Abstract
Bin/Amphipysin/Rvs (BAR) domain proteins sculpt cellular membranes, playing key roles in processes such as endocytosis, cell motility and morphogenesis. BAR domains are divided into three subfamilies: BAR and F-BAR domains generate positive membrane curvature and stabilize cellular invaginations, whereas the I-BAR domain induces negative curvature and stabilizes protrusions. We show that a previously uncharacterized member of the I-BAR subfamily, Pinkbar, is specifically expressed in intestinal epithelial cells where it localizes to Rab13-positive vesicles and to the plasma membrane at intercellular junctions. Remarkably, the BAR domain of Pinkbar does not induce membrane tubulation, but promotes the formation of planar membrane sheets. Structural and mutagenesis analysis revealed that the BAR domain of Pinkbar has a relatively flat lipid-binding interface and that it assembles into sheet-like oligomers in crystals and in solution, which may explain its unique membrane deforming activity.
The dynamic deformation of cellular membranes is essential for many physiological processes, including cell morphogenesis, motility, cytokinesis, endocytosis, and secretion 1, 2. An important group of membrane-deforming proteins are the members of the Bin/Amphiphysin/Rvs (BAR) domain family 3, 4. The BAR domain is an anti-parallel all-helical dimer, with a curved (banana-like) shape, that promotes membrane tubulation 5, 6. The BAR domain binds cellular membranes through its positively-charged concave surface, and thus induces positive membrane curvature 6. Members of the related F-BAR family had been independently characterized 6–9. However, the structural relationship with the BAR domain only emerged after determination of the first F-BAR domain structures, revealing a similar banana-shaped fold, albeit longer and less curved than that of BAR domain 10, 11. With one exception 12, the BAR and F-BAR domain-containing proteins characterized thus far are involved in the formation of positive membrane curvature, and the majority of these proteins are implicated in endocytosis 3. Recently, a cryo-electron microscopy study revealed that membrane tubulation results from the cooperative action of large numbers of F-BAR domains, which oligomerize to form helical coats on membranes 13.
Structural studies have also identified a third class of BAR domain, named the I-BAR domain because of its inverted curvature 14, 15. Sequence analysis identifies five I-BAR domain-containing proteins in vertebrates: MIM, IRSp53, ABBA, IRTKS and Pinkbar. These proteins have different isoforms, but they all contain an N-terminal I-BAR domain. Most have an additional C-terminal WH2 domain, which at least in MIM, ABBA and IRSp53 binds G-actin 15–18. Their central region is more variable and is characterized by the presence of protein-protein interaction modules, which link these proteins to the actin cytoskeleton and signaling molecules 19.
All the I-BAR domains characterized thus far induce formation of plasma membrane protrusions when expressed in cells, and generate negative membrane curvature in vitro by binding to the inner leaflet of membrane tubules through their convex lipid-binding interface 20–22. I-BAR domains bind phosphoinositide-rich membranes mainly through electrostatic interactions and induce phosphoinositide clustering in vitro. The I-BAR domains of MIM and ABBA additionally insert an amphipathic helix into the membrane. This increases the diameter of the membrane tubules that they generate and may sense lipid packing defects within the bilayer 22, 23. Cellular and genetic studies suggest that I-BAR domain-containing proteins function at the interface between the actin cytoskeleton and the plasma membrane to promote the formation of membrane protrusions during cell morphogenesis and migration 18, 24–31.
Here, we report that a previously uncharacterized I-BAR domain protein that we have named Pinkbar (for planar intestinal and kidney specific BAR domain protein) is expressed predominantly in intestinal and kidney epithelial cells where it localizes to Rab13-positive vesicles and to the plasma membrane at intercellular junctions. Our results suggest that the BAR domain of Pinkbar is unique in that it does not induce membrane protrusions or invaginations, and instead promotes the formation of planar membrane sheets. The crystal structure of this domain combined with mutagenesis and biochemical analyses suggest a molecular mechanism for the novel membrane deforming activity of Pinkbar.
RESULTS
Pinkbar is expressed in specific epithelial cells of the intestine and kidney
I-BAR domain-containing proteins can be divided into three evolutionarily distinct classes: MIM/ABBA, IRSp53/IRTKS, and a class consisting of a previously uncharacterized protein, which we have named Pinkbar (Supplementary Fig. 1a online). Generally, these proteins are ubiquitously expressed 16, 18, 19, 27. In contrast, in situ hybridization analysis of mouse tissues revealed that Pinkbar mRNA expression is restricted to the epithelial layer of the intestine and to specific regions of the kidney (Supplementary Fig. 1b, e online). The tissue-specific expression of Pinkbar mRNA is also consistent with the findings of the Affymetrix database (http://biogps.gnf.org/, gene ID 207495) 32. Immunohistochemical analysis using a specific antibody confirmed that also the Pinkbar protein was detectable in the epithelial layer of human intestine (Supplementary Fig. 1c online).
Among several cell lines tested, Pinkbar protein was only detected in human epithelial colorectal adenocarcinoma cells (Caco-2), which are morphologically and functionally similar to intestinal enterocytes 33. The expression level of Pinkbar was strongly increased when cells were polarized on filters (Supplementary Fig. 1d online). Immunofluorescence microscopy revealed that in polarized Caco-2 cells and in the epithelial cells of intestine sections Pinkbar displayed punctuate localization close to cell-cell contacts. Occasionally, Pinkbar also localized to tight junctions (Supplementary Fig. 2a, b online).
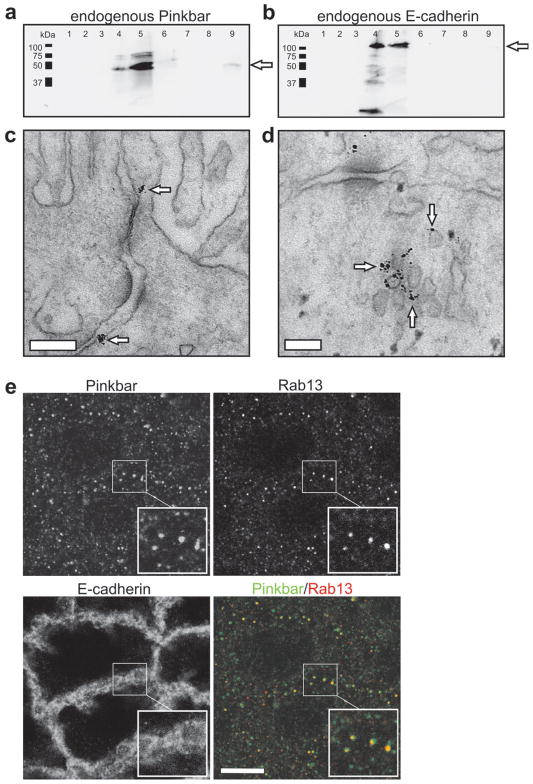
Immunoelectron microscopy and cell fractionation assays revealed that in polarized Caco-2 cells endogenous Pinkbar protein localized to intracellular vesicles and to relatively flat membrane surfaces at intercellular junctions of the apical half of the cell (Fig. 1a–d). Experiments using several wild-type Rab GTPases revealed that mCherry-Pinkbar specifically co-localizes with Rab13 at tubular and punctuate cytoplasmic structures in non-polarized and polarized Caco-2 cells, respectively (Supplementary Fig. 2c online). Importantly, many endogenous punctuate Pinkbar structures also co-localize with Rab13 in Caco-2 cells (Fig. 1e). Thus, Pinkbar localizes to Rab13-positive vesicles and to the plasma membrane at intercellular contacts in intestinal epithelial cells.
Figure 1.
Endogenous Pinkbar associates with membranes in Caco-2 cells, and co-localizes with the small GTPase Rab13. (a and b) The association of endogenous Pinkbar (MW=58.4 kDa) with cellular membranes was studied by sucrose gradient centrifugation of Caco-2 cell lysate. Fractions (1 ml) were collected from top (fraction 1) to bottom (fraction 9). Pinkbar is present at the interface of 10 % and 65 % sucrose gradient in the same fractions (4 and 5) as the membranes and the positive control, E-cadherin (MW=97.5 kDa). Protein is indicated by arrows. (c and d) Immunoelectron microscopy sections showing the localization of endogenous Pinkbar in Caco-2 cells (day 16). Pinkbar (indicated by arrows) localizes to the plasma membrane at cell-cell junctions c and to intracellular vesicle clusters d. Bars: 200 nm and 100 nm, respectively. (e) In fully polarized Caco-2 cells (day 16) endogenous Pinkbar displays partial co-localization with endogenous Rab13 in punctuate cytoplasmic structures, as detected by immunofluorescence microscopy. Bar: 10 μm.
The BAR domain of Pinkbar stabilizes planar membrane sheets
Similarly to other I-BAR domains 22, the BAR domain of Pinkbar induced strong concentration-dependent quenching of bodipy-TMR-PI(4,5)P2 fluorescence, indicating that it interacts with phosphoinositide-rich membranes and binds to PIP2-headgroups in a multivalent manner (Supplementary Fig. 3a online). A subset of BAR and I-BAR domains additionally inserts an amphipathic helix into the membrane, as detected among other methods by an increase in steady-state diphenylhexatriene (DPH) anisotropy 22, 34. Likewise, the binding of the BAR domain of Pinkbar to PI(4,5)P2-containing membranes induced a significant increase in DPH anisotropy, suggesting that the fluidity of the membrane is decreased as a result of the insertion of this domain into the acyl chain region of the bilayer (Supplementary Fig. 3b online). DPH anisotropy also suggested that the BAR domain of Pinkbar does not display significant phosphoinositide specificity (Supplementary Fig. 4a online). Deletion of nine N-terminal residues from the domain diminished the effects in DPH anisotrophy (Supplementary Fig. 4b online). Experiments on a mutant BAR domain containing only a single tryptophan residue at position 9 (Y9W/W141L/W216L) demonstrated that phosphatidylcholines brominated along the acyl chains at positions 6,7 or 9,10 did not significantly affect the tryptophan fluorescence, whereas bromination at positions 11,12 induced marked quenching of the fluorescence (Supplementary Fig. 4c online). Thus, the mutated residue Trp9 is located near positions 11 and 12 of the acyl chains, corresponding to a distance of approximately 6.3 Å from the center of the bilayer.
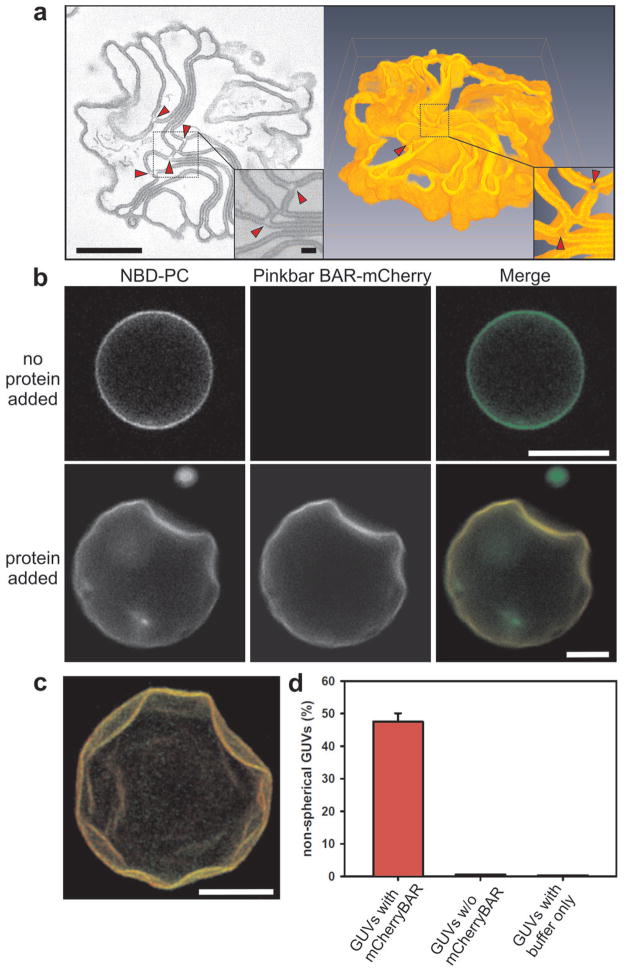
In vivo, BAR domains induce either plasma membrane invaginations or protrusions 3. In contrast, the BAR domain of Pinkbar did not localize to plasma membrane invaginations or filopodia-like protrusions nor did it induce their formation (Supplementary Fig. 3c online), suggesting that it may have different membrane deforming properties. Consistent with this idea, electron microscopy analysis of PI(4,5)P2-containing vesicles incubated with the BAR domain of Pinkbar revealed that it did not induce membrane tubulation, but promoted the formation of sheet-like membrane structures (Supplementary Fig. 5a, b online). Electron tomography analysis of these structures revealed piled stacks of membrane sheets (Fig. 2a). Increased electron density was observed in the regions forming flat membrane sheets, whereas highly curved regions of the membrane were depleted of additional density (Fig. 2a). The additional density was interpreted as resulting from clustering of the BAR domain of Pinkbar, which only occurred in association with the regions of planar membrane structure.
Figure 2.
The BAR domain of Pinkbar promotes the formation of planar membrane structures. (a) 3D electron tomography analysis of a membrane sheet network induced by the BAR domain of Pinkbar. Left: Tomographic slice from reconstruction of a 250 nm thick section, where a magnification of the selected region is shown as an inset. Bars: 400 nm and 50 nm, respectively. Arrowheads indicate highly curved membrane areas, lacking higher electron density attributable to Pinkbar. Right: 3D model obtained from the same structure in which a small number of the sheets that were not connected with other membranes within the reconstructed volume were removed. (b) Giant unilamellar vesicles (GUVs) containing fluorescent NBD-labeled phosphatidylcholine (PC) (green) were incubated with mCherry-labeled BAR domain of Pinkbar (red). In the absence of protein, the vesicles were predominantly spherical, whereas BAR domain-decorated vesicles often displayed abnormal morphology and flat surfaces. The lipid composition of the GUVs used in the assay was POPC:POPE:POPS:PIP2:NBD-PC (49:20:20:10:1) and the protein concentration was 100 nM. Bars: 10 μm. (c) Easy3D-illustration of flat indentations on the surface of a GUV produced by the BAR domain of Pinkbar. BAR-mCherry is shown in red and NBD-PC in green. Bar: 10 μm. (d) Quantification of the proportion of deformed vesicles in GUVs decorated with the BAR domain of Pinkbar, GUVs from the same experiment that did not accumulate Pinkbar, and GUVs from samples treated with buffer only. The data are from three independent experiments and the error bars represent s.d.
Light microscopy imaging of fluorescently labeled giant unilamellar vesicles (GUVs) mixed with mCherry-fused I-BAR domain suggested that the binding of the BAR domain of Pinkbar to PI(4,5)P2-containing vesicles is cooperative, as only 85–90% of the fluorescently labeled GUVs accumulated the domain on their surface. Moreover, vesicles that were heavily decorated with the BAR domain of Pinkbar typically displayed a non-spherical morphology with relatively flat surfaces (Figs. 2b, c and Supplementary Fig. 5c online). Such flat surfaces were rarely observed in vesicles that did not accumulate the BAR domain, or in control samples treated with the same buffer (Fig. 2d). These data suggest that the BAR domain of Pinkbar does not tubulate membranes, but induces the formation of planar or very gently curved membrane structures.
Crystal structure and oligomerization of the BAR domain of Pinkbar
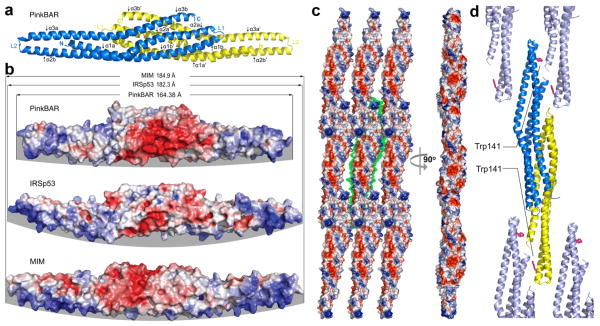
We determined the crystal structure of the BAR domain of Pinkbar to shed light on its planar membrane-inducing mechanism (Table 1 and Methods online). The structure revealed the characteristic elongated I-BAR fold, consisting of an anti-parallel dimer of three-helix bundles (Fig. 3a), featuring on one side a positively charged surface (Fig. 3b). However, the BAR domain of Pinkbar was noticeably shorter (164 Å) than those of IRSp53 (182 Å) 14 and MIM (185 Å) 15. Compared to these two proteins, the BAR domain of Pinkbar was also significantly flatter, which appeared to result from the combination of two factors: reduced bending and twisting of the helices as they emerge from the ‘core’ region where the two subunits of the antiparallel dimer overlap (Supplementary Fig. 6 online), and the presence of a downward projecting bulb at the distal ends of the dimer, which is not observed in MIM or IRSp53 (compare the distal ends of the structures in Figure 3b).
Figure 3.
Crystal structure and oligomerization of the BAR domain of Pinkbar. (a) Ribbon diagram representation of the structure of the BAR domain of Pinkbar. The two chains that form the BAR domain are colored yellow and blue. Each chain contains three α-helices, which are kinked toward their middle (see also Supplementary Fig. S6), and are thus labeled as α1a-α1b to α3a-α3b (or α1a′-α1b′ to α3a′-α3b′ for the second chain). (b) Electrostatic surface representation of the BAR domain of Pinkbar compared to those of IRSp53 and MIM. Blue and red indicate negatively and positively charged regions, respectively. Note that the BAR domain of Pinkbar is shorter and its membrane-binding surface (highlighted by a gray background) less curved than those of IRSp53 and MIM. (c) The BAR domain of Pinkbar, which forms sheet-like membrane structures in solution, also forms a planar oligomer in the crystal lattice, in which the membrane-binding interface of individual BAR domains all face in the same direction (i.e. facing the reader in the view shown on the left). The green background highlights the boundaries of a single BAR domain dimer. This planar oligomer is somewhat reminiscent of the planar BAR-BAR coats observed by EM tomography of F-BAR domains 13. (d) The conspicuously exposed residue Trp141 may be involved in the stabilization of the lateral oligomer (the ribbon diagram is shown in a similar orientation as part c, left view), as suggested by light scattering analysis in solution of wild type and Trp141Ser mutant BAR domain constructs (see Supplementary Fig. 7a, b).
The BAR domain of Pinkbar formed a planar oligomer in the crystal lattice (Fig. 3c), which had not been observed in the structures of the I-BAR domains of MIM or IRSp53. The arrangement of the BAR domain in the planar oligomer was somewhat reminiscent of the BAR-BAR coats observed by electron microscopy (EM) tomography of F-BAR domains 13, and appeared consistent with the formation of sheet-like membrane structures in solution (Fig. 2). At sites of BAR-BAR contacts, an alternating pattern of negative and positive surface charges was observed (Fig. 3c), which might contribute toward the formation of the planar oligomer. Additionally, the conspicuously exposed residue Trp141 (missing in other members of the I-BAR family; Supplementary Fig. 6c), appeared to stabilize lateral BARBAR interactions in the planar coat formed by the BAR domain of Pinkbar (Fig. 3d). Tryptophan residues tend to be well conserved and are rarely fully exposed in protein structures, and exceptions to this rule typically point to an important function, such as substrate recognition, protein-protein or protein-membrane interactions 35, 36. In the structure, Trp141 is surrounded by various positively charged residues that in other I-BAR proteins have been implicated in membrane binding 18, 21. Using light scattering, we determined that in solution the BAR domain of Pinkbar existed in equilibrium between dimers and higher molecular weight oligomers, but only dimers formed when Trp141 was mutated to Ser (Supplementary Fig. 7a, b online). This result suggested that Trp141-mediated lateral contacts that help stabilize the planar BAR-BAR coat in the crystal lattice are also implicated in oligomerization of the BAR domain in solution.
Lipid-binding interface of Pinkbar
The phosphoinositide-binding interface of the BAR domain was mapped by systematic mutagenesis. Because the BAR domain of Pinkbar interacts with PI(4,5)P2-rich vesicles through electrostatic interactions (Supplementary Fig. 8a online), we mutated clusters of arginines and lysines to alanines (see Supplementary Fig. 8b online). Consistent with earlier work on other I-BAR domains 18, 21, none of the single mutants completely abolished PI(4,5)P2-clustering, suggesting that the BAR domain of Pinkbar does not contain a specific PIP2-binding pocket. Three mutants around Trp141 (K109A/K116A, R127A/K135A, and R145A/K146A/R147A) had relatively strong effects on PI(4,5)P2-clustering, whereas the single mutant I124S had an intermediate effect (Fig. 4a). The impaired lipid-binding activity of the most strongly affected mutant, R145A/K146A/R147A, was further confirmed by a vesicle co-flotation assay (Supplementary Fig. 9 online). All the mutants that affected PIP2-clustering localized to the relatively flat surface of the BAR domain of Pinkbar (Fig. 4b), corresponding to the convex surface implicated in lipid-binding in classical I-BAR domains 18, 21. The mutants on the opposite side of BAR domain did not significantly affect PI(4,5)P2 clustering (Fig. 4a).
Figure 4.
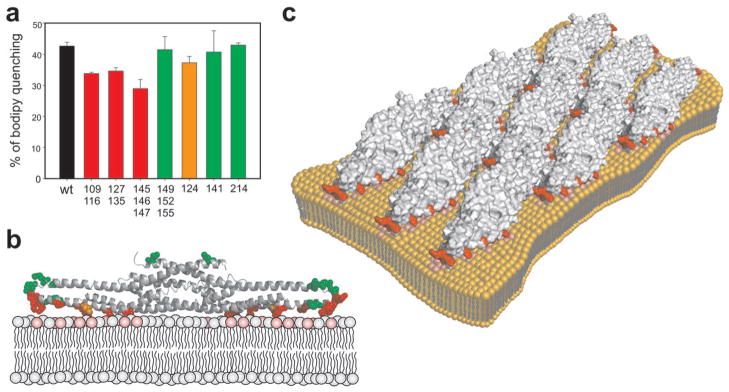
Mechanism of membrane binding and deformation by the BAR domain of Pinkbar. (a) Effect of mutations of the BAR domain of Pinkbar on the clustering of Bodipy-TMR-PI(4,5)P2 measured by fluorometric assay. Double or triple mutations of the positively charged residues (K109A/K116A, R127A/K135A, and R145A/K146A/R147A) had the strongest effect on membrane binding (red). A moderate effect was observed for mutant I124S (orange). The triple mutant K149/R152/K155 and the single mutants W141S and L214S had no significant effect on PI(4,5)P2 clustering (green). The composition of the liposomes used in the assay was POPC:POPE:POPS:PIP2:bodipy-PIP2 (50:20:20:9.5:0.5) and the concentration of protein 1.6 μM. Each bar represents the mean of three individual measurements and the error bars represent s.d. (b) The residues mutated in this study are shown as ball and stick in a ribbon diagram of the structure of the BAR domain of Pinkbar and colored as in a. Note that the residues that had the strongest effects on bodipy quenching cluster along the membrane-binding surface of the BAR domain. (c) The membrane-binding surfaces of individual BAR domains all face in the same direction in the planar oligomer of the crystal lattice and thus the lipid-binding interface in the oligomer is relatively flat.
Importantly, in the planar oligomer observed in crystal structure (Fig. 3c), individual BAR domains were oriented such that their membrane-binding interfaces all faced in the same direction (Fig. 4c). Thus, not only was the membrane-binding interface of Pinkbar more flat that those of other I-BAR proteins, but this effect appeared to be additionally accentuated by oligomerization into a planar oligomer. In support to this, live-imaging assays revealed that the oligomerization-deficient W141S mutant was significantly less efficient in deforming GUVs than the wild-type BAR domain of Pinkbar, despite displaying no defects in binding to or inducing clustering of PI(4,5)P2-rich vesicles (Supplementary Figs. 7c and d online, and Fig. 4a).
DISCUSSION
BAR domain proteins sense/generate membrane curvature, and are therefore central players in many processes involving plasma membrane protrusions or invaginations. Our investigation of a previously uncharacterized BAR domain protein, which we have named Pinkbar, revealed that this protein is expressed in specialized epithelial cells of the intestine and kidney. In colon sections and in cultured intestinal epithelial cells, Pinkbar localized to vesicles and to the plasma membrane at the cell-cell junctions. Many Pinkbar-containing vesicles stained positive for the small GTPase Rab13. Like Pinkbar, Rab13 is highly expressed in the intestinal epithelial cells, where it promotes tight junction integrity 37, 38. Because the Pinkbar-containing vesicles display positive membrane curvature and Rab13 vesicles are involved in recycling tight junction components, it appears unlikely that Pinkbar is involved in sculpting the membrane of these structures. Instead, these vesicles may be involved in transporting Pinkbar to cell-cell junctions. Pinkbar also accumulates to the plasma membrane at cell-cell junctions that display a relatively flat geometry. Thus, it is possible that through its specific membrane binding/deforming interface, Pinkbar may sense/stabilize specific planar membrane structures at cell-cell junctions. In support to this proposal, the related I-BAR domain proteins IRSp53 and MIM were shown to contribute to assembly and maintenance of intercellular junctions in other types of epithelial cells 39, 40. Thus, Pinkbar may be involved in the formation of specific membrane structures at the intercellular junctions of enterocytes that may e.g. regulate the intestinal permeability or nutrient absorption.
The BAR domain of Pinkbar does not induce membrane tubulation in vitro or in cells, but instead deforms phosphoinositide-rich membranes into planar structures. The crystallographic and mutagenesis data suggest a mechanism for this activity. First, the lipid-binding interface of the BAR domain of Pinkbar is significantly less curved than those of other BAR domains characterized thus far, including other members of the I-BAR subfamily 14, 15 that generate membrane tubules 22. Second, the BAR domain of Pinkbar forms a planar oligomer in the crystal lattice, which appears to exist also in solution.
A previous study 13 suggested that F-BAR domain oligomerization, cooperatively stimulated by interaction with the membrane, mediates the formation of three-dimensional helical coats of F-BAR domains and plays a critical role in the mechanism of membrane deformation. In the case of Pinkbar, the oligomeric state of the BAR domain appears to be somewhat more stable, as oligomers can form even in the absence of phospholipid membranes. Importantly, the oligomerization-deficient W141S mutant was significantly less efficient in generating planar membrane structures and/or indentations on GUVs compared to the wild-type BAR domain. Thus, oligomerization may be a general mechanism by which BAR domain- containing proteins deform cellular membranes. We note, however, that the W141S mutant was still capable of generating planar membrane structures in vitro, although less efficiently compared to the wild-type domain. This result is consistent with the proposed mechanism of coat formation of F-BAR domains in which multiple interactions, including lateral and tip-to-tip contacts, contribute to stabilize helical F-BAR domain coats on membranes 13.
The N-terminal α-helix of the BAR domain of Pinkbar inserts into the bilayer. Because amphipathic α-helices can sense and generate positive membrane curvature 1,2, this helix in Pinkbar may work together with the slightly negatively curved geometry of the membrane-binding interface of the BAR domain to stabilize flat membrane structures. Similarly, deletion of the N-terminal membrane-inserting α-helix of the I-BAR domain of MIM results in generation of more extensive negative membrane curvature 22. However, further cryo-electron microscopy studies on the BAR domain of Pinkbar will be needed to address the role of membrane insertion and to uncover the exact mechanism of coat formation.
All the members of the canonical BAR domain family characterized to date display a banana-like fold and bind phospholipid-rich membranes through their concave surfaces to generate positive membrane curvature 3, 6, 41. Similarly, most F-BAR domain proteins generate positive membrane curvature and are involved in endocytic processes in cells 3, 8, 42–44. A recent study, however, revealed an unexpected functional plasticity within this family, showing that the F-BAR domain of srGAP2 induces negative membrane curvature in vitro and plasma membrane protrusions in cells 12. Here, we provide evidence that significant functional plasticity also exists within the I-BAR domain family, whose members can either generate negative membrane curvature or stabilize planar membrane sheets, depending on the specific structural properties of their I-BAR domains. To our knowledge, this study represents the first example of a protein domain involved in the stabilization/generation of planar membrane structures.
METHODS
Protein expression and purification
For biochemical and cellular studies, the fragment corresponding to Pinkbar residues 1–239 was subcloned into pGFP-N1, pmCherry-N1, pHAT1 45, and pBSIIKS vectors. The C-terminal fragments corresponding to residues 240–522, 327–522, 1–499, and 421–522 were cloned into the pHAT1 vector. Site-directed mutagenesis was performed as described 21. The Pinkbar constructs were expressed as His-tag fusion proteins in E. coli BL21 (DE3) cells (Invitrogen, Carlsbad, CA) and purified on Ni-NTA Superflow column (Sigma-Aldrich) as described 21. Although crystals of construct 1–239 were obtained, their quality could not be improved and the slightly shorter construct 1–220 was thus used in crystallization experiments. The 1–220 fragment was cloned into NdeI and EcoRI sites of vector pTYB12 (New England BioLabs, Ipswich, MA). The mutant Trp141Ser was obtained using the QuickChange XL site-directed mutagenesis kit (Stratagene). Proteins were expressed in BL21(DE3) cells, grown in Terrific Broth medium at 37°C until the OD at 600 nm reached 1.0–1.2. Expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and carried out for 5 hours at 20°C. Cells were harvested by centrifugation, resuspended in chitin affinity column equilibration buffer (20 mM Hepes pH 7.5, 500 mM NaCl, 1 mM EDTA and 100 μM PMSF) and lysed using a microfluidizer apparatus (Microfluidics). Standard purification on a chitin affinity column (New England Biolabs) was followed by ion-exchange chromatography on a MonoS 5/50 GL column (GE Healthcare) using a 100–500 mM NaCl gradient. To prevent protein precipitation during dialysis, protein samples were diluted to a NaCl concentration of 100 mM prior to ion exchange purification.
Crystallization and structure determination
The BAR domain of Pinkbar was dialyzed against 20 mM Hepes pH 7.5, 250 mM NaCl and concentrated to 1.5 mg/ml. Above this concentration, the BAR domain formed crystalline precipitates revealed by inspection under a light microscope. The addition of 5% or 10% glycerol limited the formation of crystal seeds and the individual crystals that emerged spontaneously grew over a period of 2–3 days in hanging drops equilibrated against the same buffer at 20°C (Supplementary Fig. 6a). The crystals obtained with 5% glycerol diffracted the X-rays to higher resolution, and a dataset was collected to 2.25 Å resolution at beamline 17-BM of the Advanced Photon Source (APS) at Argonne National Laboratory (Table 1). The structure was determined by molecular replacement, using a poly-Ser model of the core region (lacking the distal ends of helices 2 and 3) of the I-BAR domain of IRSp53 as a search model 14. Molecular replacement and refinement were carried out with the program Phenix 46, and model building was performed with the program Coot 47.
Preparation of lipid vesicles and lipid assays
Vesicles were prepared as previously described 22. Briefly, lipids at the desired concentrations were mixed, dried under a stream of nitrogen and hydrated in 20 mM Hepes, pH 7.5, 100 mM NaCl to yield multilamellar vesicles at a lipid concentration of 1 mM. To obtain small unilamellar vesicles, large multilamellar liposomes with a lipid composition of POPC:POPE:POPS:PIP2 (50:20:20:10) were extruded through a polycarbonate filter (100 nm pore size) using a mini extruder (Avanti Polar Lipids). Giant unilamellar vesicles (GUV) were prepared as described 22. The lipid composition of the GUVs was POPC:POPE:POPS:PIP2:NBD-PC (49:20:20:10:2). A vacuum dried mixture of the desired lipids was dissolved in diethylether:methanol (9:1, v/v, final total lipid concentration 1 mM), spread (4–8 μl) on the surfaces of two Pt electrodes and then dried under a stream of nitrogen. Remains of the organic solvent were removed by evaporation at RT for 30 min. An AC field (sinusoidal wave function with a frequency of 10 Hz and amplitude of 0.2 V) was applied before adding 1 ml of 200 mM sucrose dissolved in 5 mM Hepes buffer, pH 7.5, 200 mM sucrose. During the first minute of hydration the voltage was increased to 1 V. The AC field was turned off after 2.5 h. GUVs were mixed with 200 mM glucose in 5 mM Hepes buffer pH 7.5 in 1:5 ratio prior to imaging. The GUVs containing NBD-PC (1 %) fluorescent label were imaged as described above. The total volume of the samples during imaging was 300 μl and protein was added to the GUVs to a final concentration of 0.1–2 μM. Co-sedimentation assays and clustering of bodipy-TMR-PI(4,5)P2 by the BAR were performed as described 21,22.
Electron Microscopy and Tomography
Samples for transmission electron microscopy were prepared by mixing 10 or 20 μM protein with 166 μM multilamellar vesicles in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl. Sample preparation and microscopy were done as described 21 with the exception that all the EM-micrographs were obtained from 120 nm-thick sections. For 3D electron tomography, a 250 nm-thick section was prepared as described above and imaged with an electron microscope (Tecnai 20 FEG; FEI Corp.) operating at 200 kV. For collection of tilt series, the specimen was tilted ±63° at 1° intervals around two orthogonal axes 48. Images were recorded with a 4k × 4k Ultrascan 4000 CCD camera (Gatan Corp.) at a nominal magnification of 19,000× (0.58 nm/pixel). The acquired images were binned by 2 providing a pixel size of 1.15 nm in the reconstruction. The alignment and reconstruction of tilt series were done with the IMOD program package 49 using 10 nm colloidal gold particles as fiducial markers. The tomographic reconstruction was visualized and modeled with the Amira software using volume rendering (Visage Imaging Inc.).
Immuno Electron Microscopy
Fully polarized (day 16) Caco-2 cells grown on filters were immunostained according to 50. The anti-Pinkbar antibody (Atlas Antibodies) was used in 1:50 dilution and Nano-Gold α-rabbit IgG (Nanoprobes) in 1:60 dilution. Best silver enhancement was achieved with 5 minute incubation.
Supplementary Material
Acknowledgments
Supported by NIH National Institute of Mental Health grant MH087950 to R.D. and grants from the Finnish Cancer Foundation and Academy of Finland to P.L. H.Z. was supported by the Academy of Finland, and A.P. and J.S. were supported by fellowships from VGSB and GPMB graduate schools, respectively. Use of IMCA-CAT beamline 17-BM was supported by the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. The Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38.
Footnotes
AUTHOR CONTRIBUTIONS
A.P., M.B., H.Z., J.S., G.S., M.J., J.H., E.K., H.V. and R.D. performed the experiments. A.P., J.P., E.J., M.S., E.I., R.D. and P.L designed and supervised the project. A.P., R.D. and P.L. prepared the manuscript.
Accession code. Coordinates have been deposited with the protein data bank under accession code 3OK8.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Campelo F, Fabrikant G, McMahon HT, Kozlov MM. Modeling membrane shaping by proteins: focus on EHD2 and N-BAR domains. FEBS Lett. 2010;584:1830–1839. doi: 10.1016/j.febslet.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 6.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 7.Aspenstrom P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr Biol. 1997;7:479–487. doi: 10.1016/s0960-9822(06)00219-3. [DOI] [PubMed] [Google Scholar]
- 8.Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Tsujita K, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada A, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 11.Henne WM, et al. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Guerrier S, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost A, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millard TH, et al. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SH, et al. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J Biol Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- 17.Woodings JA, Sharp SJ, Machesky LM. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem J. 2003;371:463–471. doi: 10.1042/BJ20021962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saarikangas J, et al. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J Cell Sci. 2008;121:1444–1454. doi: 10.1242/jcs.027466. [DOI] [PubMed] [Google Scholar]
- 19.Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Suetsugu S, et al. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- 21.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saarikangas J, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 23.Bhatia VK, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suetsugu S, et al. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Disanza A, et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol. 2006;8:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- 26.Lim KB, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. 2008;283:20454–20472. doi: 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- 27.Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 28.Kim MH, et al. Enhanced NMDA receptor-mediated synaptic transmission, enhanced long-term potentiation, and impaired learning and memory in mice lacking IRSp53. J Neurosci. 2009;29:1586–1595. doi: 10.1523/JNEUROSCI.4306-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawallisch C, et al. The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J Biol Chem. 2009;284:9225–9236. doi: 10.1074/jbc.M808425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan BK, et al. Cdc42- and IRSp53-dependent contractile filopodia tether presumptive lens and retina to coordinate epithelial invagination. Development. 2009;136:3657–3667. doi: 10.1242/dev.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinones GA, Jin J, Oro AE. I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J Cell Biol. 2010;189:353–367. doi: 10.1083/jcb.200910136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology. 1989;96:736–749. [PubMed] [Google Scholar]
- 34.Gallop JL, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda Y, et al. Thermal unfolding of chitosanase from Streptomyces sp N174: role of tryptophan residues in the protein structure stabilization. Biochim Biophys Acta. 1999;1429:365–376. doi: 10.1016/s0167-4838(98)00243-x. [DOI] [PubMed] [Google Scholar]
- 36.Clark EH, East JM, Lee AG. The role of tryptophan residues in an integral membrane protein: diacylglycerol kinase. Biochemistry. 2003;42:11065–11073. doi: 10.1021/bi034607e. [DOI] [PubMed] [Google Scholar]
- 37.Morimoto S, et al. Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J Biol Chem. 2005;280:2220–2228. doi: 10.1074/jbc.M406906200. [DOI] [PubMed] [Google Scholar]
- 38.Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19:971–983. doi: 10.1091/mbc.E07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massari S, et al. LIN7 mediates the recruitment of IRSp53 to tight junctions. Traffic. 2009;10:246–257. doi: 10.1111/j.1600-0854.2008.00854.x. [DOI] [PubMed] [Google Scholar]
- 40.Saarikangas J, et al. Missing-in-metastasis (MIM/MTSS1) promotes actin assembly at intercellular junctions and is required for kidney epithelia integrity. J Cell Sci. 2011;124:1245–1255. doi: 10.1242/jcs.082610. [DOI] [PubMed] [Google Scholar]
- 41.Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J Mol Biol. 2005;351:653–661. doi: 10.1016/j.jmb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Reider A, et al. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henne WM, et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 46.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 49.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 50.Uchiyama K, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.