal regions) and chrY for each subject were counted and compared to phenotypic sex – 21 subjects (3 cases and 18 controls) with sex discrepancies were removed. Twelve subjects (9 cases and 3 controls) were removed for excessive missing genotypes (≥0.05). After application of these filters, there were 1,471 subjects (738 cases and 733 controls).
Fifth, the final set of SNPs was selected for analysis. Beginning with the 508,286 SNPs delivered by Perlegen, 76 duplicated SNPs, 3,803 SNPs with minor allele frequencies <0.01, 9,279 SNPs with missingness >0.05362 (the 99.5th percentile in this dataset), and 2,272 SNPs that led to ≥2 disagreements in 36 pairs of duplicated samples were removed. SNPs were not excluded based solely on deviations from Hardy-Weinberg Equilibrium (37) given the ancestries of the subjects and as there are informative reasons for departures from HWE (38). After application of these filters, there were 492,900 SNPs for analysis (44 SNPs were excluded for multiple reasons). We investigated batch effects and found none.
Control of Population Stratification
Cases and controls are of mixed ancestry. Using the US census “race” categories, 56.3% of subjects described themselves as only White, 29.6% as only Black/African-American, and 14.1% selected some other or more than one racial category (i.e., American Indian/Alaska Native, Asian, Black/African American, Native Hawaiian/Other Pacific Islander; White, and Hispanic/Latino). GWAS in a sample of mixed ancestry can yield false positive findings due to population stratification if there are sufficiently large differences in the phenotype and genotype distributions within the strata defined by ancestry (39, 40).
There were two critical decisions in regard to the analysis of these GWAS data. First, rather than stratifying the analysis by self-reported "race" (an imperfect proxy for ancestry) (41), the entire cohort was analyzed together in order to maximize statistical power. Second, the available statistical methods for population stratification control were evaluated. At least five major methods for population stratification control have been described – genomic control (42), structured association (43), principal components (44), multi-dimensional scaling (36), and partial least squares relating phenotype to ancestry-informative markers (45, 46). These methods were evaluated extensively using these GWAS data – see Supplemental Methods Part 2 and Lee et al. (in preparation) for details. Briefly, the central criteria in evaluating these methods were control of Type 1 error while preserving statistical power. To avoid bias, these comparisons were based on lists of p-values alone and were blinded to all SNP rs numbers and annotation information.
The consensus among the statistical geneticists involved in these evaluations was to use principal components (44, 47) in order robustly to control for stratification effects while preserving statistical power. The consensus was to use seven principal components as covariates in all logistic regression models (see Supplemental Methods Part 2). Moreover, as has been reported elsewhere (18, 44), the principal components method can capture both subtle and extensive variation due to both genomic and experimental features and, with the availability of >105 genetic markers, self-reported “race” is no longer required as a proxy for ancestry (41). The principal components method is computationally efficient and uses the genotyping matrix to infer continuous axes of genetic variation (eigenvectors) which then serve as covariates. The potential weakness of this method is lowered statistical power in some genomic regions – if a SNP is highly stratified, then it could contribute strongly to a principal component and induce collinearity. This concern is mitigated by the fact that large numbers of SNPs contribute to each principal component. All autosomal GWAS SNPs were used as input to EigenSoft † (44, 47) and default parameters were used except that the outlier removal option was turned off in order to generate estimates for all subjects.
Single Marker Analyses
All autosomal and pseudo-autosomal SNPs that passed quality control checks were tested for association with SCZ using logistic regression under a log-additive mode of inheritance (1 df, each SNP coded as the number of minor alleles). All regression models included the first seven principal components as covariates in order to adjust for stratification and any artifacts detectible in the GWAS data. Of the different inheritance models that could be considered, log-additive model appear to minimize the number of comparisons while detecting effects under a range of unknown inheritance models (48). For chrX SNPs, a sex term was added as a covariate and the number of copies of the minor allele were coded as 0 or 2 for males and 0, 1, or 2 for females. Analyses of chrY SNPs were necessarily limited to males.
Control of False Discoveries
Given the ~500,000 statistical comparisons in a GWAS, highly significant findings (p<10−6) are expected by chance. To control the risk of false discoveries, q-values (49, 50) were computed for all p-values for single-marker tests of association. A q-value is an estimate of the proportion of false discoveries among all significant markers. In other words, a q-value is the false discovery rate (FDR) for the corresponding p-value. As argued elsewhere (51, 52), the use of q-values is preferable to more traditional multiple testing controls because q-values: a) provide a better balance between the competing goals of finding true positives versus controlling false discoveries; b) allow more similar comparisons across studies because proportions of false discoveries are much less dependent on the number of tests conducted; c) are relatively robust against the effects of correlated tests (49, 53–60); and d) provide a more nuanced picture regarding the possible relevance of the tested markers rather than an all-or-nothing conclusion about whether a study produces significant results. The q-value threshold for declaring significance was 0.10 – i.e., the top 10% of the significant findings are, on average, allowed to be false discoveries (see references (51, 56) for a thorough consideration of these issues). FDR thresholds <0.10 result in a disproportional drop in power to detect true effects.
Multi-Marker Analyses
The score test of Schaid et al. (61) implemented in the R (62) (v2.5.1) function "haplo.score" (v1.3.1) † was used for multi-marker association analyses. This test uses unphased genotypes and probabilistic haplotype assignments in order to estimate haplotype-phenotype association within a given genomic region. Overlapping sliding windows over the genome (500 kb segments plus 200 kb segments centered on boundaries between the 500 kb blocks) were used to estimate linkage disequilibrium (LD) and to form haplotypes via the Gabriel method (63) implemented in HaploView (64). LD blocks were estimated in European subjects, the predominant ancestry group in this sample. The resulting 74,411 haplotypes have a median length of 8.2 kb and inter-quartile range of 2.7–20.0 kb. These haplotypes contained 335,539 SNPs (69.23% of all autosomal SNPs) and covered 1.368 mb (47.78% of the autosomal genome). As haplotype frequencies differ by ancestry (65), an assumption of haplo.score – that all subgroups share the same haplotype frequencies – was violated. Therefore, GWAS multi-marker analyses were run in separate subgroups, and a combined test statistic is reported.
Statistical Power
Quanto (66, 67) was used to generate an illustrative approximation of statistical power. An FDR framework was used to determine the α level – assuming that ~25 of 492,900 GWAS SNPs have true effects (i.e., the proportion of SNPs without true effects p0=0.99995), two-tailed α=4.4 × 10−6. Additional assumptions were: 738 cases and 733 controls, lifetime morbid risk of SCZ of 0.0072 (1, 2), and a log additive genetic model. For statistical power of 0.80 (β=0.20), the minimum detectible genotypic relative risks are 1.83, 1.56, and 1.50 for minor allele frequencies of 0.10, 0.25, and 0.40. While the logistic model employed in our study includes seven EigenSoft principal components as covariates, the power analyses remain approximately correct as the additional covariates subtract only a very modest number of degrees of freedom from the test statistic (see Supplemental Methods). Moreover, the use of principal components could have reduced power if they accounted for variance due to case-control status.
Software
PLINK (36), SAS (v9.1.3) (68), JMP (69), and UCSC Genome Graphs (70) were used for data management, quality control, statistical analyses, and graphics. Statistical analyses conducted using PLINK (36) and the SAS LOGISTIC procedure (68) produced identical results.
Bioinformatics
All genomic locations are based on NCBI Build 35 (71) (UCSC hg17) (70). Pseudo-autosomal region 1 (PAR1) is assumed to be located on chrX:1–2,692,881 and chrY:1–2,692,881 and PAR2 on chrX:154,494,747-154,824,264 and chrY:57,372,174-57,701,691 (72). SNP annotations were created using the TAMAL database (73) based chiefly on UCSC genome browser files (70), HapMap (65), and dbSNP (71) using a set of SAS programs. These annotation files are available for download and there is a searchable repository for genomewide studies in neuropsychiatry at the same site †. All gene names referenced are the standard names set by the HUGO Gene Nomenclature Committee.
Fisher’s method (74) was used to combine lists of p-values for the same SNPs (e.g., from different GWAS) or for the same haplotypes (i.e., from AFR and EUR subjects in this GWAS). The test statistic for K independent p-values is. .
Project Context
A list of completed and proposed genotyping projects using the CATIE samples is available on-line ‡. Future manuscripts using these GWAS data will investigate copy number variation and genetic influence on treatment response, tardive dyskinesia, and neurocognition. The NIMH controls are being used in multiple other projects.
Results
Sample Description
Table 1 presents descriptive data for cases and controls. Cases and controls were well-matched for age and ancestry but not for sex (due to insufficient numbers of African-American males in the control pool as described in the Methods). There were large differences between cases and controls in education, marital status, and employment consistent with the adverse effects of a chronic mental illness with onset in early adulthood. Cases had been ill for a mean of 14 years and the mean PANSS scores (75) are consistent with a moderately ill sample. As described previously (76), CATIE subjects who provided DNA samples had lower symptom severity (PANSS total 74 vs. 77), lesser current drug/alcohol abuse/dependence (29% vs. 36%) and less likely to describe themselves as African-American (29% vs. 40%) in comparison to CATIE subjects who did not provide a DNA sample.
Table 1.
Descriptive data for cases with schizophrenia and controls included in GWAS.
| Subject Descriptor | Cases | Controls | Test | |
|---|---|---|---|---|
| Number of subjects genotyped | 738 | 733 | - | |
| Mean age in years (SD) † | 40.9 (11.1) | 41.0 (11.6) | F1,1469 =0.09 p=0.77 | |
| Proportion male (N) † | 0.74 (544) | 0.67 (493) | ||
| Ancestry proportions (inferred from
self-report) † African only (N) European only (N) Other (N) |
0.29 (217) 0.57 (417) 0.14 (104) |
0.30 (219) 0.56 (411) 0.14 (103) |
|
|
| Proportion with high school degree or more (N) | 0.74 (543) | 0.93 (684) | ||
| Proportion married (N) | 0.11 (78) | 0.57 (415) | ||
| Proportion employed (N) | 0.06 (43) | 0.75 (548) | ||
| Mean years since first antipsychotic prescribed | 14.2 (10.8) | - | ||
| Mean PANSS total score | 74.0 (17.2) | - | ||
| Mean PANSS positive symptom score | 17.8 (5.5) | - | ||
| Mean PANSS negative symptom score | 19.9 (6.4) | - |
Matching variable.
SNP Description
The analysis SNP set had 492,900 SNPs including 484,664 autosomal SNPs, 8,084 SNPs on chrX, 143 SNPs on chrXY/PAR1, 9 SNPs on chrY, and 0 SNPs on PAR2 and chrM (mitochondrial DNA). Table 2 depicts summary missingness and MAF estimates from the GWAS analysis set in the entire sample and separately for cases and controls; on average across the genome, SNP missingness appears to be acceptably low. The average marker density over the genome was 1 SNP every 6.2 kb (=3,077,088,087 bases / 492,900 SNPs). The median inter-marker distance was 2.8 kb with an interquartile range 0.8–7.1 kb and the 99th percentile was 39.7 kb.
Table 2.
Distribution descriptors for missingness and MAF in GWAS SNP set †.
| Property | SNP Location | All subjects | Cases | Controls |
|---|---|---|---|---|
| Missingness | chr1-22 | 0.0048 (0.0014–0.0116) | 0.0041 (0.0014–0.0108) | 0.0041 (0.0014–0.0123) |
| chrX | 0.0082 (0.0027–0.0177) | 0.0081 (0.0027–0.0163) | 0.0082 (0.0027–0.0177) | |
| PAR1 | 0.0082 (0.0048–0.0180) | 0.0081 (0.0041–0.0149) | 0.0082 (0.0041–0.0177) | |
| chrY ‡ | 0.0010 (0.0005–0.0039) | 0.0000 (0.0000–0.0018) | 0.0020 (0.0000–0.0071) | |
| MAF | chr1-22 | 0.212 (0.102–0.347) | 0.213 (0.103–0.347) | 0.212 (0.100–0.346) |
| chrX | 0.241 (0.121–0.371) | 0.241 (0.122–0.369) | 0.240 (0.119–0.370) | |
| PAR1 | 0.240 (0.073–0.372) | 0.241 (0.071–0.375) | 0.244 (0.073–0.374) | |
| chrY ‡ | 0.218 (0.055–0.462) | 0.252 (0.052–0.465) | 0.181 (0.057–0.456) |
Data are medians (25th-75th percentiles). MAF=minor allele frequency.
Males only.
Single Marker Association Tests
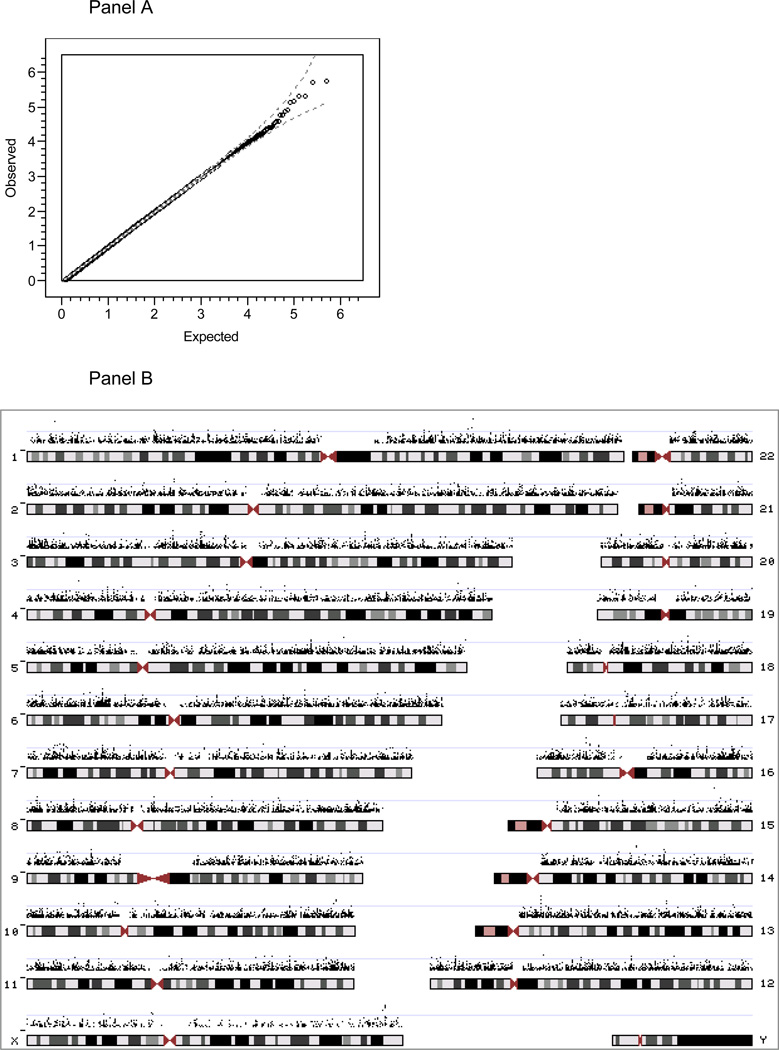
We used logistic regression to test for association of the 492,900 SNPs in the GWAS dataset with case/control status (with seven principal components (44) included as covariates to account for population stratification). The minimum p-value obtained was 1.71 × 10−6. There were 26,738 p-values <0.05 – six p-values in the 10−6 bin, 42 in the 10−5 bin, 486 in the 10−4 bin, 4,845 in the 10−3 bin, and 21,359 in the interval [0.01–0.05). The GWAS results are depicted in Figure 1. Panel A shows the QQ plot (77) for GWAS for case-control status. The QQ plot suggests that the observed p-values do not strongly depart from the p-value distribution expected by chance. Panel B shows the −log10(p) for the 26,738 p-values < 0.05 (5.425%) in the context of the human genome in order to make spatial clustering evident. To facilitate comparisons with other datasets, inclusion in meta-analyses, and further investigation of these findings, all p-values are available in Supplemental Table 2 and the data used to create Figure 1b are contained in Supplemental Table 3. SNPs that failed quality control are contained in Supplemental Table 4.
Figure 1.
Results of single-marker association tests of case-control status for 492,900 SNPs. Panel A. shows the QQ plot (77) of the observed p-values, −log10 (p), versus those expected by chance, , where p is the asymptotic p-value from the additive test that the SNP coefficient is zero, i is the rank for each SNP p-value (1=smallest, L=largest), and L is the number of SNPs. The dashed lines show the expected 95% probability interval for ordered p-values. Panel B depicts −log10 (p) for the 26,738 SNPs with p-values < 0.05 in the context of the human genome in order to make spatial clustering more evident.
For an FDR threshold of 0.10, the proportion of all SNPs without true effects (p0) can be estimated from the GWAS results (60) and was found to be p0= 0.9999904. This result is consistent with the presence of ~5 SNPs with true effects in these GWAS data for SCZ. Q-values were calculated for all p-values under the conservative assumption that p0=1, and no SNPs reached genomewide significance as the minimum q-values of 0.45 (rs10911902 and rs16977195) did not exceed the pre-specified FDR threshold of 0.10 (Table 3). Nearly identical results were obtained using p0=0.9999904 (data not shown).
Table 3.
Top 25 results from GWAS for SCZ in the CATIE study †
| SNP data | Gene data | Logistic regression tests for association | Allele frequency data | HapMap Allele FreqsComments ‡ | Context | SNP annotations | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chromosome | Position | Transcripts in which SNP locate | Odds Ratio | Wald test | p-value | q-value | Rank | AFR odds ratio | EUR odds ratio | Cases | Controls | CATIE AFR | CATIE EUR | AFR/YRI | EUR/CEU | SNPs with p<0.0 | Proxy SNP ≤0.00 | CNV | Conserved p95 | Regulatory potential | Transfactor binding site | |
| rs4846033 | 1 | 11,722,830 | 0.348 | −4.594 | 4.36E-06 | 0.56 | 3 | 0.401 | 0.146 | 0.025 | 0.053 | 0.103 | 0.009 | 0.179 | 0.009 | 1 | Isolated finding | No | no | no | Yes | no | |
| rs10911902 | 1 | 183,363,974 | 0.560 | −4.770 | 1.85E-06 | 0.45 | 2 | 0.627 | 0.523 | 0.099 | 0.171 | 0.046 | 0.173 | 0.000 | 0.200 | 2 SNPs, 0.4 kb | No | no | no | Yes | Yes | ||
| rs9309325 | 2 | 60,492,482 | 0.727 | −4.076 | 4.58E-05 | 0.85 | 22 | 0.704 | 0.770 | 0.382 | 0.455 | 0.414 | 0.414 | 0.433 | 0.458 | 6 SNPs, 7 kb | Yes | Yes | Yes | Yes | no | ||
| rs1569351 | 3 | 60,287,457 | FHIT | 0.697 | −4.225 | 2.39E-05 | 0.85 | 11 | 0.609 | 0.729 | 0.337 | 0.415 | 0.173 | 0.470 | 0.192 | 0.433 | 12 SNPs, 31 kb | Yes | Yes | no | Yes | no | |
| rs4568102 | 3 | 72,070,679 | 4.118 | 4.103 | 4.08E-05 | 0.85 | 19 | 4.947 | 0.663 | 0.035 | 0.008 | 0.062 | 0.003 | 0.117 | 0.000 | 1, 2 | 2 SNPs, 5 kb | Yes | no | no | Yes | no | |
| rs1380272 | 4 | 21,431,975 | KCNIP4 | 0.522 | −4.397 | 1.10E-05 | 0.74 | 7 | 0.425 | 0.610 | 0.057 | 0.107 | 0.074 | 0.088 | 0.033 | 0.102 | 6 | 4 SNPs, 18 kb | Yes | no | no | Yes | no |
| rs1495716 | 4 | 177,103,725 | GPM6A | 1.423 | 4.223 | 2.41E-05 | 0.85 | 12 | 1.915 | 1.277 | 0.365 | 0.296 | 0.208 | 0.391 | 0.142 | 0.425 | 2 SNPs, 8 kb | No | no | no | no | no | |
| rs9295938 | 6 | 31,061,084 | C6orf205 | 1.642 | 4.138 | 3.51E-05 | 0.85 | 16 | 1.288 | 1.769 | 0.143 | 0.097 | 0.072 | 0.143 | 0.033 | 0.192 | 7 SNPs, 17 kb | No | no | no | no | no | |
| rs9400690 | 6 | 114,453,545 | 1.534 | 4.052 | 5.08E-05 | 0.85 | 24 | 1.588 | 1.635 | 0.193 | 0.151 | 0.057 | 0.230 | 0.000 | 0.267 | 3 SNPs, 6 kb | No | no | no | no | no | ||
| rs16917897 | 10 | 52,856,709 | D45864 PRKG1 Y07512 BC062 | 2.750 | 4.078 | 4.55E-05 | 0.85 | 21 | 2.864 | 1.650 | 0.049 | 0.019 | 0.099 | 0.005 | 0.119 | 0.000 | 1, 6 | Isolated finding | No | no | no | Yes | no |
| rs297257 | 10 | 127,298,704 | 0.689 | −4.129 | 3.64E-05 | 0.85 | 17 | 0.727 | 0.705 | 0.452 | 0.498 | 0.805 | 0.318 | 0.883 | 0.367 | 3 SNPs, 12 kb | Yes | no | no | Yes | no | ||
| rs9512730 | 13 | 26,975,144 | 1.515 | 4.586 | 4.52E-06 | 0.56 | 4 | 1.724 | 1.385 | 0.274 | 0.211 | 0.212 | 0.262 | 0.229 | 0.263 | 6 SNPs, 38 kb | Yes | no | no | no | no | ||
| rs942348 | 13 | 26,976,147 | 0.718 | −4.194 | 2.74E-05 | 0.85 | 14 | 1.641 | 1.399 | 0.451 | 0.521 | 0.311 | 0.547 | 0.220 | 0.500 | no | no | Yes | no | ||||
| rs17070578 | 13 | 78,431,983 | 1.903 | 4.069 | 4.72E-05 | 0.85 | 23 | 1.560 | 1.589 | 0.080 | 0.052 | 0.029 | 0.092 | 0.008 | 0.117 | 6 | 3 SNPs, 9 kb | No | no | no | no | no | |
| rs17095545 | 14 | 58,555,364 | 1.648 | 4.204 | 2.62E-05 | 0.85 | 13 | 1.501 | 1.746 | 0.153 | 0.099 | 0.205 | 0.094 | 0.217 | 0.100 | Isolated finding | No | no | no | no | no | ||
| rs7144633 | 14 | 58,582,146 | 1.784 | 4.309 | 1.64E-05 | 0.81 | 10 | 1.246 | 1.987 | 0.117 | 0.076 | 0.095 | 0.102 | 0.100 | 0.100 | Isolated finding | No | no | no | no | no | ||
| rs16977195 | 15 | 84,785,244 | AGBL1 | 6.005 | 4.785 | 1.71E-06 | 0.45 | 1 | 2.028 | 6.801 | 0.035 | 0.006 | 0.003 | 0.028 | 0.000 | 0.017 | 1 | 2 SNPs, 0.6 kb | No | no | no | Yes | no |
| rs234993 | 16 | 20,555,145 | ACSM1 BUCS1 | 0.524 | −4.036 | 5.44E-05 | 0.85 | 25 | 0.263 | 0.596 | 0.049 | 0.091 | 0.034 | 0.083 | 0.017 | 0.042 | 2 SNPs, 0.4 kb | No | no | no | Yes | no | |
| rs151222 | 16 | 20,581,993 | ACSM1 BUCS1 | 0.477 | −4.524 | 6.08E-06 | 0.57 | 5 | 0.310 | 0.549 | 0.045 | 0.093 | 0.038 | 0.080 | 0.017 | 0.042 | 3 SNPs, 2 kb | No | no | no | no | no | |
| rs17455133 | 18 | 49,719,513 | 0.643 | −4.142 | 3.44E-05 | 0.85 | 15 | 0.357 | 0.713 | 0.138 | 0.211 | 0.090 | 0.216 | 0.075 | 0.233 | 19 SNPs, 60 kb | Yes | no | no | Yes | no | ||
| rs2824301 | 21 | 17,738,145 | 0.720 | −4.103 | 4.09E-05 | 0.85 | 20 | 1.917 | 1.386 | 0.421 | 0.484 | 0.267 | 0.525 | 0.192 | 0.552 | 6 SNPs, 47 kb | Yes | no | no | Yes | no | ||
| rs10521865 | X | 146,501,918 | 0.550 | −4.125 | 3.70E-05 | 0.85 | 18 | 0.809 | 0.304 | 0.028 | 0.078 | 0.013 | 0.071 | 0.000 | 0.067 | 1, 3, 4, 6 | Isolated finding | No | no | no | Yes | no | |
| rs2159767 | X | 147,086,567 | 0.752 | −4.496 | 6.94E-06 | 0.57 | 6 | 0.288 | 0.753 | 0.430 | 0.549 | 0.274 | 0.618 | 0.144 | 0.678 | 5 | 2 SNPs 13 kb | Yes | no | no | Yes | no | |
| rs2536589 | X | 147,099,127 | 0.757 | −4.377 | 1.21E-05 | 0.74 | 8 | 0.285 | 0.762 | 0.412 | 0.529 | 0.291 | 0.639 | 0.156 | 0.678 | 5 | no | no | Yes | no | |||
| rs952515 | X | 147,142,998 | 0.759 | −4.321 | 1.56E-05 | 0.81 | 9 | 0.337 | 0.711 | 0.424 | 0.546 | 0.273 | 0.625 | 0.144 | 0.678 | 5 | 7 SNPs, 24 kb | Yes | no | no | Yes | no | |
All allele frequency data were standardized to the same reference alleles. Yellow boxes highlight issues of concern. Although SNPs with overall MAF<0.01 were dropped In the QC process, subgroups may have MAF<0.01. Some MAF>0.5 as the reference allele may be different in subgroups. EUR=European ancestry, AFR=African ancestry.
1=rare allele, 2=direction of effects different in AFR and EUR subgroups, 3=low mean SNP quality scores, 4=greater SNP missingness in cases than controls, 5=frequency difference of >10% versus HapMap 6=suboptimal clustering of genotype groups on manual inspection of scatterplots
Additional data about the SNPs with the 25 smallest p-values are shown in Table 3. First, the allele frequency differences between CATIE AFR and EUR subjects were generally similar (median difference 0.09, inter-quartile range 0.05–0.25) suggesting that one precondition for population stratification artifacts was not universally met. Moreover, the SNP with the greatest MAF difference (rs297257, |MAFAFR-MAFEUR|=0.49) yielded similar odds ratio estimates in AFR and EUR subjects (0.73 and 0.71) despite the allele frequency difference. Second, critically, the odds ratios in Table 3 were generally of similar direction and magnitude for the overall sample (including seven principal components) as well as in logistic regressions without covariates for AFR and EUR subjects separately (note that significance tests were not conducted for the stratified logistic regressions). There was one exception as noted in Table 3 (rs4568102). These findings support the approach to population stratification control used here, and suggest that the overall odds ratios were generally unlikely to be caused by stratification artifacts despite different allele frequencies in subjects of EUR and AFR ancestries.
Third, SNP allele frequencies in CATIE were reassuringly similar to those in the HapMap panels with the possible exception of three chrX SNPs in CATIE AFR subjects. Fourth, some results in Table 3 may be problematical due to lack of robustness of odds ratio estimates for uncommon alleles (rs4846033, rs4568102, rs16917897, rs16977195, rs10521865), sub-optimal performance of the calling algorithm upon manual review (rs1380272, rs16917897, and rs17070578), and differential missingness in cases and controls (rs10521865). The existence of a high LD “proxy” SNP that also shows association reduces the chance of a false positive due to genotyping artifact.
Fifth, most of the top 25 findings were not located within the transcript of a known gene and, for the 11 genes listed in Table 3, searches of PubMed and SZGene identified no published studies of schizophrenia although prior linkage studies implicated these genomic regions. Sixth, two pairs of SNPs in the top 25 were located near one another and two additional SNPs clustered with over 10 SNPs that were nominally significant and in relatively close proximity. Finally, Table 3 also presents SNP annotations; two SNPs were in copy number variant regions, one SNP was modestly conserved, one SNP was in a predicted transfactor binding site, and 17 of the top 25 SNPs were predicted to be in regions with regulatory potential.
Spatial clustering of SNPs with significant associations may be an indication that a genomic region does not represent a spurious finding, particularly if the degree of linkage disequilibrium is not very high. As a descriptive approach prior to multi-marker analyses, SNP clusters with p-values <0.05 and with distances to the next significant marker ≤15 kb were identified. For the 26,738 SNPs with p-values <0.05, 11,638 occurred in isolation, and there were 4,427 clusters containing 2–4 SNPs, 550 clusters containing 5–9 SNPs, 60 clusters containing 10–14 SNPs, and 12 clusters containing 15–22 SNPs. Additional data about the latter set of clusters (15–22 SNPs, p<0.05, and inter-marker distances of ≤15 kb) are shown in Table 4. The overlap of the genes in these regions with genes previously studied for SCZ is limited to a single study of FMO3 (78). Several regions would appear intriguing, particularly chr7:31,322,276-31,375,664 and chr18:49,714,826-49,774,618.
Table 4.
Clusters containing ≥15 SNPs (all p<0.05 and inter-marker distances of <15 kb).
| Chr. | Start | End | Distance | SNPs in cluster (all p<0.05) |
SNPs with p<0.001 |
Minimum p | RefSeq genes |
|---|---|---|---|---|---|---|---|
| 1 | 167,784,811 | 167,850,858 | 66,047 | 16 | 0 | 1.9E–03 | FMO3 FMO6 |
| 2 | 10,878,985 | 10,902,509 | 23,524 | 16 | 0 | 2.4E-02 | PDIA6 |
| 6 | 4,012,045 | 4,070,654 | 58,609 | 17 | 1 | 5.1E-04 | PECI C6orf146 C6orf201 |
| 6 | 30,052,958 | 30,154,225 | 101,267 | 22 | 0 | 8.4E-03 | HLA-A29.1 HCG9 ZNRD1 PPP1R11 RNF39 |
| 6 | 30,429,340 | 30,500,227 | 70,887 | 22 | 0 | 3.3E-03 | - |
| 6 | 32,447,818 | 32,497,626 | 49,808 | 15 | 0 | 4.2E-03 | BTNL2 |
| 7 | 31,322,276 | 31,375,664 | 53,388 | 22 | 8 | 1.4E-04 | LOC223075 CCDC129 |
| 11 | 126,178,796 | 126,246,694 | 67,898 | 15 | 0 | 5.9E-03 | KIRREL3 |
| 16 | 80,722,907 | 80,789,170 | 66,263 | 19 | 1 | 6.6E-04 | MPHOSPH6 |
| 18 | 49,714,826 | 49,774,618 | 59,792 | 19 | 5 | 3.4E-05 | - |
| 21 | 24,343,408 | 24,426,048 | 82,640 | 19 | 0 | 1.2E-03 | - |
| 21 | 43,307,905 | 43,360,473 | 52,568 | 22 | 0 | 5.8E-03 | PKNOX1 CBS |
Positive Controls & Bioinformatic Comparisons
It is highly desirable to compare GWAS findings to “positive controls” (i.e., genomic regions with very strong prior evidence for association like APOE*ε4 and Alzheimer’s disease (79) or FTO/intron 1 and body mass index (15)). At the time of this writing in 10/2007, there are no such regions for SCZ although there are a few genomic regions with multiple positive but inconsistent findings.
First, the GWAS platform used here had at least one SNP in 25,287 “known genes” (80). The median number of SNPs per gene was 4, inter-quartile range 2–11, and range 1–1,088. The number of SNPs per gene is strongly related to gene size (Spearman ρ=0.79) meaning that larger genes have more SNPs on average.
Second, the GWAS findings were compared to a list of 525 candidate genes for SCZ that had been investigated at least once in a published report †. This list includes studies with positive or negative findings and thus represents candidate genes that at least one set of investigators believed to be relevant to the etiology of SCZ. The number of studies per gene varied widely – from 302 genes studied in single reports to genes that had been studied >50 times (52 reports for HTR2A, 57 reports for DRD2, 65 reports for DRD3, and 68 reports for COMT). In the GWAS results, there were 534 SNPs with p-values <0.001 (an arbitrary choice) that implicated 249 genes (one SNP could be located in multiple genes) – of these, only 6 (2.41%) had been the subject of a prior study (ARMC3, CACNA1A, FEZ1, NRG1, PIWIL2, and VDR). Although a GWAS platform can provide very good or excellent coverage over the genome on average, there may be important candidate genes with sub-optimal SNP coverage. Of the 525 candidate genes previously studied in the literature, 84 genes had no SNPs that passed quality control, including eight genes that had been studied in ≥10 reports (APOE, CNTF, CYP2D6, DRD1, DRD4, GRIN1, TH, and TNF).
Third, we focused more closely on a consensus set of 15 candidate genes for SCZ with the best evidence of association – 12 candidate genes were selected from a review (8), CSF2RA and IL3RA were from a published SCZ GWAS (23), and PLXNA2 was from a large-scale candidate gene study for SCZ (81) (Table 5). Of the 249 genes with one or more p-values <0.001 in this GWAS, only 1 was on this list of 15 candidate genes (NRG1). Although the GWAS platform we used had generally good coverage across the genome (1 SNP/6.2 kb on average), six of these 15 genes had inadequate coverage and nine genes had SNP densities better than the GWAS average (2.2–5.0 SNPs/kb). For this subset of the GWAS data, the proportion of SNPs without true effects (p0) (60) was estimated at 0.997 for an FDR threshold of 0.10 and the minimum q-value was 0.30. In comparison to all GWAS SNPs, these results suggest that these candidate regions may be enriched for genetic variants influencing susceptibility to SCZ.
Table 5.
GWAS results for a selected set of candidate genes for SCZ.
| Adequate coverage |
Gene name |
Gene product |
Gene size |
Size percentile |
Number of SNPs |
SNP Density (kb/SNP) |
P-values <0.05 |
P-values <0.001 |
Minimum p-value |
|---|---|---|---|---|---|---|---|---|---|
| No | AKT1 | v-akt murine thymoma oncogene homolog 1 | 24,250 | 53 | 2 | 12.1 | 1 | 0 | 0.0316 |
| No | CSF2RA | colony stimulating factor 2 receptor, alpha | 41,089 | 68 | 0 | - | - | - | - |
| No | IL3RA | interleukin 3 receptor, alpha | 46,220 | 71 | 1 | 46.2 | 0 | 0 | 0.41 |
| No | PRODH | proline dehydrogenase 1 | 23,772 | 53 | 3 | 7.9 | 1 | 0 | 0.023 |
| No | RGS4 | regulator of G-protein signaling 4 | 7,228 | 26 | 1 | 7.2 | 0 | 0 | 0.84 |
| No | ZDHHC8 | zinc finger, DHHC-type containing 8 | 16,165 | 43 | 0 | - | - | - | - |
| Yes | COMT | catechol-O-methyltransferase | 27,222 | 56 | 9 | 3.0 | 2 | 0 | 0.016 |
| Yes | DAOA | D-amino acid oxidase activator | 25,168 | 54 | 9 | 2.8 | 0 | 0 | 0.12 |
| Yes | DISC1 | disrupted in schizophrenia 1 | 414,456 | 98 | 99 | 4.2 | 4 | 0 | 0.0011 |
| Yes | DRD3 | dopamine receptor D3 | 50,200 | 72 | 13 | 3.9 | 0 | 0 | 0.31 |
| Yes | DTNBP1 | dystrobrevin binding protein 1 | 140,231 | 91 | 28 | 5.0 | 0 | 0 | 0.064 |
| Yes | HTR2A | serotonin receptor 2A | 62,663 | 77 | 29 | 2.2 | 0 | 0 | 0.15 |
| Yes | NRG1 | neuregulin 1 | 1,124,806 | 99 | 293 | 3.8 | 19 | 1 | 0.00091 |
| Yes | PLXNA2 | plexin A2 | 216,898 | 95 | 70 | 3.1 | 4 | 0 | 0.013 |
| Yes | SLC6A4 | serotonin transporter | 37,800 | 65 | 8 | 4.7 | 0 | 0 | 0.52 |
None of the results in Table 5 survive FDR multiple comparison correction. Nonetheless, we investigated a few of these results further (Supplemental Figure 7). Of the 146 SNPs in the vicinity of DISC1 (minimum p-value 0.001), the significant findings clustered around the chr1 (1;11) (q42;q14.3) translocation break point (13) as follows: rs2738875 (p=0.001) – 5.0 kb – translocation break point – 3.0 kb – rs11122342 (p=0.62) – 0.6 kb – rs6672782 (p=0.016) – 0.1 kb – start of haplotype HEP1 – 0.8 kb – rs11588937 (p=0.83) – 8.6 kb – end of haplotype HEP1 – 8.0 kb – rs12744978 (p=0.048). These data are not conclusive, but the locations of the significant DISC1 SNPs coincide very closely with the DISC1 breakpoint (13) and the HEP1 haplotype implicated in the etiology of SCZ (82) and reduced prefrontal cortex gray matter density (83). Of the 401 SNPs in the vicinity of NRG1 (minimum p-value 0.0009), there were 15 SNPs (6 with p<0.05 including the most significant NRG1 SNP, rs16879809) in a cluster at the 3’ end of NRG1. This cluster was 875 kb from the 5’ portion of NRG1 that been of particular interest (9, 84, 85). Three of 53 SNPs in the “HapIce” haplotype (9) had p-values between 0.01–0.05. The SNP rs6994992 (84, 85) was not genotyped in this GWAS but had been done in these samples previously: rs6994992 was weakly associated with SCZ but in the opposite direction than has been reported (86). Of the 13 SNPs in the vicinity of COMT, two consecutive exonic SNPs 1 kb apart both had p=0.02. These were rs4633 (synonymous) and rs4680 (val158met) which has been widely studied as a genetic risk factor for SCZ and other disorders (87). Finally, of the 84 SNPs in the vicinity of PLXNA2, the significant SNPs were 80 kb away from those highlighted in the initial report (81) and whose most notable SNP (rs752016) was not significant in this GWAS (p=0.90). It is important to stress that none of these findings met genome-wide significance and the overlap with prior studies could have been merely due to chance.
Fourth, these findings were compared with the findings of two published GWAS. The Affymetrix 500K “A” chips were also used in these studies facilitating direct comparisons. For a SCZ GWAS (23), the list of p-values was not available from the authors but the most significant association was for the PAR1 SNP rs4129148 (P= 3.7×10−7) (23). This finding was not replicated in the CATIE GWAS (p=0.41). Indeed, only 5 of 143 SNPs in the pseudo-autosomal region had association p-values <0.05 (none within 600 kb of rs4129148) and there were no pseudo-autosomal region p-values <0.001. In comparing the CATIE GWAS with the Wellcome Trust bipolar disorder GWAS (18), the QQ plot for the 370,216 common to both studies is shown in Supplemental Figure 8. This comparison was motivated by suspicions that SCZ and bipolar disorder might share etiological genetic risk factors; however, there was no substantial divergence of the observed from the expected p-value distributions.
Multi-Marker Analyses
The QQ plot for combined p-values from genomewide multi-marker analyses conducted on AFR and EUR subjects is shown in Supplemental Figure 9; there was no marked deviation of the observed from the expected p-value distributions. Moreover, there was no overlap of the 12 genes listed in Table 5 or the 525 genes that have been investigated in studies of SCZ with the 100 haplotypes with p<0.001.
Discussion
The present genomewide association study (GWAS) for schizophrenia (SCZ) had multiple notable features. It is the second published GWAS for SCZ, and the first for which all individual genotype and phenotype data were made available to the research community under an “open source” philosophy.
Moreover, this project was conceptualized from the beginning as a two-stage study (56, 88). The present report constitutes the first stage and, as anticipated, there are no “slam-dunk” findings that meet genomewide significance; however, it is very possible that there exist true findings in these results that may not be impressive in any single study but that only emerge by comparing multiple large GWAS.
It is hoped that the process of gene-finding for SCZ will mirror that of type 2 diabetes mellitus (T2DM). In 2006, three genes had accumulated strong evidence in support of association – after the publication of six T2DM GWAS in 2007 (18–22, 89), there are now eight (and perhaps as many as 11) genes with highly compelling support (90). Notably, several of the initial T2DM GWAS had QQ plots very similar to that in Figure 1a (19, 20) and only after “aggressive data sharing” (22) across studies and additional genotyping in thousands of additional samples did multiple high confidence findings emerge (90). Indeed, some of the findings that eventually proved to be highly significant had initial p-value ranks in the hundreds or even thousands.
Therefore, these data from the CATIE GWAS will be part of an inclusive meta-analysis of individual phenotype and genotype data from all available SCZ GWAS that will be done in Q1 2008 under the auspices of the Psychiatric GWAS Consortium. It is anticipated that there will be well over 10,000 cases plus controls available for meta-analysis – particularly large-scale projects include the GAIN SCZ samples (91) and a consortium lead by Dr. Pamela Sklar at the Broad Institute). Drs. van den Oord and Sullivan hope to conduct large-scale Stage 2 genotyping to confirm and refine results from the SCZ meta-analysis. The CATIE samples are being used for deep re-sequencing of candidate genes under an award from the Medical Re-sequencing/Allelic Spectrum project supported by the NHGRI and can be used to discover variants not previously identified.
Additional notable features of this project include that, at the time genotyping was conducted, the GWAS platform used here had the best genomic coverage for common genomic variation with r2≥0.8 for 86% of genome in subjects with European ancestry, 79% for East Asian ancestry, and 49% for African ancestry (92). The CATIE sample was ascertained from diverse sites across the continental United States in order to accrue a “real-world” sample of patients in treatment for chronic SCZ (27) and a rich set of phenotypes are available for all CATIE subjects – i.e., treatment response in a randomized, double-blind clinical trial (24), multiple assessments of tardive dyskinesia (93), and repeated assessments of neurocognition (94).
The principal finding of the present study was that no SNP or multi-marker combination of SNPs achieved genomewide significance. Moreover, there was no important overlap of our findings with those from published GWAS for SCZ (23) or bipolar disorder (18). However, some findings with uncorrected p-values <0.05 overlapped with regions of DISC1 and COMT that have been highlighted in prior studies. Of the 146 SNPs in the vicinity of DISC1 (minimum genewise p-value 0.001), the significant findings clustered around the chr1 translocation break point (13) and the HEP1haplotype implicated in the etiology of SCZ (82) and associated with reduced prefrontal cortex gray matter density (83). This observation is bedeviling – these statistical signals could easily be due to chance (even the best finding would not survive multiple comparison correction for SNPs in DISC1 alone much less for the 492,900 SNPs in this GWAS) and yet the location of the signals is intriguing. Whether the observed overlap is merely due to chance (14) or reflects genetic influences on liability to SCZ will require the meta-analyses described above.
There are three broad explanations for the pattern of findings observed in this SCZ GWAS. The first and most optimistic possibility is that there are true findings relevant to the etiology of SCZ imbedded in these results but that it will require careful and rigorous comparisons with other GWAS datasets along with additional genotyping in new samples to delineate true from false positive findings.
Statistical power is a particular concern. The sample size of 738 SCZ cases and 733 group-matched controls and a genomewide set of 492,900 SNPs provided the capacity to detect genetic effects of moderate size for reasonably common polymorphisms (i.e., minor allele frequencies exceeding 10%). True genetic effects influencing case-control status might not have been detected in this study for reasons predictable from the design of this study. Non-detection could have occurred if the genotypic effect size was below the detection threshold, if the effect was located in a genomic region for which there was poor SNP coverage, if the effect was a genetic variant other than a SNP (e.g., a copy number variant) and if there was low LD with genotyped SNPs, or in the presence of excessive phenotypic or locus heterogeneity. Additionally, it is possible that true positive findings could have been obscured by the use of principal components analysis in this heterogeneous sample particularly for SNPs in strongly stratified genomic regions.
The second possibility is that there are true genetic effects for SCZ but that assumptions fundamental to GWAS are incorrect. It is possible that current definitions of SCZ (28, 95) lack validity. If true, attempting to identify genetic variants associated with “caseness” in a GWAS may prove fruitless as case classifications based on signs and symptoms are poorly correlated with genetic etiological factors. It is possible that the fundamental model is incorrect – GWAS are predicated under the common disease/common variant model whereby prevalent human diseases are caused by polymorphisms of relatively modest effects. If SCZ is caused by multiple rare variants (causation due to multiple quite uncommon genetic variants of very strong effect) (96) then the GWAS design is inappropriate to the fundamental genetic phenomenon. It is possible that liability to SCZ is mostly or entirely due to interactions between genomic regions or between genomic regions and environmental factors and that these must be explicitly modeled in order to be detected. Each of these instances provides an explanation why true effects were not detected.
Finally, the least optimistic possibility is that, despite the indirect evidence from family, adoption, and twin studies (4), there exist no true genetic effects causal to SCZ. This option does not appear consistent with a sizeable body of genetic epidemiological data; however, data from quasi-experimental studies do not constitute proof of the involvement of specific genetic variants in the etiopathology of SCZ.
It could prove possible to determine of which of these three possibilities are operative for SCZ on near horizon. In the meantime, SCZ genetics needs no more false claims (14) – there is an urgent need to identify replicable findings associated with SCZ. Consistent with this belief, these analyses have been interpreted with caution and the individual phenotype and genotype data made available to the scientific community †.
Supplementary Material
Acknowledgments
Acknowledgements
Dr. Sullivan was supported by R01s MH074027 and MH077139, Dr. Zou by GM074175, and Dr. Wright by P30 HD003110. The CATIE project was funded by NIMH contract N01 MH90001. We thank Dr. Nick Patterson for assistance with the EigenSoft program.
We are indebted to the "Molecular Genetics of Schizophrenia II" (MGS-2) collaboration for their exceptional collegiality in making the control samples available to the research community. Control subjects were from the National Institute of Mental Health Schizophrenia Genetics Initiative, and phenotypes and DNA samples were collected by the MGS-2 collaboration whose investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI).
Financial Disclosures
Eli Lilly funded the GWAS genotyping done at Perlegen Sciences. Dr. Sullivan reports receiving research funding from Eli Lilly in connection with this project. Dr. Stroup reports having received research funding from Eli Lilly and consulting fees from Janssen Pharmaceutica, GlaxoSmithKline, and Bristol-Myers Squibb. Dr. Lieberman reports having received research funding from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceutica, and Pfizer and consulting and educational fees from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, Eli Lilly, Forest Pharmaceuticals, GlaxoSmithKline, Janssen Pharmaceutica, Novartis, Pfizer, and Solvay.
Footnotes
http://slep.unc.edu (accessed 28JUN2007)
‡ http://www.schizophreniaforum.org/res/sczgene (accessed 28JUN2007)
http://www.nimhgenetics.org (accessed 28JUN2007)
Available at dbGaP, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap (accessed 28JUN2007)
http://www.nimhgenetics.org (accessed 28JUN2007)
http://genepath.med.harvard.edu/~reich/Software.htm, v1.0 (accessed 28JUN2007)
http://cancercenter.mayo.edu/mayo/research/schaid_lab/software.cfm, v1.3.1 (accessed 28JUN2007)
https://slep.unc.edu/evidence (downloads tab, accessed 28JUN2007)
https://slep.unc.edu/evidence (downloads tab, accessed 02OCT2007)
http://www.schizophreniaforum.org/res/sczgene (accessed 28JUN2007)
http://www.nimhgenetics.org (accessed 28JUN2007)
References
- 1.Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Medicine. 2005;2(5):e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Bjork C, Hultman CM, Scolnick EM, Sklar P, Sullivan PF. Recurrence risks for schizophrenia in a Swedish national cohort. Psychological Medicine. 2006;36:1417–1426. doi: 10.1017/S0033291706008385. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman II, Shields J. Schizophrenia: The Epigenetic Puzzle. Cambridge, UK: Cambridge University Press; 1982. [Google Scholar]
- 4.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 5.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th ed. London: Longman Group Ltd.; 1996. [Google Scholar]
- 6.Risch N. Linkage strategies for genetically complex traits: I. Multilocus models. American Journal of Human Genetics. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- 7.Neale B, Ferreira M, Medland S, Posthuma D. Statistical Genetics: Gene Mapping through Linkage and Association. London: Taylor and Francis; 2007. [Google Scholar]
- 8.Sullivan PF. The genetics of schizophrenia. PLoS Medicine. 2005;2:614–618. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. American Journal of Human Genetics. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15(12):1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 11.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse Dysbindin gene, is associated with schizophrenia. American Journal of Human Genetics. 2002;71(2):337–348. doi: 10.1086/341750. Erratum in American Journal of Human Genetics 2002 Oct;72(4):1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet. 2006;79(5):903–909. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human Molecular Genetics. 2000;9(9):1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan PF. Spurious genetic associations. Biological Psychiatry. 2007;61:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science. 2007 doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 20.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science. 2007 doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007 doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 22.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of Genome-Wide Association Signals in U.K. Samples Reveals Risk Loci for Type 2 Diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry. 2007;12(6):572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 24.Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM, et al. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull. 2003;29(1):15–31. doi: 10.1093/oxfordjournals.schbul.a006986. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman JA. Comparative effectiveness of antipsychotic drugs. A commentary on: Cost Utility Of The Latest Antipsychotic Drugs In Schizophrenia Study (CUtLASS 1) and Clinical Antipsychotic Trials Of Intervention Effectiveness (CATIE) Arch Gen Psychiatry. 2006;63(10):1069–1072. doi: 10.1001/archpsyc.63.10.1069. [DOI] [PubMed] [Google Scholar]
- 26.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 28.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Fourth Edition ed. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 29.First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV Axis I Disorders--Administration Booklet. Washington, DC: American Psychiatric Press, Inc.; 1994. [Google Scholar]
- 30.Affymetrix. GeneChip® Mapping 500K Assay Manual. Santa Clara, CA: 2006. [Google Scholar]
- 31.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307(5712):1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 33.Affymetrix. BRLMM: an Improved Genotype Calling Method for the GeneChip® Human Mapping 500K Array Set. Santa Clara, CA: Apr 14, 2006. [Google Scholar]
- 34.GAIN Collaborative Research Group. New models of collaboration in genome-wide association studies: the Genetic Association Information Network (GAIN) Submitted. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan PF, Purcell S. In: Statistical Genetics: Gene Mapping through Linkage and Association. Neale B, Ferreira M, Medland S, Posthuma D, editors. London: Taylor and Francis; 2007. [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–883. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(6):967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36(5):512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 40.Heiman GA, Hodge SE, Gorroochurn P, Zhang J, Greenberg DA. Effect of population stratification on case-control association studies. I. Elevation in false positive rates and comparison to confounding risk ratios (a simulation study) Hum Hered. 2004;58(1):30–39. doi: 10.1159/000081454. [DOI] [PubMed] [Google Scholar]
- 41.Shields AE, Fortun M, Hammonds EM, King PA, Lerman C, Rapp R, et al. The use of race variables in genetic studies of complex traits and the goal of reducing health disparities. American Psychologist. 2005;60:77–103. doi: 10.1037/0003-066X.60.1.77. [DOI] [PubMed] [Google Scholar]
- 42.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 43.Pritchard JK, Stephens M, Rosenberg NA, Donnelley P. Association mapping in structured populations. American Journal of Human Genetics. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 45.Epstein MP, Allen AS, Satten GA. A simple and improved correction for population stratification in case-control studies. Am J Hum Genet. 2007;80(5):921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, Sullivan PF, Zou F, Wright FA. Comment on "A Simple and Improved Correction of Population Stratification". American Journal of Human Genetics. doi: 10.1016/j.ajhg.2007.10.014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;22:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol. 2007;31(4):358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 49.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- 50.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19(10):537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 52.van den Oord EJCG. Controlling false discoveries in genetic studies. American Journal of Medical Genetics (Neuropsychiatric Genetics) doi: 10.1002/ajmg.b.30650. In press. [DOI] [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society (Series B) 1995;57:289–300. [Google Scholar]
- 54.Brown BW, Russell K. Methods of correcting for multiple testing: operating characteristics. Stat Med. 1997;16(22):2511–2528. doi: 10.1002/(sici)1097-0258(19971130)16:22<2511::aid-sim693>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 55.Fernando RL, Nettleton D, Southey BR, Dekkers JC, Rothschild MF, Soller M. Controlling the proportion of false positives in multiple dependent tests. Genetics. 2004;166(1):611–619. doi: 10.1534/genetics.166.1.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Oord EJ, Sullivan PF. A framework for controlling false discovery rates and minimizing the amount of genotyping in the search for disease mutations. Hum Hered. 2003;56(4):188–199. doi: 10.1159/000076393. [DOI] [PubMed] [Google Scholar]
- 57.Tsai CA, Hsueh HM, Chen JJ. Estimation of false discovery rates in multiple testing: application to gene microarray data. Biometrics. 2003;59(4):1071–1081. doi: 10.1111/j.0006-341x.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- 58.van den Oord EJ. Controlling false discoveries in candidate gene studies. Mol Psychiatry. 2005;10(3):230–231. doi: 10.1038/sj.mp.4001581. [DOI] [PubMed] [Google Scholar]
- 59.Sabatti C, Service S, Freimer N. False discovery rate in linkage and association genome screens for complex disorders. Genetics. 2003;164(2):829–833. doi: 10.1093/genetics/164.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meinhausen N, Rice J. Estimating the proportion of false null hypotheses among a large number of independently tested hypotheses. Annals of Statistics. 2006;34:373–393. [Google Scholar]
- 61.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- 63.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 64.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 65.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155(5):478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 67.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 68.SAS Institute Inc. SAS/STAT® Software: Version 9. Cary, NC: SAS Institute, Inc.; 2004. [Google Scholar]
- 69.SAS Institute Inc. JMP User's Guide (Version 6) Cary, NC: SAS Institute, Inc.; 2005. [Google Scholar]
- 70.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34(Database issue):D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34(Database issue):D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blaschke RJ, Rappold G. The pseudoautosomal regions, SHOX and disease. Curr Opin Genet Dev. 2006;16(3):233–239. doi: 10.1016/j.gde.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 73.Hemminger BM, Saelim B, Sullivan PF. TAMAL: An integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics. 2006;22:626–627. doi: 10.1093/bioinformatics/btk025. [DOI] [PubMed] [Google Scholar]
- 74.Fisher RA. Statistical Methods for Reesearch Workers. 11th edition. London: Oliver and Boyd; 1950. [Google Scholar]
- 75.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan PF, Keefe RSE, Lange LA, Lange EM, Stroup TS, Lieberman JA, et al. NCAM1 and neurocognition in schizophrenia. Biological Psychiatry. 2007;Volume 61(7):902–910. doi: 10.1016/j.biopsych.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 77.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7(10):781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 78.Sachse C, Ruschen S, Dettling M, Schley J, Bauer S, Muller-Oerlinghausen B, et al. Flavin monooxygenase 3 (FMO3) polymorphism in a white population: allele frequencies, mutation linkage, and functional effects on clozapine and caffeine metabolism. Clin Pharmacol Ther. 1999;66(4):431–438. doi: 10.1053/cp.1999.v66.a102203. [DOI] [PubMed] [Google Scholar]
- 79.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 80.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22(9):1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 81.Mah S, Nelson MR, Delisi LE, Reneland RH, Markward N, James MR, et al. Identification of the semaphorin receptor PLXNA2 as a candidate for susceptibility to schizophrenia. Mol Psychiatry. 2006 doi: 10.1038/sj.mp.4001785. [DOI] [PubMed] [Google Scholar]
- 82.Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12(23):3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 83.Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62(11):1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 84.Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5’ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103(17):6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hall J, Whalley HC, Job DE, Baig BJ, McIntosh AM, Evans KL, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9(12):1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 86.Crowley JJ, Keefe RS, Perkins DO, Stroup TS, Lieberman JA, Sullivan PF. The neuregulin 1 promoter polymorphism rs6994992 is not associated with chronic schizophrenia or neurocognition. doi: 10.1002/ajmg.b.30727. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, et al. A highly significant association between a COMT haplotype and schizophrenia. American Journal of Human Genetics. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 89.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007 doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 90.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8(9):657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 91.Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39(9):1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 92.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38(6):659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 93.Miller D, McEvoy JP, Davis SM, Caroff SN, Saltz BL, Chakos MH, et al. Clinical correlates of tardive dyskinesia in schizophrenia: baseline data from the CATIE schizophrenia trial. Schizophr Res. 2005;80(1):33–43. doi: 10.1016/j.schres.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 94.Keefe RS, Bilder RM, Harvey PD, Davis SM, Palmer BW, Gold JM, et al. Baseline neurocognitive deficits in the CATIE schizophrenia trial. Neuropsychopharmacology. 2006;31:2033–2046. doi: 10.1038/sj.npp.1301072. [DOI] [PubMed] [Google Scholar]
- 95.World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research. Geneva: World Health Organization; 1993. [Google Scholar]
- 96.McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.