Abstract
The toxin RelE is a ribosome-dependent endoribonuclease implicated in diverse cellular processes, including persistence. During amino acid starvation, RelE inhibits translation by cleaving ribosomal A-site mRNA. Although RelE is structurally similar to other microbial endoribonucleases, the active-site amino acid composition differs substantially and lacks obvious candidates for general acid-base functionality. Highly conserved RelE residues- Lys52, Lys54, Arg61, Arg81, and Tyr87- surround the mRNA scissile phosphate and specific 16S ribosomal RNA contacts further contribute to substrate positioning. We used a single-turnover kinetic assay to evaluate the catalytic importance of individual residues in the RelE active site. Within the context of the ribosome, RelE rapidly cleaves A-site mRNA at a rate similar to traditional ribonucleases. Single-turnover rate constants decreased between 102 - 106-fold for the RelE active-site mutants Lys52, Lys54, and Arg61, Arg81. RelE may principally promote catalysis via transition-state charge stabilization and leaving-group protonation, in addition to achieving inline substrate positioning in cooperation with the ribosome. This kinetic analysis complements structural information to provide a foundation for understanding the molecular mechanism of this novel endoribonuclease.
Keywords: toxin-antitoxin, RelE, mRNA cleavage, ribosome, ribonuclease, transphosphorylation, catalysis
Prokaryotic lifestyle changes are initiated in response to environmental cues by pathways that sense physiological differences and initiate downstream events. Toxin-antitoxin (TA) modules have recently emerged as central to the regulation of diverse cellular processes that modulate bacterial activities, including the general stress response, biofilm formation, and persistence (1-3). TA systems are abundant genetic modules that encode a toxin protein that interferes with normal cell physiology and a cognate antitoxin that inactivates the toxin under normal environmental conditions (1, 3, 4). In type II TA systems, the antitoxins and toxins are proteins that tightly associate in an inactive complex that autoregulates operon transcription (1, 3). The antitoxins are proteolytically degraded upon environmental stresses, including nutrient starvation, which liberates free toxin and relieves TA operon down-regulation (5, 6).
A major class of type II toxins acts as mRNA interferases, which cleave mRNA (7, 8). One of the most well-characterized type II toxins is the endoribonuclease RelE, which cleaves translating mRNA in the ribosomal A-site upon amino acid starvation (5, 9-13). RelE is normally in a tightly associated complex with the antitoxin RelB, which is nucleolytically inactive. Upon amino acid starvation, Lon protease degrades RelB to liberate the active RelE protein (5, 6). RelE demonstrates strict ribosome-dependence and cleaves translating mRNA between the second and third nucleotide of ribosomal A-site codons, preferentially upstream of purines (10, 13-15). The relaxed codon sequence specificity may enable RelE to globally modulate translation rates and facilitate the fast adaption of cells to environmental changes.
Limited similarity of RelE toxins to standard ribonucleases (RNases) have hindered efforts to understand the mechanism of RelE-mediated phosphodiester bond cleavage (4, 8, 16, 17). RelE is most similar to the broad class of metal-independent RNases that yield a 2′, 3′-cylic phosphate and 5′-hydroxyl upon phosphodiester bond breakage (18). Phosphodiester bond cleavage by these RNases typically proceeds via general-acid-base chemistry mediated by a glutamate-histidine pair that readily promotes catalysis under physiological pH conditions (18). These active-site residues are replaced with basic residues in RelE, yet structural studies of E.coli and archaeal RelE proteins revealed that despite the lack of sequence similarity, RelE shares a microbial RNase fold with these endoribonucleases (17, 19, 20).
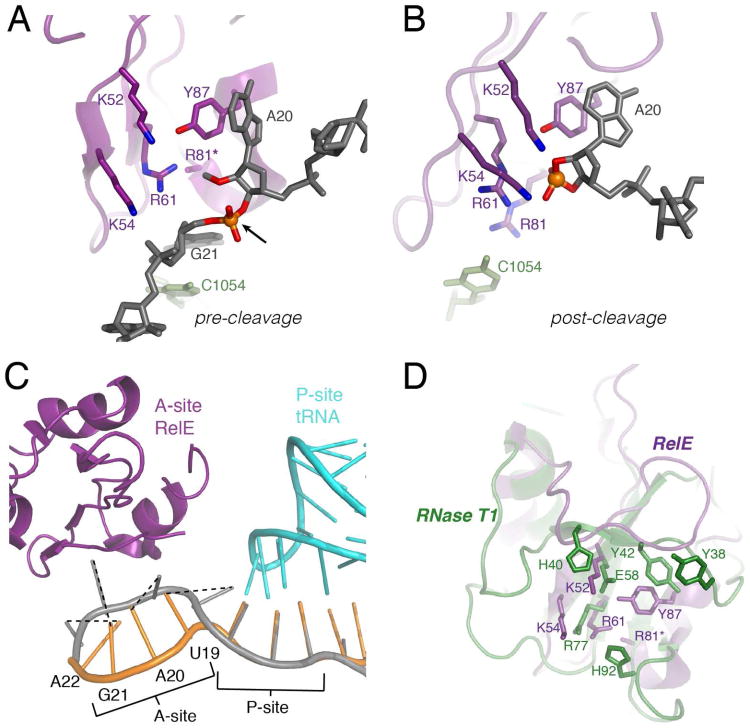
The co-crystal structures of E. coli RelE with the ribosome-bound mRNA substrate in the pre- and post-cleavage states provided the first opportunity to examine the RelE active site with substrate and with product (Figure 1 A-C) (12). The mRNA is sequestered over 7 Å from its normal A-site path into the highly positively charged RelE active site and is additionally stabilized with contacts with the 16S ribosomal RNA (Figure 1C) (12). The distorted mRNA configuration exposes the scissile phosphate and aligns the 2′-hydroxyl for an in-line nucleophilic attack. These structures also confirmed that although the RelE active-site residues overlay well with other RNase active sites, the side chain identities differ (Figure 1D, Figure S1) (12, 21, 22). The most conserved residues in RelE - Arg61, Arg81, Tyr87, Lys52, and Lys54 - are within hydrogen-bonding distance of the scissile phosphate, its 5′- and 3′- nucleotides, or the mRNA 2′-hydroxyl (Figure 1 A-B) (12).
Figure 1. Structural insights into the RelE cleavage mechanism.
The active site of (A) pre-cleavage and (B) post-cleavage co-crystal structures with RelE (purple) with a modified mRNA substrate (2′-O-methyl at nucleotide 20) or cyclic phosphate product (grey) and nearby ribosomal 16S rRNA (light green). Heteroatoms that may be important for catalysis are highlighted in blue (nitrogen) and red (oxygen). The arrow denotes the scissile phosphate, shown as an orange sphere. In the pre-cleavage structure, R81(*) is mutated to an alanine (PDB IDs: 3KIQ, 3KIU). (C) RelE induces a conformational change in the A-site mRNA. Overlay of mRNA in the 70S ribosomal A-site and P-site when RelE is bound (grey) or a cognate tRNA (not shown) is bound (orange) in the A-site. Dashed lines denote the corresponding mRNA nucleobases between each state (PDB ID: 2J00). (D) The structure of E.coli RelE from (A) (purple) superimposed on the structure of RNase T1 (green, PDB 1RGA).
Even with the RelE structures, the fundamental question remains of how the largely basic side chains that constitute the RelE active site promote phosphodiester bond cleavage. On the basis of the co-crystal structure, Neubauer et al made preliminary tests of several active-site mutants, but observed only modest effects on mRNA cleavage (12). Despite these small effects, Neubauer et al proposed a general acid-base mechanism where Arg81 and Ty87 act as the general acid-base pair, Arg61 provides transition state charge stabilization and Lys52 and Lys54 may contribute to phosphate charge stabilization and substrate binding (12). However, the biochemical results were not consistent with the structural predictions or this proposed mechanism. In other RNases, the measured mutational rate effects for catalytic side chains can be on the order of 103 -105-fold (23-26).
Here we report the kinetic analysis of the ribosome-dependent mRNA cleavage by E. coli toxin RelE using a single-turnover cleavage assay to directly monitor RelE cleavage and substrate association. This kinetic analysis served as the framework to examine how specific RelE active-site residues contribute to catalysis and substrate binding. These results provide biochemical data to complement the structural information regarding RelE function within the ribosome. The detailed enzymatic analysis of RelE also has applicability to other non-canonical endoribonucleases.
Experimental Procedures
RelE and RelB Overexpression and Purification
The relBE locus from E. coli K-12 MG1655 with an N-terminal 6× His-tag was cloned into pET22-b between the NdeI and BamHI sites under T7 RNA polymerase control. Internal deletion mutants in E.coli relb were constructed with site-directed mutagenesis to disrupt the antitoxin's strong interactions with RelE and aid in RelE purification. RelB mutants used were: Δ3 (deletion of Ala19-Glu21), Δ6 (deletion of Ala19-Gly24) and Δ9 (deletion of Ala19-Pro27). Wild-type RelE was overexpressed and purified using the Δ9-His6×-RelB:RelE construct. RelE mutants were generated with site-directed mutagenesis and overexpressed in the background of the Δ9 (K52A, K54A, Y87A, K52A/Y87F), Δ6 (R61A), or Δ3 (Y87F) RelB strains. The RelBE complexes were all expressed in E. coli BL21 (DE3) and purified as follows. A 5 mL overnight culture was diluted into 600 mL LB with 100 μg/mL ampicillin and grown to OD600 0.8 at 37°C before induction with 1 mM IPTG. After 3 hours, cells were harvested via centrifugation and the pellet was resuspended in Lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 5 mM 2-mercaptoethanol, 0.2 mg/mL lysozyme) (11) and lysed by sonication at 4 °C. Lysate was cleared by centrifugation at 4 °C and incubated with Nickel-NTA agarose resin (Qiagen) (1 hour, 4 °C). Resin was washed with Lysis buffer with 35 mM imidazole before RelE was selectively eluted by denaturation in 100 mM NaH2PO4, 10 mM Tris-HCl, 9.8 M urea, 1 mM 2-mercaptoethanol, pH 8.0 (11). RelE proteins were purified to apparent homogeneity as monitored by SDS-polyacrylamide gel electrophoresis (PAGE) with Coomassie Brilliant Blue staining. Purified protein was dialyzed into 50 mM Bicine pH 8.4, 8 M urea before refolding via dialysis in 50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 300 mM KCl, 7 mM MgCl2, 1 M urea, 1 mM dithiothreitol. Refolded protein was concentrated under Argon using a 5,000 MWCO ultrafiltration membrane and filtered (0.22 μm) before storage in 50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 1 mM dithiothreitol, 20% glycerol. His6×-RelB was expressed in the context of the relBE locus with the relE start codon (ATG) mutated to isoleucine (ATT). The protein was overexpressed and purified as done for the RelBE complex, however, RelB was eluted from the Ni-NTA resin using 250 mM imidazole under native conditions. Purified RelB was also stored in 50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 1 mM dithiothreitol, 20% glycerol.
Circular Dichroism Spectroscopy
Circular dichroism spectra were obtained using an Applied Photophysics Chirascan™ spectrometer and a 2 mm path length quartz cuvette. Samples were prepared at 10 μM as described (11). Scans were baseline corrected to buffer alone and each spectra represents the average from at least three scans (260 – 202 nm) with 0.5 nm step size collected at 4 °C and 20 °C.
mRNA and Ribosome Complex Preparation
The E.coli 70S ribosomes were prepared as described (27). The deprotected mRNA oligonucleotide (5′GGCAAGGAGGUAAAAAUGUAGAAAAACAAU-3′) were 5′-32P-labeled as described (28). Ribosome complexes (RC) (400 nM tRNAfmet, 100 nM mRNA oligonucleotide, trace 5′-32P labeled mRNA oligonucleotide, 300 nM 70S ribosomes) were formed at 37 °C in reaction buffer (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 1 mM dithiothreitol) for 30 minutes before dilution with reaction buffer to final concentration 40 nM mRNA oligonucleotide. Ribosome complexes containing the phosphorothioate mRNA substrates were prepared in the same manner. The mRNA oligonucleotides containing phosphorothioate substitutions between the second and third nucleotide of the A-site codon (UApsG) were constructed using a splinted T4 RNA Ligase reaction with a 12-nucleotide oligonucleotide harboring the substitution and a 5′ phosphate (5′-P-UApsGAAAAACAAU-3′) ligated to an 18-nucleotide RNA (5′-GGCAAGGAGGUAAAAAUG-3′) using a 22-mer nucleotide DNA splint(29). Prior to ligation, the RP and SP diastereomers were separated using reversed-phase HPLC on a C18 column (30).
Single-turnover Kinetic Measurements
Single-turnover kinetic measurements were acquired for wild-type RelE using a rapid-quench-flow apparatus (KinTek Corporation). Reactions were initiated by adding equal volume RelE (0.1 μM – 20 μM final) to the RC (20 nM RNA final) at 20 °C and quenching with excess chemical quench (80% formamide, 50 mM Tris-MES pH 6.5, 65 mM EDTA, 0.2 mg/mL bromophenol blue and 0.2 mg/mL xylene cyanol). Cleavage rate constants for Y87F, K52A, and K54A were measured using the same quench-flow apparatus methods. The remaining RelE mutants were measured using manual bench-top quench methods, which were similarly initiated upon addition of equal volume RelE to RC at 20 °C and reaction aliquots were stopped with excess chemical quench. Single-turnover reactions in the absence of the ribosomes were performed using the bench-top reaction procedures. Wild-type RelE (10 μM final) alone, or pre-mixed with 6-fold excess RelB, was added to mRNA substrate (20 nM final mRNA oligonucleotide and trace 5′-32P labeled mRNA oligonucleotide) in reaction buffer at 20 °C and reaction aliquots were stopped with excess chemical quench. Reactions in the absence of ribosomes were monitored up to 1.5 hours after reaction initiation. For all reactions, the extent of A-site mRNA cleavage was determined by separating substrate and product RNAs by 15% denaturing PAGE and visualization with a STORM phosphorimager (Molecular Dynamics). Individual band intensities were quantified with ImageQuant (GE Healthcare) and the fraction cleaved was quantified as the fraction product oligonucleotide of the total.
Individual time courses for cleaved product accumulation for reactions were fit to the double exponential equation (1) using Prism 6:
| (1) |
where Afast and Aslow are the amplitudes of the two phases, kfast and kslow are the rate constants of the phase (s−1), t is reaction time (s), and Y0 is the initial fraction reacted. Due to the slow reaction progression for R61A and K52A/Y87F, it was not possible to measure the complete reaction and these time courses were best fit to the single exponential equation (2):
| (2) |
where A is the amplitude, k is the rate constant of the phase (s−1), t is the reaction time (s), and Y0 is the initial fraction reacted. The observed rates for the fast and slow phases were plotted as a function of RelE enzyme concentration (μM). The data points from each replicate were fit independently by the hyperbolic equation (3):
| (3) |
where kobs is the observed rate constant, [E] is RelE enzyme concentration (μM), k2 is the cleavage rate constant at saturating enzyme concentration (s−1) and K1/2 is the apparent dissociation constant (μM). The mean and standard error from at least three independent determinations are reported.
Equilibrium Pulse-Chase with RelB
To confirm that RelB quenches the single-turnover RelE cleavage reactions, RC or RelE were incubated with a 6-fold molar excess of RelB antitoxin over RelE prior to initiating reactions. For RelB rapid quench-flow experiments, RelE and RC were reacted for the programmed reaction time in addition to the travel time through the exit line (t1). Unquenched reactions were collected into a tube with 75 μl chemical quench or RelB (6-fold molar excess) followed by addition of 150 μl chemical quench after 10-reaction half-lives, t2. For bench-top RelB quench experiments, RelE and RC were mixed for reaction time, t1, followed by addition of either excess chemical quench or 6-fold excess RelB. After the quench delay time, t2, excess chemical quench was added to reaction. Reactions were analyzed as described above.
Results
We set out to characterize the kinetics of wild-type RelE cleavage activity and a series of active-site mutants. Estimates of wild-type multiple-turnover activities for different A-site substrates have been measured, but the kinetic characterization of RelE catalysis has been limited (10, 12, 16, 17). In order to provide a framework for understanding how RelE functions at both a microscopic and global level, we have used a single-turnover assay to directly measure mRNA cleavage rate constants. We then used this kinetic framework to examine the effects of RelE active site modifications on mRNA cleavage.
mRNA Cleavage by RelE
Efficient overexpression of RelE protein for kinetic assays was achieved by co-expression with N-terminally His-tagged RelB antitoxin (11, 16). RelE protein was purified from RelB under strong denaturing conditions and circular dichroism far-UV analysis of RelE was consistent with refolding (Figure S2) (11).
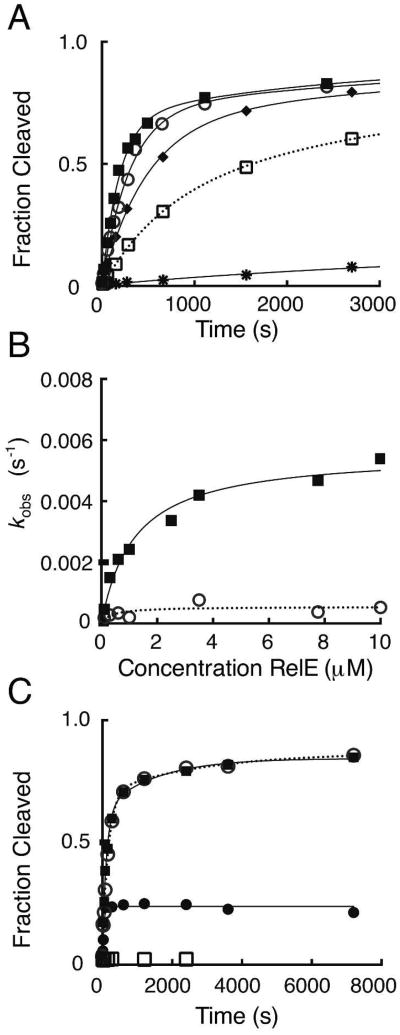
We developed a single-turnover kinetic assay to directly monitor RelE cleavage of mRNA. Cleavage activity was measured with RelE enzyme in excess of the ribosomal-complex (RC) substrate comprised of 70S E. coli ribosomes, an mRNA containing an A-site UAG codon, and a P-site tRNA. The addition of excess RelE enzyme to RC resulted in selective A-site codon cleavage between the second and third nucleotide, as described previously (10, 12). No cleavage attributable to RelE was detected in the absence of ribosomes (data not shown). For the ribosome-RelE reaction, time courses for accumulation of cleaved mRNA product reached endpoints of approximately 85 % and were best described by a double exponential (Figure 2A). In most reactions, the second rate did not significantly contribute to the total reaction amplitude, but on the basis of the systematic residuals resulting from single exponential fits, a double exponential function was selected. The two-state kinetic behavior was consistently observed in all time courses for wild-type and mutant RelE reactions. It may result from different substrate or enzyme populations. In all cases, the relative magnitude of the effects is similar for the fast and slow rate constants, but for clarity, the fast rate constant is used for mechanistic interpretations. The reaction rate was dependent on the enzyme concentration at low RelE concentrations (0.1 -3 μM), but reached a maximum at higher concentrations, with the rate constant, k2 = 380 s−1 (Figure 2A, B). The values for k2 and K1/2 were obtained from fitting the hyperbolic dependence of kobs versus RelE enzyme concentration (Table 1, Figure 2, Table S1).
Figure 2. Wild-type RelE rapidly cleaves ribosomal RNA.
(A) Measurement of single-turnover rates for wild-type RelE. Reactions contained 20 nM RC and increasing concentrations of wild-type RelE enzyme: 0.1 μM (*), 0.3 μM (□), 0.6 μM (◆), 1 μM (○), or 10 μM (■). The fractions of mRNA substrate cleaved during the first 1.2 seconds of each reaction is shown, but reactions were measured at least until 3 seconds and were fit as described in Experimental Procedures. (B) Dependence of cleavage rates on wild-type RelE enzyme concentration. The rates, kobs (s-1), for the fast (■) and slow phases (○) for a single replicate are plotted as a function of RelE concentration (μM). The rate constants (k2) and dissociation constants (Kd) were extracted from hyperbolic fits of each replicate and the mean and SEM for each constant is listed in Table 1 and Table S1.
Table 1. RelE single-turnover cleavage rate and dissociation constants.
| RelE | k2, fast (s-1)a | Fold Changeb | Kd, fast (μM)a |
|---|---|---|---|
|
| |||
| Wild-type | 380 ± 25 | 3.1 ± 0.4 | |
| K52A | 0.18 ± 0.01 | 2.1 × 103 | 0.48 ± 0.13 |
| K54A | 0.14 ± 0.01 | 2.7 × 103 | 1.5 ± 0.3 |
| R61A | 0.00014 ± 0.00001 | 2.7 × 106 | 0.88 ± 0.07 |
| R81A | 0.0064 ± 0.0001 | 6.0 × 104 | 0.81 ± 0.13 |
| Y87F | 3.0 ± 0.1 | 1.3 × 102 | 1.1 ± 0.3 |
| Y87A | 0.0021 ± 0.0002 | 1.8 × 105 | 1.8 ± 0.3 |
| K52A/Y87F | 0.00037 ± 0.00006 | 1.0 × 106 | 1.0 ± 0.4 |
Values are the mean ± SEM from at least three independent determinations.
Values calculated as the change in cleavage rate constant for each mutant relative to wild-type.
To determine whether reactions were in rapid equilibrium, we used a pulse-chase approach that exploited the nanomolar affinity interaction between RelE and the antitoxin RelB (6, 19). Direct protein-protein interaction with RelB prevents RelE from entering the ribosomal A-site and cleaving the mRNA substrate (19). Pre-incubation of either RelE or RC with a 6-fold molar excess of antitoxin RelB resulted in negligible levels of RNA cleavage. For the pulse-chase experiment, RelE and RC were allowed to react for time, (t1), before pulsing with excess RelB. Any free RelE or RelE that rapidly dissociated from RC should be irreversibly sequestered into a complex with RelB. This complex formation would prevent any additional product accumulation before the chemical quench was added at time, t2. In the single-turnover reaction, RelB quenched the RelE cleavage reaction to an extent equal to that of the chemical quench indicating that the rate of RelE dissociation (k-1) is faster than the rate of cleavage (k2) for RC-bound RelE. Therefore, the RelE reaction is in rapid equilibrium and K1/2 is equivalent to the dissociation constant, where Kd = 3.1 ± 0.4 μM for the wild-type RelE-RC enzyme-substrate interaction.
We next examined RelE cleavage of an RNA harboring a phosphorothioate substitution at the scissile phosphate. This substrate was previously found to be cleaved to almost the same extent as the unmodified RNA under conditions with 5-fold excess RelE enzyme; however, the reaction rate was not measured (12). If the chemical step is rate-limiting in our assay, the intrinsic difference in chemical reactivity between the oxygen- and sulfur-containing substrates should be reflected in a modest decrease in rate constant of approximately 4 - 10-fold (31). These thio-effects would be masked under conditions where reactions steps other than chemistry, such as a conformational change in the mRNA, were rate-limiting. RelE cleaved the phosphorothioate diastereomers to endpoints comparable to the parent mRNA substrate, but both were less efficiently cleaved (Table 2). The 39-fold and 900-fold differences in rate constants for RP and SP substrates, respectively, provide evidence that the chemical step is at least partially rate-limiting in this RelE assay. Thus, the single-turnover assay provides a framework in which RelE mutants can be considered for their effects on substrate-binding and chemical catalysis. However, the magnitude of the thio-effects are larger than expected for a classical general acid-base mechanism and may be indicative of transition state interactions between RelE and the non-bridging phosphate oxygens (32-34).
Table 2. Thio-effects on wild-type and Y87A RelE cleavage rate constants.
| Substrate | O | S (RP) | S (SP) | ||
|---|---|---|---|---|---|
|
|
|||||
| RelE | k fast (s-1)a | k fast (s-1)a | Thio-effectb | k fast (s-1)a | Thio-effectb |
| Wild type | 380 ± 25 | 9.7 ± 0.1 | 39-fold | 0.42 ± 0.18 | 900-fold |
| Y87A | 0.0021 ± 0.0002 | 0.00027 ± 0.00006 | 8-fold | 0.00037 ± 0.00014 | 6-fold |
O, unsubstituted mRNA; S (RP) or S (Sp), sulfur substitution at the non-bridging oxygen between mRNA nucleotides 20 and 21.
Values are the mean ± SEM from at least three independent determinations of the rate constant at a saturating enzyme concentration.
Values were calculated as the change in cleavage rate for sulfur-substituted mRNA substrates relative to the unsubstituted substrate.
Kinetic characterization of RelE active-site mutants
Using the single-turnover assay developed for wild-type RelE, we investigated the contributions of the active-site residues -Lys52, Lys54, Arg61, Arg81, and Tyr87- to the RelE cleavage mechanism. In addition to being highly conserved, these residues are positioned to directly participate in catalysis, and were proposed to be involved in the reaction mechanism (Figure 1) (12). Single alanine or phenylalanine substitutions were made for these five residues to examine their contributions to RelE catalysis. Additionally, a double mutant, K52A/Y87F was tested to investigate interactions, such as a hydrogen-bonding network, between these two residues that are in close proximity to each other and the 2′-hydroxyl. All RelE active-site mutants were purified following the same scheme used for wild-type protein. Circular dichroism analysis for each mutant was similar to that of wild-type RelE and consistent with refolding (Supporting Information, Figure S2) (11).
All seven mutants were active in the single-turnover kinetic assay and cleaved the RNA between the second and third nucleotide of the A-site UAG codon. Mutant cleavage reactions had similar kinetic behavior to wild-type and all rates reached a plateau at high RelE concentration (Figure 3, Figure S3). The cleavage rate constant effects for these mutants span four orders of magnitude, emphasizing that not all RelE active-site residues contribute equally to RelE phosphodiesterase activity (Table 1, Table S1). Using the RelB pulse-chase experiment described for wild-type RelE, reactions were confirmed to be in rapid-equilibrium, and therefore all K1/2 values report on the dissociation constant (Kd) for each RelE-RC interaction (Table 1, Figure 3, Table S1). None of the mutations tested significantly impacted the binding affinity of RelE for the ribosome-bound mRNA substrate (Table 1, Table S1).
Figure 3. Analysis of single-turnover reaction kinetics of mutant RelE proteins.
(A) Measurement of single-turnover rates for R81A RelE. Reactions contain 20 nM RC and increasing concentrations of R81A RelE enzyme: 0.1 μM (*), 0.3 μM (□), 0.6 μM (◆), 2.5 μM (○), or 10 μM (■). The first 3,000 seconds of the time course are shown and full time courses were fit as done for wild-type. (B) Dependence of cleavage rates on R81A RelE enzyme concentration. The rates, kobs (s-1), for the fast (■) and slow phases (○) for a single replicate are plotted as a function of RelE concentration (μM). The rate constants (k2) and dissociation constants (Kd) extracted from hyperbolic fits of each replicate and the mean and SEM for each constant is listed in Table 1 and Table S1. (C) Antitoxin RelB pulse-chase quench experiments with R81A RelE. Reactions contain 20 nM RC and 10 μM R81A RelE and were quenched either chemically (○), enzymatically with 6-fold excess RelB (■), or the RC was pre-mixed with RelB prior to addition of RelE (□). Product formation for the pulse-chase reaction with antitoxin RelB is plotted for the total time (reaction time, t1, + quench delay time or chase, t2) where RelB was added after t1 = 78 seconds (•).
Arg61 and Arg81, two of the most highly conserved residues in RelE, were implicated in catalysis by translation inhibition and cellular toxicity studies (16, 17, 19). Furthermore, Arg61 and Arg81 assume comparable positions to the catalytic residues responsible for stabilizing the scissile phosphate and for 5′-leaving group protonation in other RNases (12, 18, 21, 22). Alanine substitution reduced RelE cleavage rate constants by 2.7 × 106-fold for R61A and 6.0 × 104-fold for R81A. These effects are at least an order of magnitude more than most other single alanine mutants tested.
The structurally conservative Y87F mutation yielded the most catalytically active mutant among those tested with just a 130-fold effect on the reaction rate constant. However, replacement of Tyr87 with alanine severely inhibited cleavage activity, down 1.8 × 105-fold relative to wild-type. As with the other mutants tested, each had a Kd similar to wild-type (Table 1). The Y87A substitution should abolish the stacking interactions with nucleotide 2 and was expected to reduce the efficient sequestering of mRNA into the RelE active site (Figure 1). To confirm that chemistry remained rate-limiting for this mutant, the Y87A cleavage rate for the phosphorothioate substrates was measured. Thio-effects were measured for each substrate (5-fold for RP and 6-fold for SP), suggesting that chemistry remains at least partially rate-limiting for this mutant (Table 2). However, these effects were substantially lower than those observed for wild-type and the apparent stereospecificity of wild-type was not observed with the Y87A mutant. The magnitude of these effects is closer to what it is expected for the intrinsic thio-effect, and may suggest that the reaction mechanism differs in the Y87A mutant.
The terminal amino groups from Lys52 and Lys54 are near Tyr87 and within hydrogen bonding distance of the RNA 2′-hydroxyl and phosphate. We found that under single-turnover conditions the K52A and K54A mutations each decreased the RelE rate constant by 2 × 103-fold (Table 1). The proximity of Lys52 to the conserved Tyr87 and to the 2′-hydroxyl may be critical for deprotonation at the 2′-hydroxyl. To investigate any interactions between these critically positioned residues, in particular if Tyr87 contributes to the positioning and activation of Lys52, the effect of the Lys52 mutation was measured in the context of the Y87F mutant. A modest synergistic effect for this secondary mutation renders RelE minimally active, with a 1.0 × 106-fold drop in rate from wild-type, suggesting distinct mechanistic contributions by each residue.
Discussion
RelE is a specific endoribonuclease that only cleaves mRNA in the context of the ribosome. Because of its non-standard active-site residues, RelE uses an alternative strategy from more traditional endoribonucleases for efficient substrate recognition and catalysis. Features of the RelE active site that are conserved, yet catalytically unconventional, provide substrate positioning, general acid-base function, and charge stabilization during the reaction. The unusual RelE active-site composition may lead to a requirement for substrate positioning in order to achieve catalytic activation. The obligate ribosome-dependence may derive from the ability of the ribosomal A-site to provide scaffolding that promotes productive binding interactions or the formation of the appropriate microenvironment that activates RelE catalytic side chains (12, 35).
RelE does not efficiently cleave free mRNA, but in the context of the ribosome, RelE rapidly cleaves ribosomal mRNA in the A-site (Figure 2) (10). The single-turnover catalytic rate for RelE observed in this study, k2 = 380 s-1, approaches that of the prototypical RNase A, which cuts free oligonucleotide with a kchem ∼ 1,000 s-1 (36). Yet under these conditions, there is negligible RelE-mediated cleavage of mRNA in the absence of ribosomes (data not shown) and the ribosome is estimated to enhance the RelE reaction by at least 108-fold. This specificity may result from non-productive substrate binding to free mRNA because the global ribosomal contacts may drive productive RelE-mRNA association. In RNase T1, the downstream mRNA is engaged by residues, Tyr38, His92, and Phe100, but for RelE, the ribosome must provide contacts to hold the nucleotides in place and maintain backbone distortion (12, 18, 21). This distortion promotes favorable substrate geometry for the intramolecular in-line nucleophilic attack by the mRNA 2′-hydroxyl (18). However, no ribosomal RNA or ribosomal proteins directly contact the mRNA nucleophile or leaving group, which are instead surrounded by RelE basic side chains.
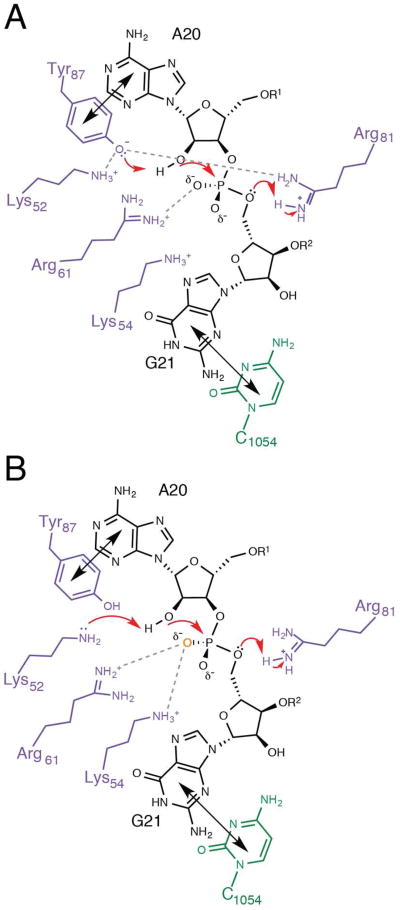
The biochemical evaluation of RelE active-site side chains by Neubauer et al led to the proposal of a general acid-base mechanism that invoked Arg81 and Tyr87 as the general acid-base pair, and Arg61 as transition state charge stabilization, with Lys52 and Lys54 providing additional transition state charge stabilization (Figure 4A) (12). However, the biochemical data presented with the structure were not consistent with this proposal. It is likely that significant catalytic contributions from each active-site side chain were overlooked because the extent of mRNA cleavage was measured after a single 15-minute time point with a small excess of substrate (12). Given a cleavage rate constant of 380 s-1, 15 minutes corresponded to approximately 500,000 single-turnover reaction half-lives for wild-type RelE. Under such conditions only the most debilitating RelE mutations would show even the slightest effect and the magnitude of the effects could not be defined.
Figure 4. Mechanism of phosphodiester bond cleavage by RelE.
(A) Previously proposed mechanism for RelE catalysis with RelE (purple), mRNA (black), and 16S rRNA C1054 (green) (12) . Tyr87 and C1054 stack with the second and third nucleotides, respectively (shown with black double arrows) to position the substrate for nucleophilic attack. The positively charged environment shifts the pKa of Tyr87, which could act a general base, while Arg61 stabilizes the transition state and Arg81 protonates the leaving group, acting as general acid. Dashed lines indicated proposed interactions. (B) Revised model for RelE catalysis, depicted as in (A). Here, Lys52 is activated as general base by the positively charged microenvironment, while Arg61 and Lys54 stabilize the transition state, and Arg81 acts as general acid. The large phosphorothioate effects suggest strong interactions between RelE and the pro-Sp non-bridging oxygen (orange), possibly from Arg61 and the neighboring Lys54.
In order to discern differences between wild-type and mutant RelE that are mechanistically important, we used a single-turnover assay to isolate RelE catalysis. By directly measuring mRNA cleavage rate constants, mutational effects can be ascribed to each RelE active-site residue. In addition to requiring in-line positioning, phosphodiester bond cleavage is promoted by the combination of nucleophile deprotonation, leaving group protonation and stabilization of the transition state (18, 37). In solution although a dianionic intermediate is expected during base-catalyzed reactions, monoanionic or neutral intermediates are possible for acid-catalyzed reactions, whereby non-bridging phosphate oxygens are protonated (38, 39). These different protonation states of the phosphorane intermediate are also possible in the context of enzymatic reactions. The reaction mechanism and catalytic functions have not been readily assigned for the non-canonical RelE reaction. Together with the co-crystal structures, the measured mutational effects provide a foundation for considering catalytic contributions of individual residues and future mechanistic investigations of RelE.
While the single-turnover effects do not explicitly implicate a particular RelE residue for a specific catalytic role, they do demonstrate that individual active-site residues make significant mechanistic contributions. The interpretation of these effects in the context of the available structural information has led to a revised mechanistic proposal for RelE-mediated cleavage (Figure 4B). In the revised mechanism, Lys52 is activated by the positively charged microenvironment to act as a general base, while Arg61 and Lys54 contact and stabilize the negatively charged phosphate and Arg81 acts a general acid. Well-conserved positively charged residues Arg61 and Arg81 are situated near the phosphate and 5′-oxygen leaving group and each significantly affects the chemical rate constant. As such, their proposed roles in transition state charge stabilization and leaving group protonation, respectively, appear indispensible to RelE (12). The positive charges and Tyr87 engage the RNA substrate in the distorted conformation required for the intramolecular in-line nucleophilic attack at the phosphate (12). Large effects (103) upon mutation of either lysine adjacent to the 2′-oxygen are indicative that both lysines contribute catalytically. A pKa shift of one amine, possibly Lys52, in the context of a highly positively charged microenvironment may enable efficient proton abstraction at the 2′-hydroxyl. Direct contacts to the phosphate oxygens, in particular the pro-SP oxygen, from Lys54 and Arg61, may polarize the phosphate bonds and further promote catalysis. Each RelE active-site substitution negatively impacted chemistry, but did not perturb the substrate binding affinity, indicating that these residues stabilize the transition state, but not the ground state.
The large catalytic effect measured for R61A (106-fold) is consistent with its proposed role in transition state stabilization. The universal conservation of Arg61 and the analogous role in charge stabilization attributed to structurally equivalent residues in RNases, Arg77 in RNase T1 and Arg71 in RNase Sa2, further reinforce the importance of this arginine in promoting nucleophilic attack by stabilizing transition-state charge build-up on the scissile phosphate (Figure 1, Figure S1) (12, 21, 22). Furthermore, the guanidinium group of RelE Arg61 is located at the juncture between other RelE side chains (Lys54, Arg81, and Tyr87), and the 2′-hydroxyl and non-bridging phosphate oxygens on the substrate (Figure 1) (12). From this position, Arg61 may coordinate active-site organization and substrate positioning, further accounting for the considerable rate effect upon mutation (Figure 4B).
The other active-site arginine in RelE, Arg81, was evoked as a general acid and the significant rate effect for its mutation reinforces the assignment of a critical catalytic function (Table 1). Leaving-group protonation in other protein phosphodiesterase enzymes is often achieved by histidine because of its near-neutral pKa, but there is precedent for arginine-mediated general acid activity in enzymes such as X-family DNA polymerases and fumarate reductase (18, 40-42). The positive charge of both this arginine and Arg61 may help lower the pKa of neighboring active-site residues and aid general base activity. In phosphodiesterase reactions, leaving group protonation may be the rate-limiting step (37, 43-45). Evidence from recent kinetic isotope effect measurements and computational calculations for enzyme and solution base-catalyzed reactions suggest that RNA transphosphorylation proceeds via a late transition state with rate-limiting expulsion of the 5′-oxygen leaving group (38, 39). Therefore, general acid catalysis is particularly important in RNase active sites for both protein and RNA enzymes. In some instances, stabilization of the developing charge at the 5′-oxygen may be more critical than optimal general base activity (43).
A primary challenge in understanding RelE-mediated phosphodiesterase activity involves nucleophile activation. Deprotonation of the 2′-hydroxyl in RNases is often initiated by glutamate or histidine side chains (Figure S1) (18). Yet, there are examples of alternative general base side chains - lysine, arginine, and tyrosine- in diverse enzymes such as oxidative decarboxylases, dehydrogenases, and epimerases (40, 46-48).
In RelE, Tyr87 has generated speculation regarding its role as a general base, but our results are inconsistent with it functioning in this role. Loss of a catalytically essential group in an RNase active site can abolish all measurable activity, as seen in Barnase upon mutation of general base Glu73 (23). However, in RelE the phenyl ring at position 87 appears nearly sufficient to maintain catalytic efficiency, with only a 130-fold effect (Table 1). Tyr87 structurally aligns with tyrosines in RNase active sites, such as Tyr42 in T1 and Tyr103 in Barnase (Figure 1, Figure S1). (12, 21, 22). Substitution of Tyr103 in Barnase with phenylalanine yielded similar results to Tyr87 in RelE, with almost no difference in cleavage or binding (24). In T1, mutation to alanine diminished binding, but not activity (49). Conserved tyrosine and other aromatic residues in RNase active sites are primarily thought to help position the nucleotide adjacent to the cut site and possibly aid specificity (18, 21, 22, 24).
The large catalytic effects and structural proximity to the 2′-hydroxyl suggest that Lys52 and/or Lys54 may play a role as the general base. Lys52 structurally coincides with the glutamate general base in RNase active sites (Figure 1D, Figure S1) (12, 21, 22). Across bacterial RelE proteins, Lys52 and Lys54 are well-conserved, but they are replaced by the analogous residue arginine in many archaeal examples (4, 12, 50). A study examining archaeal RelE translational inhibition activity identified the residue, Arg58 (Lys52 in E. coli RelE) as functionally important and the single-turnover cleavage rate constant decreased by three orders of magnitude upon mutation of either lysine (Table 1(17). The solution pKa of each active-site side chain is far from neutrality, but shifts up to 5 pKa units have been measured for lysine and arginine in other contexts (40, 51, 52). The RelE microenvironment created by the positively-charged residues could sufficiently shift the pKa of a lysine side chain to enhance its general-base functionality as seen in other enzymes (40, 51, 52). Therefore, RelE may be inactivated with increasing pH as a result of deprotonation of active-site residues critical for maintaining this microenvironment.
On the basis of the identical rate effects, we cannot exclude the possibility that Lys54 acts as general base in RelE. However, Lys54 may be better situated to interact with Lys52 and help to lower its pKa while also offering phosphate stabilization as proposed (12). Each lysine was proposed to function in additional charge stabilization at the phosphate, similar to RNase Sa2 (12, 22). In RNase Sa2, two arginine residues polarize the phosphate-oxygen bonds and prime the phosphate for intramolecular nucleophilic attack (Figure S1) (22). The large thio-effects for wild-type RelE may result from the lower propensity to hydrogen-bond with or protonate a sulfur atom, and therefore reflect strong polarization of the phosphate oxygen bonds in the transition state or formation of a triester as proposed for RNase T1 (32-34). Specifically the pro-SP oxygen may be engaged in interactions with RelE (Figure 4B). This non-bridging oxygen can be protonated to form a phosphate triester, similar to the acid-catalyzed solution reaction that proceeds via a monoanionic intermediate (38). This proton can be donated from various sources, including the general acid and 2′-hydroxyl, resulting in a phosphate triester that is 103 - 105-fold more reactive than the diester substrate (32-34). The higher reactivity of a triester substrate could lessen the requirement for a robust general base in RelE. Alternatively, the positive side chains could merely neutralize the charged phosphate. Lys54 of RelE, in particular is situated similarly to Lys27 in Barnase and His40 in RNase T1, which are critical residues for electrostatic catalysis in their respective enzymes (Figure 1D, Figure S1) (12, 21, 23, 53, 54).
Although the RelE active-site positive charges would be expected to aid ground state binding of the negatively charged RNA substrate, their loss is only detrimental to the transition state, suggesting this interaction may be largely facilitated by global interactions between RelE and the ribosomal substrate. There is a charge redistribution expected for phosphoryl transfer reaction transition states that proceed via a negatively charged pentacoordinate intermediate, which RelE may preferentially stabilize over the ground state (37, 43, 55). Furthermore, RelE side chains could contribute to stabilizing the catalytically competent distorted mRNA conformation. The differential stabilization of the highly negatively-charged or distorted phosphorane intermediate may be responsible for the observed chemical rate effects in the absence of binding effects.
Conserved and catalytically important features of the RelE active site include residues that offer substrate positioning, general acid functionality, and charge stabilization (4, 12, 16). RelE, which is a highly efficient and specific nuclease, may represent a shift in the RNase general acid-base catalytic paradigm. The strategy used by RelE relies heavily on catalytic side chains that may require activation through a positive charge network and that promote catalysis predominantly by leaving group protonation, charge stabilization, and possibly triester formation. This may render RelE largely dependent on optimal substrate geometry and overall active-site organization, which are only provided in the context of the ribosomal A-site thereby preventing indiscriminate nuclease activity. Unlike the numerous RNases such as RNase A, T1, and Barnase, that are poised for RNA cleavage with a variety of free RNA substrates, RelE can only efficiently act in the context of its ribosomal “cofactor” (10, 18).
A number of other nucleases have unusual active sites that are similar to RelE. MqsR is another bacterial toxin that may require the ribosome for mRNA cleavage and Colicin E5 targets the anticodon loop of specific tRNA molecules (56-58). Both nucleases cleave very specific RNA structures where it can be expected that substrate recognition and positioning are critical for function. Each active site is similarly composed of basic lysine and arginine side chains in addition to glutamine (both MqsR and E5) and tyrosine (MqsR) (56, 58). Since all three enzymes act on highly structured RNA substrates and share active-site composition features, these nucleases may use a similar mechanistic strategy to RelE.
These and other examples of non-canonical nuclease active sites similar to the widespread RelE toxin suggest that it may represent an alternative solution to nuclease activity that provides efficient catalysis, while ensuring strict substrate specificity. RelE relies on features of the ribosomal-bound substrate, to bind and orient the substrate. The efficiency of RelE-mediated cleavage and ribosome dependence enable RelE to specifically modulate translation rates and regulate cellular responses to environmental changes.
Supplementary Material
Acknowledgments
We thank all members of the Strobel lab for advice and encouragement, particularly, Dr. David A. Hiller for helpful discussions and critical commentary on this manuscript.
Funding: This work was supported by the National Institutes of Health Grant GM054839-15.
Abbreviations
- RNase
ribonuclease
- RC
ribosomal complex substrate
- PAGE
polyacrylamide gel electrophoresis
Footnotes
Supporting Information: Additional figures and tables for kinetic experiments, circular dichroism analysis, and structural discussions of ribonuclease active site organization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annual review of microbiology. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J Mol Biol. 2009;394:183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol. 2011;9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 8.Guglielmini J, Van Melderen L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mobile genetic elements. 2011;1:283–290. doi: 10.4161/mge.18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 11.Cherny I, Overgaard M, Borch J, Bram Y, Gerdes K, Gazit E. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry. 2007;46:12152–12163. doi: 10.1021/bi701037e. [DOI] [PubMed] [Google Scholar]
- 12.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goeders N, Dreze PL, Van Melderen L. Relaxed cleavage specificity within the RelE toxin family. J Bacteriol. 2013 doi: 10.1128/JB.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley JM, Cruz JW, Ouyang M, Woychik NA. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5′ end of coding regions in vivo. J Biol Chem. 2011;286:14770–14778. doi: 10.1074/jbc.M110.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 17.Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 18.Yang W. Nucleases: diversity of structure, function and mechanism. Quarterly reviews of biophysics. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li GY, Zhang Y, Inouye M, Ikura M. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem. 2009;284:14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Zegers I, Haikal AF, Palmer R, Wyns L. Crystal structure of RNase T1 with 3′-guanylic acid and guanosine. J Biol Chem. 1994;269:127–133. [PubMed] [Google Scholar]
- 22.Bauerova-Hlinkova V, Dvorsky R, Perecko D, Povazanec F, Sevcik J. Structure of RNase Sa2 complexes with mononucleotides--new aspects of catalytic reaction and substrate recognition. The FEBS journal. 2009;276:4156–4168. doi: 10.1111/j.1742-4658.2009.07125.x. [DOI] [PubMed] [Google Scholar]
- 23.Mossakowska DE, Nyberg K, Fersht AR. Kinetic Characterization of the Recombinant Ribonuclease from Bacillus-Amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis. Biochemistry. 1989;28:3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- 24.Meiering EM, Serrano L, Fersht AR. Effect of Active-Site Residues in Barnase on Activity and Stability. Journal of Molecular Biology. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 25.Yakovlev GI, Mitkevich VA, Shaw KL, Trevino S, Newsom S, Pace CN, Makarov AA. Contribution of active site residues to the activity and thermal stability of ribonuclease Sa. Protein science: a publication of the Protein Society. 2003;12:2367–2373. doi: 10.1110/ps.03176803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro R, Fox EA, Riordan JF. Role of lysines in human angiogenin: chemical modification and site-directed mutagenesis. Biochemistry. 1989;28:1726–1732. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- 27.Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-Dependent Conformational Change of Elongation-Factor Tu Preceding Gtp Hydrolysis on the Ribosome. Embo J. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder SP, Strobel SA. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 29.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Differential analogue binding by two classes of c-di-GMP riboswitches. Journal of the American Chemical Society. 2011;133:15578–15592. doi: 10.1021/ja204650q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frederiksen JK, Piccirilli JA. Separation of RNA phosphorothioate oligonucleotides by HPLC. Methods in enzymology. 2009;468:289–309. doi: 10.1016/S0076-6879(09)68014-9. [DOI] [PubMed] [Google Scholar]
- 31.Herschlag D, Piccirilli JA, Cech TR. Ribozyme-catalyzed and nonenzymatic reactions of phosphate diesters: rate effects upon substitution of sulfur for a nonbridging phosphoryl oxygen atom. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- 32.Herschlag D. Ribonuclease Revisited - Catalysis Via the Classical General Acid-Base Mechanism or a Triester-Like Mechanism. Journal of the American Chemical Society. 1994;116:11631–11635. [Google Scholar]
- 33.Loverix S, Winqvist A, Stromberg R, Steyaert J. Mechanism of RNase T-1: concerted triester-like phosphoryl transfer via a catalytic three-centered hydrogen bond. Chem Biol. 2000;7:651–658. doi: 10.1016/s1074-5521(00)00005-3. [DOI] [PubMed] [Google Scholar]
- 34.Stivers JT, Nagarajan R. Probing enzyme phosphoester interactions by combining mutagenesis and chemical modification of phosphate ester oxygens. Chemical reviews. 2006;106:3443–3467. doi: 10.1021/cr050317n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamal MZ, Mohammad TAS, Krishnamoorthy G, Rao NM. Role of Active Site Rigidity in Activity: MD Simulation and Fluorescence Study on a Lipase Mutant. Plos One. 2012;7 doi: 10.1371/journal.pone.0035188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song H, Ismagilov RF. Millisecond kinetics on a microfluidic chip using nanoliters of reagents. Journal of the American Chemical Society. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herschlag D, Eckstein F, Cech TR. The Importance of Being Ribose at the Cleavage Site in the Tetrahymena Ribozyme Reaction. Biochemistry. 1993;32:8312–8321. doi: 10.1021/bi00083a035. [DOI] [PubMed] [Google Scholar]
- 38.Gu H, Zhang S, Wong KY, Radak BK, Dissanayake T, Kellerman DL, Dai Q, Miyagi M, Anderson VE, York DM, Piccirilli JA, Harris ME. Experimental and computational analysis of the transition state for ribonuclease A-catalyzed RNA 2′-O-transphosphorylation. Proc Natl Acad Sci U S A. 2013;110:13002–13007. doi: 10.1073/pnas.1215086110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong KY, Gu H, Zhang S, Piccirilli JA, Harris ME, York DM. Characterization of the reaction path and transition states for RNA transphosphorylation models from theory and experiment. Angewandte Chemie. 2012;51:647–651. doi: 10.1002/anie.201104147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris TK, Turner GJ. Structural basis of perturbed pK(a) values of catalytic groups in enzyme active sites. Iubmb Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 41.Mowat CG, Moysey R, Miles CS, Leys D, Doherty MK, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase. Biochemistry. 2001;40:12292–12298. doi: 10.1021/bi011360h. [DOI] [PubMed] [Google Scholar]
- 42.Doherty MK, Pealing SL, Miles CS, Moysey R, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Identification of the active site acid/base catalyst in a bacterial fumarate reductase: A kinetic and crystallographic study. Biochemistry. 2000;39:10695–10701. doi: 10.1021/bi000871l. [DOI] [PubMed] [Google Scholar]
- 43.Bevilacqua PC. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 44.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 45.Oivanen M, Kuusela S, Lonnberg H. Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Bronsted acids and bases. Chemical reviews. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 46.Sekimoto T, Matsuyama T, Fukui T, Tanizawa K. Evidence for lysine 80 as general base catalyst of leucine dehydrogenase. J Biol Chem. 1993;268:27039–27045. [PubMed] [Google Scholar]
- 47.Aktas DF, Cook PF. A lysine-tyrosine pair carries out acid-base chemistry in the metal ion-dependent pyridine dinucleotide-linked beta-hydroxyacid oxidative decarboxylases. Biochemistry. 2009;48:3565–3577. doi: 10.1021/bi8022976. [DOI] [PubMed] [Google Scholar]
- 48.Schlippe YVG, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 49.Loverix S, Doumen J, Steyaert J. Additivity of protein-guanine interactions in ribonuclease T1. Journal of Biological Chemistry. 1997;272:9635–9639. doi: 10.1074/jbc.272.15.9635. [DOI] [PubMed] [Google Scholar]
- 50.Francuski D, Saenger W. Crystal structure of the antitoxin-toxin protein complex RelB-RelE from Methanococcus jannaschii. J Mol Biol. 2009;393:898–908. doi: 10.1016/j.jmb.2009.08.048. [DOI] [PubMed] [Google Scholar]
- 51.Harms MJ, Schlessman JL, Sue GR, Garcia-Moreno B. Arginine residues at internal positions in a protein are always charged. Proc Natl Acad Sci U S A. 2011;108:18954–18959. doi: 10.1073/pnas.1104808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci U S A. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baudet S, Janin J. Crystal-Structure of a Barnase-D(Gpc) Complex at 1.9-a Resolution. Journal of Molecular Biology. 1991;219:123–132. doi: 10.1016/0022-2836(91)90862-z. [DOI] [PubMed] [Google Scholar]
- 54.Steyaert J, Hallenga K, Wyns L, Stanssens P. Histidine-40 of Ribonuclease-T1 Acts as Base Catalyst When the True Catalytic Base, Glutamic Acid-58, Is Replaced by Alanine. Biochemistry. 1990;29:9064–9072. doi: 10.1021/bi00490a025. [DOI] [PubMed] [Google Scholar]
- 55.Ruben EA, Schwans JP, Sonnett M, Natarajan A, Gonzalez A, Tsai Y, Herschlag D. Ground state destabilization from a positioned general base in the ketosteroid isomerase active site. Biochemistry. 2013;52:1074–1081. doi: 10.1021/bi301348x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown BL, Grigoriu S, Kim Y, Arruda JM, Davenport A, Wood TK, Peti W, Page R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS pathogens. 2009;5:e1000706. doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoue-Ito S, Yajima S, Fushinobu S, Nakamura S, Ogawa T, Hidaka M, Masaki H. Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5. J Biochem. 2012;152:365–372. doi: 10.1093/jb/mvs077. [DOI] [PubMed] [Google Scholar]
- 58.Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006;34:6074–6082. doi: 10.1093/nar/gkl729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.