Abstract
The KCNH voltage-dependent potassium channels (ether-á-go-go, EAG; EAG-related gene, ERG; EAG-like channels, ELK) are important regulators of cellular excitability1-3 and have key roles in diseases such as cardiac long QT syndrome type 2 (LQT2)4, epilepsy5, schizophrenia6 and cancer7. The intracellular domains of KCNH channels are structurally distinct from other voltage-gated channels. The amino-terminal region contains an eag domain, which is comprised of a Per-Arnt-Sim (PAS) domain and a PAS-cap domain8, while the carboxy-terminal region contains a cyclic nucleotide-binding homology domain (CNBHD) which is connected to the pore through a C-linker domain. Many disease-causing mutations localize to these specialized intracellular domains, which underlie the unique gating and regulation of KCNH channels9. It has been suggested that the eag domain may regulate the channel by interacting with either the S4-S5 linker or the CNBHD8,10. Here we present a 2-Å resolution crystal structure of the eag domain-CNBHD complex of the mouse EAG1 (mEAG1) channel. It displays extensive interactions between the eag domain and the CNBHD, indicating that the regulatory mechanism of the eag domain involves primarily the CNBHD. Surprisingly, the structure reveals that a number of LQT2 mutations at homologous positions in hERG, and cancer-associated mutations in EAG channels, localize to the eag domain-CNBHD interface. Furthermore, mutations at the interface produced dramatic effects on channel gating demonstrating the important physiological role of the eag domain-CNBHD interaction. Our structure of the eag domain-CNBHD complex of mEAG1 provides unique insights into the physiological and pathophysiological mechanisms of KCNH channels.
KCNH channels have two large intracellular regions which underlie the specialized gating and regulation of this channel family9 (Fig. 1a). The CNBHD in the carboxy-terminal region shares similarity with the cyclic nucleotide-binding domains of cyclic nucleotide-gated (CNG) and hyperpolarization-activated cyclic nucleotide-modulated (HCN) channels11,12. The CNBHD of KCNH channels, however, does not bind cyclic nucleotides and regulates KCNH channel gating in a cyclic nucleotide-independent manner13. The amino-terminal region contains an eag domain, not present in CNG and HCN channels, which also plays a key role in channel gating. Comprised of a 110-amino acid PAS domain preceded by a highly conserved 25-amino acid PAS-cap (Fig. 1a and supplementary Fig. 1), the eag domain regulates activation and inactivation in EAG1, and deactivation in hERG1 channels9,14. Moreover, many of the human mutations that cause LQT2 and cancer localize to the conserved eag domain15-17. In addition, high expression of a primate-specific isoform of ERG (KCNH2-3.1), missing most of the eag domain, was shown to correlate with risk for schizophrenia6. The mechanism underlying KCNH channel regulation by the eag domain remains unclear. Two competing hypothesis have emerged. Some evidence suggests that the eag domain interacts with the S4-S5 linker and directly regulates the movement of the S4 voltage sensor10,18. By contrast, other evidence favors the CNBHD as the interaction partner of the eag domain9,14,19. Using fluorescence we found that the eag domain interacts directly with the CNBHD with an affinity of 13.2 ± 2.3 μM (supplementary Fig. 2).
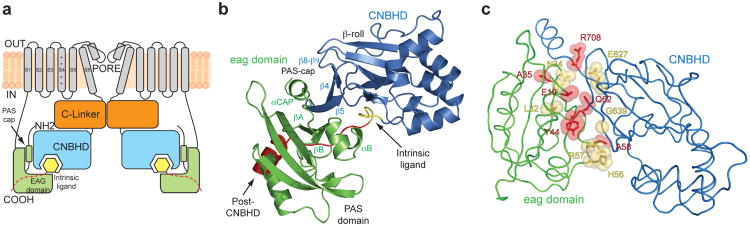
Figure 1. Structure of the eag domain-CNBHD complex of mEAG1.
a, Cartoon of a cross section of a KCNH channel. Transmembrane domains are in grey, the amino-terminal eag domains are in green, the C-linkers are in orange and the CNBHD domains in blue. The intrinsic ligand motifs are highlighted in yellow, and the post-CNBHD in red. b, Structure of the eag domain-CNBHD complex. Color corresponds to panel a. c, Disease-causing mutations at the interface of eag domain and CNBHD. LQT2 mutations are shown in yellow. Cancer-associated mutations are shown in red. Y44 is involved in both LQT2 and cancer and is shown in red.
To uncover the mechanism for how the eag domain regulates the channel, we solved the X-ray crystal structure of the eag domain-CNBHD complex to 2.0 Å resolution (supplementary Table 1). There were eight chains in the asymmetric unit, four eag domains and four CNBHDs. The spatial arrangement of the eag domains and CNBHDs within the asymmetric unit showed three different eag domain-CNBHD interactions, termed interaction A, B and C (supplementary Fig. 3). Interaction A, however, had significantly greater buried surface area than the other interactions (supplementary Table 2) and was confirmed by mutagenesis experiments in the intact channel (see Fig. 3 below). Therefore interaction A was designated as the biological unit.
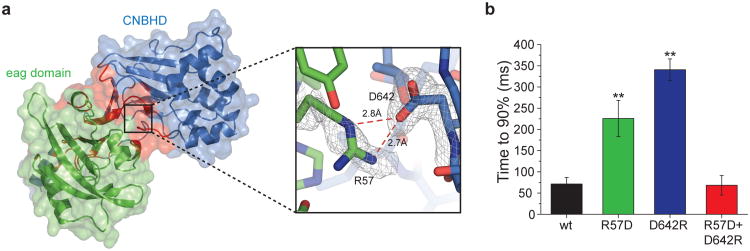
Figure 3. A salt bridge between the eag domain and the CNBHD.
a, Cartoon and surface representation of the primary interaction interface (interaction A) of the eag domain-CNBHD complex (left). Eag domain is shown in green, CNBHD in blue, and interface residues in red. A detailed illustration of the R57-D642 salt bridge in shown on the right, overlaid with 2Fo-Fc composite ‘omit’ map contoured at 1σ (grey mesh). b, Summary of time to 90% maximum current amplitude for wild-type and mutants (error bars indicate SEM, n=6-10 cells, P < 0.005, Student's t-test).
Fig. 1b shows the structure of the eag domain-CNBHD complex in the biological unit. The structure of the CNBHD (residues 552-720) includes the last 35 amino acids of the C-linker, the CNBHD, and 22 amino acids of the post-CNBD region (Fig. 1b and supplementary Fig. 1). The structure of the eag domain (residues 16-136) includes the PAS domain and the last 10 amino acids of the PAS-cap domain. These structures share many similarities to structures of CNBHDs and eag-domains crystallized alone. The CNBHD is similar to the isolated structures of mEAG1 (RMSD=0.53 Å)12, mosquito ERG (RMSD=1.05 Å)20, and zebrafish ELK (RMSD=1.39 Å)11. The PAS domain is similar to the isolated structures of mEAG1 (RMSD=0.32 Å)21, hERG1 (RMSD=0.71 Å)8,21, and fruit-fly ELK (RMSD=0.62 Å)21, and also to the solution NMR structures of hERG119,22.
The interface between the eag domain and CNBHD in the co-crystal structure, however, is entirely novel. When we mapped the positions of disease-causing mutations in KCNH channels onto the structure of the eag domain-CNBHD complex, an unexpected trend emerged: many LQT2 mutations (at homologous positions in hERG1) and cancer-associated mutations (in hEAG1) are located at the interface between the eag domain and CNBHD (Fig. 1c, supplementary Fig. 4). These mutations, which alter channel gating as well as trafficking to the membrane9,14, result in prolongation of the cardiac action potential (in hERG1) and were documented in several primary cancer types (in hEAG1)15. For example, position N34 in the eag domain, which forms hydrogen bonds with V634 of the CNBHD, was shown to cause LQT2 (hERG1 N33T). In addition, Y44 of the eag domain forms an interaction with I637 and G639 of the CNBHD. Y44H in hEAG1 is a mutation known to be correlated with large intestine carcinoma15, and mutation at the equivalent position in hERG1 (Y43C) causes LQT223 (Fig. 1c). G639 is a known LQT2 position (hERG1 G800W)23. These observations suggest that perturbation of the interaction between the eag domain and the CNBHD underlies part of the pathophysiology of KCNH-related diseases. Moreover, the clustering of so many mutations at the eag domain-CNBHD interface is consistent with this interface found in the crystal representing the physiological architecture of KCNH channels.
The interface between the eag domain and the CNBHD is extensive, with an average buried solvent accessible surface area of ∼1400 Å2 (supplementary Table 2). The interface consists of three sub-regions: (i) the intrinsic ligand motif of the CNBHD interacts with the αB helix in the PAS domain, (ii) the βA and βB strands of the PAS domain interact with the post-CNBHD segment of the CNBHD, and (iii) an amphipathic helix (αCAP) in the PAS-cap domain forms an interaction with the β-roll of the CNBHD (Fig. 1a and b).
The interaction of the PAS domain (residues 57-61) directly with the intrinsic ligand motif of the CNBHD was particularly surprising (Fig. 2a and 2b). Previous structures of isolated CNBHD from mEAG112 and zebrafish ELK (zELK)11 showed a short loop (residues 697-701) following the αC helix occupies the space filled by cyclic nucleotides in canonical cyclic nucleotide-binding domains (supplementary Fig. 1). This short loop was therefore called the “intrinsic ligand”. Mutations in this intrinsic ligand were shown to regulate channel activity, shifting the voltage dependence and kinetics of activation and deactivation. Interestingly, mutations in the eag domain of KCNH channels result in similar alterations9,14,16. The interaction between the eag domain and the intrinsic ligand, and the functional similarities between mutations in these domains, suggests that the intrinsic ligand motif may be a critical determinant of the regulation of KCNH channels by the eag domain.
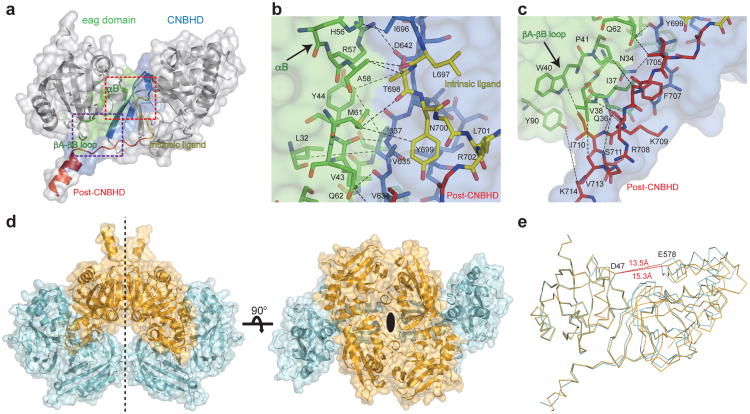
Figure 2. The various interfaces between the eag domain and CNBHD, and different confomers in the asymmetric unit.
a, Cartoon and surface representation of the eag domain-CNBHD complex. Interaction surface of the eag domain and the CNBHD is shown in green and blue, respectively. The intrinsic ligand and post-CNBHD region are in yellow and red, respectively. The non-interacting parts of the structure are in grey. Red and purple rectangles define regions detailed in b (intrinsic ligand) and c (post-CNBHD region). d, Cartoon and surface representation of the asymmetric unit complexes related by non-crystallographic symmetry in the same conformation. Two-fold symmetry axis indicated. e, Comparison of the structures of the two confomers of the complex, color coded as in d. Dashed red lines show distances between D47 of the eag domain and E578 of the CNBHD.
Another surprising site of interaction of the eag domain is the post-CNBHD region (beginning with residue 702). This region, immediately C-terminal to the intrinsic ligand, has been shown to mediate the regulation of KCNH channel function by a variety of cellular signaling events, including phosphorylation, and interaction with kinases, integrins, and Ca2+-calmodulin24-26. The eag domain-CNBHD structure shows that the post-CNBHD region interacts with the βA-βB loop of the eag domain (Fig. 2a and 2c, supplementary Fig. 1). The interaction between the eag domain and the post-CNBHD region suggests that KCNH channel regulation through the post-CNBHD region may involve its interaction interface with the eag domain.
The crystal contains two different conformations of the eag domain-CNBHD complex. The asymmetric unit is composed of four copies of the complex, related by a two-fold non-crystallographic symmetry (Fig. 2d). This results in two pairs of the complex (‘type I’ and ‘type II’ complexes) that are in a similar conformation within each pair (average RMSD = 0.16 Å), but a different conformation and B-factor distribution between pairs (RMSD = 0.96 Å) (Fig. 2d, supplementary Fig. 5). Alignment of the two conformations reveals that the angle between the eag domain and the CNBHD differs by 5° and the distance between the two domains differs by 1.8 Å (between D47 of the eag domain and E578 of the CNBHD) (Fig. 2e and supplementary video 1). In addition, the αP helix (residues 649-651, supplementary Fig. 1) undergoes a significant rearrangement between the two conformations, possibly due, at least in part, to variation in the local crystal contacts (Fig. 2e and supplementary video 1). These two conformations, if present in the intact channel, suggest that the eag domain-CNBHD complex is not static and represent a possible rearrangement associated with the gating regulation of the channel by the eag domain.
To determine the functional role of the eag domain-CNBHD interaction, we mutated the interaction interface between the eag domain and the CNBHD in intact mEAG1 channels. The interface contains a salt bridge between R57 in the αB helix of the PAS domain and D642 in the β6 strand of the CNBHD (Fig. 3a and supplementary Fig. 4). This salt bridge is present in both conformations of the biological unit and is not present in the other eag domain-CNBHD interactions in the crystal. Notably, R57 and D642 are conserved throughout the KCNH family (supplementary Fig. 1), and mutation of the R57 position in hERG1 (hERG1 R56Q) causes LQT216. mEAG1 channels with mutations in the salt bridge were expressed in Xenopus oocytes and their currents were recorded. Individually the eag domain R57D mutation and the CNBHD D642R mutation each produced a slowing of activation with voltage steps to +50 mV (supplementary Fig. 6). This resulted primarily from a new slow component to the activation time course and produced a three to five fold delay of the time to 90% current amplitude when compared to wild-type channels (Fig. 3b) (R57D, 225 ± 42 ms; D642R, 340 ± 25 ms; wild type, 71 ± 15 ms, P < 0.01, n = 9-10). No changes in other biophysical properties of the channels were noted (supplementary Table 3). Importantly, in the double charge reversal mutant, R57D+D642R, the time course of channel activation was similar to wild-type (68 ± 23 ms, P > 0.05, n = 6), suggesting the salt bridge was restored (Fig. 3b). These results indicate that the R57-D642 salt bridge is present in the intact channel, consistent with interaction A being the biological unit. Additionally, these results indicate that the eag domain regulates the activation of mEAG1 channels by interaction with the CNBHD.
A third important region of interaction with the CNBHD is with the PAS-cap domain. The PAS-cap domain constitutes the first 25 amino acids of the eag domain. This domain is highly conserved within the KCNH family (supplementary Fig. 1) and has been shown to be crucial for proper activation and inactivation gating in KCNH channels9,27. Strikingly, our structure of the eag domain-CNBHD complex reveals that an amphipathic helix (αCAP) of the PAS-cap domain of mEAG1 interacts directly with the CNBHD (Fig. 4a). The structure shows that αCAP is nestled between a hydrophobic patch of the PAS domain and the β4-β5 strands and β8-β9 loop of the CNBHD. The structure of the PAS-cap domain was previously assessed using NMR of isolated eag domain from hERG110,19,22. Structural alignment reveals that the PAS-cap domain in our eag domain-CNBHD complex is in a very different orientation from the NMR structures (Fig. 4b). The orientation, however, is very similar to recent X-ray crystallography structures of isolated eag domains from hERG1 and ELK21. We propose that the spatial organization of the PAS-cap domain within the eag domain-CNBHD complex recapitulates the native conformation of this domain in the intact channel, and that the PAS-cap domain exerts its functional effects through its interaction with the CNBHD.
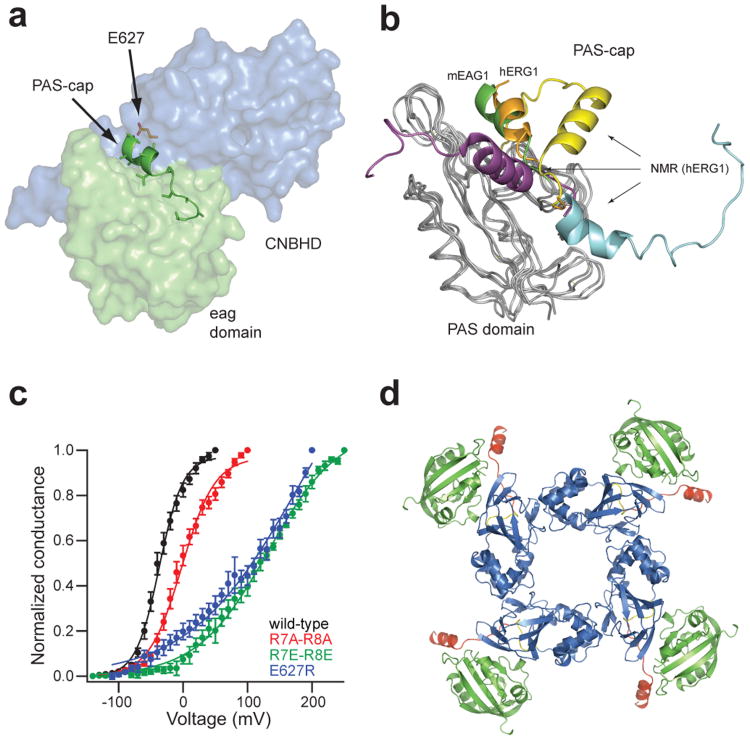
Figure 4. PAS-cap interaction with the CNBHD, and predicted tetramer of the eag domain-CNBHD complex.
a, Cartoon representation of the PAS-cap region. The PAS domain and the CNBHD are shown as surface (green and blue, respectively) with the PAS-cap regions shown as cartoon and position E627 as orange sticks. b, Alignment of the eag domain (green) with representative hERG1 eag domain NMR structures. The X-ray structures of the eag domain-CNBHD complex of mEAG1 and the isolated eag-domain of hERG1 (PDB code 4HP9) are shown as green and orange cartoons, respectively, and the NMR structures are in yellow (PDB code 2L0W), cyan (PDB code 2L4R), and purple (PDB code 2L1M). The PAS domains are represented as grey ribbons. c, Normalized conductance plotted as a function of the test voltages for mEAG1 wild-type and mutant channels (error bars indicate SEM, n=3-68 cells, P < 0.001, Student's t-test). d, Alignment of the eag domain-CNBHD complex with the CNBD of mHCN2 (PDB 1Q5O), color coded as in Fig. 1b.
Although the PAS-cap domain itself does not contain any known LQT2 mutations and only one cancer-associated mutation (hEAG1, E19D), its surroundings are rich in pathological loci, including hERG1 LQT2 mutation E788D28 (E627 in EAG1) in the β4 strand of the CNBHD9,15. These mutations may alter the gating properties of the channel by destabilizing the PAS-cap domain interaction with the CNBHD. To test this hypothesis, we measured the currents from mEAG1 channels mutated in the PAS-cap domain (R7A-R8A and R7E-R8E) and in the CNBHD region where the PAS-cap is bound (E627A and E627R). These positions are highly conserved within the KCNH family (supplementary Fig. 1). The voltage dependence of activation of the PAS-cap mutations R7A-R8A and R7E-R8E were significantly shifted to more depolarized potentials, compared to wild-type channels (Fig. 4c, supplementary table 4). The CNBHD mutations also showed a robust depolarizing shift, similar to that of the R7E-R8E mutant (Fig. 4c, supplementary table 4). Together, these results suggest that the interaction between the PAS-cap domain and the CNBHD regulates KCNH channel gating.
To model the orientation of the eag domain-CNBHD complex in the context of a four-fold symmetrical ion channel, we aligned the CNBHD of the complex with the homologous C-linker/CNBD of the mHCN2 channel (PDB code 1Q5O), which shares 35% sequence similarity with the CNBHD of mEAG1 (supplementary Fig. 1). The C-linker/CNBD in the mHCN2 structure assembles as a tetramer with a four-fold axis of rotational symmetry, in which the C-linker domains form most of the inter-subunit interactions29. While our structure of the mEAG1 CNBHD lacks most of C-linker, the alignment resulted in a concentric arrangement of the eag domain-CNBHD complex, where the eag domains are positioned at the periphery of the C-linker/CNBHD tetramer (Fig. 4d). The post-CNBHD region points away from the center and may be accessible for interaction with other proteins, such as kinases and Ca2+-calmodulin, thought to regulate KCNH channels through sites in the post-CNBHD26. Mutagenesis studies in hERG1 channels suggest that this arrangement may exhibit domain swapping where the eag domain of one subunit interacts with the CNBHD of a neighboring subunit9.
The eag domain and the CNBHD have been shown to be important regulatory domains of the KCNH channel family. Mutations found in these regions lead to LQT2 and are associated with cancer. The structure of the eag domain-CNBHD complex of mEAG1 demonstrates that the interaction interface between the two domains serves as a hotspot for many of these mutations and provides a framework to better understand the mechanisms of gating and regulation of KCNH channels. Furthermore, understanding the molecular interactions within the KCNH channel complex, and their disruption in disease, may enable design of novel therapies for these devastating conditions.
Methods summary
The mouse EAG1 eag domain (residues 6-136) and CNBHD (residues 552-724) were subcloned into a pETM11 vector containing an amino-terminal hexa-histidine affinity tag followed by a TEV cleavage site. The proteins were expressed in BL21 (DE3) Escherichia coli cells as previously described11, purified on a Ni2+ affinity column and then on an ion-exchange (CNBHD) or size exclusion column (eag domain), following an overnight cleavage with TEV at 4°C. The proteins were concentrated to about 475 μM, flash-frozen and store at −80°C until use. The eag domain and the CNBHD were mixed in a 1:1 molar ratio prior to experimentation. Crystals were grown at 20°C using sitting-drop vapor diffusion by mixing a 2:1 (v/v) ratio of protein mixture and a reservoir solution containing 18% (w/v) PEG 3350, 1.8% (v/v) Tacsimate, 10 mM MnCl2 and 90 mM HEPES, pH 7.5. The crystals diffracted to 1.995 Å and belong to space group P65 (supplementary Table 1, supplementary Fig. 7). The structure was solved by molecular replacement using the mouse EAG1 CNBHD (PDB accession 4F8A) as the search model. The final model was refined to a resolution of 1.995 Å with Rwork = 0.167 and Rfree = 0.197. Electrophysiology recordings were performed using the inside-out configuration of the patch-clamp technique on Xenopus oocytes expressing wild-type and mutant channels as previously described11.
Methods
Cloning, expression and purification
Mouse EAG1 (mEAG1) (accession codes NP034730.1 and GI:6754422) eag domain (residues 6-136) and CNBHD (residues 552-712, 552-724 and 552-752) were subcloned using 5′ NcoI and 3′ HindIII sites into a pETM11 vector containing an amino-terminal hexa-histidine affinity tag followed by a Tobacco Etch Virus protease (TEV) cleavage site. The internal NcoI restriction site in the CNBHD was removed via a silent mutation. Final CNBHD boundaries were selected based of small-scale expression and purification experiments (residues 552-724). All constructs had a GAM(G) cloning artifact sequence introduced at the N-terminus. Full-length cDNA of mEAG1 was subcloned into the high-expression pGH19 vector for expression in Xenopus oocytes. The proteins were expressed in BL21 (DE3) Escherichia coli cells as previously described1. Bacterial cultures were grown to mid-log phase and induced with 1 mM IPTG overnight at 18 °C. The cultures were spun down and resuspended in 150 mM NaCl, 20 mM Tris-HCl and 1 mM TCEP, pH 8.0, containing 1 mM AEBSF, 2.5 μg ml–1 DNAse and cOmplete protease inhibitor tablets (Roche). Cells were lysed with an EmulsiFlex C-5 homogenizer (Avestin), and the lysate was cleared by centrifugation at 131,000×g for 45 min at 4 °C. The proteins were then purified on a Ni2+ affinity resin column (HisTrap HP, GE Healthcare). The hexa-histidine tag was removed by TEV cleavage overnight at 4 °C. The CNBHD was further purified by anion exchange chromatography (HiTrap Q HP, GE Healthcare) after 10-fold dilution with buffer containing 20 mM Tris-HCl, 1 mM TCEP, pH 8.0. The protein eluted with a shallow linear NaCl gradient as one well-separated peak. The TEV digested eag domain was concentrated and further purified by gel filtration (Superdex 200, GE Healthcare) with buffer containing 150 mM NaCl, 20 mM Tris-HCl, 5 mM DTT, pH 8. Both proteins were concentrated to about 475 μM using 10 kD MWCO concentrators (Millipore), flash-frozen in liquid N2 and store at -80°C until use. Fluorescein labeling of the endogenous cysteines of the eag domain (FL-eag) was done as previously described2. Briefly, TEV digested, gel filtered purified protein was incubated with 100 μM fluorescein-5-maleimide (Invitrogen) for 1 hour at room temperature, in the above gel filtration buffer, without DTT. The labeled protein was separated from unincorporated dye by gel filtration and concentrated to 50 μM.
FSEC3 and fluorescence anisotropy
For FSEC experiments, FL-eag was mixed in a 1:1 molar ratio with purified CNBHD and the mixture was loaded on a Superdex 75 10/300 GL column (GE Healthcare) mounted on an HPLC system (Shimadzu) with a fluorescence detector set for detection of fluorescein fluorescence. Fluorescence anisotropy was recorded using a Fluorolog 3 spectrophotometer (HORIBA, Jobin Yvon). 100 nM FL-eag was placed in a quartz cuvette, and anisotropy was measured with increasing concentrations of CNBHD. Anisotropy experiments with 492 nm excitation and 514 nm emission were performed as previously described4. To estimate binding affinity, plots of the anisotropy versus total CNBHD concentration were fit using the following first-order reaction scheme:
where R, L, and RL are concentrations of the free receptor and ligand and receptor-ligand complex, respectively. Rt and Lt are total receptor and ligand concentrations, Kd is the ligand-binding affinity, and α and β are a scaling factor and an offset factor, respectively.
Crystallization and structure determination
Crystals of the eag domain-CNBHD complex were grown at 20°C using sitting-drop vapor diffusion by mixing a 2:1 ratio (Mosquito, TTP LABTECH) of protein mixture and a reservoir solution, containing 18% (w/v) PEG 3350, 1.8% (v/v) Tacsimate, 10 mM MnCl2 and 90 mM HEPES, pH 7.5. This condition produced crystals within 3 days, which grow to maximum size of about 300 μm × 80 μm × 80 μm after 21 days. For diffraction data collection, crystals were immersed in liquid nitrogen after cryoprotection in 20% glycerol. Data were collected at 110 °K on beamline 8.2.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley). Integration, scaling and merging of the diffraction data were done with the Mosflm program5. The crystals belonged to space group P65 and the unit cell had dimensions of a = b = 162.38 Å and c = 100.44 Å with α = β = 90° and γ = 120°. The structure was solved by molecular replacement using the programs Phaser6 and Phenix7 with the mEAG1 CNBHD (PDB accession 4F8A) as the search model. Data collection and refinement statistics are summarized in supplementary Table 1. Each asymmetric unit contained eight protein subunits forming four eag domain-CNBHD complexes. Electron density was visible for most of the protein chains except for a few disordered terminal residues and the initial 10 and 23 residues of the eag domains of ‘type I’ and ‘type II’ dimers, respectively. Iterative model building and refinement were carried out in Phenix with manual adjustments using COOT8. The final model was refined to a resolution of 1.995 Å with Rwork = 0.167 and Rfree = 0.197. The majority (98.2%) of the residues are in the most favored region in the Ramachandran plot. All structural illustrations were prepared with PYMOL (http://www.pymol.org).
Electrophysiology
The cDNA encoding the mEAG1 channel in the pGH19 vector was kindly provided by G. Robertson (University of Wisconsin-Madison, Madison, WI). The cRNA was transcribed using the T7 MEGAscript kit (Ambion). Xenopus laevis oocytes were defolliculated and injected with the cRNA as previously described9. Following manual removal of the vitelline membrane, currents were recorded in the inside-out patch configuration10 with an EPC-10 patch-clamp amplifier (HEKA Elektronik). Patch pipettes were pulled from borosilicate glass and had resistances of 0.4 – 0.8 MΩ after fire polishing. The intracellular (bath) and extracellular (pipette) solutions contained 130 mM KCl, 10 mM HEPES, 0.2 mM EDTA, pH 7.2. For the experiments examining the eag domain-CNBHD salt bridge (with the R57D, D642R mutations) the mEAG1 currents were elicited by applying a series of 1-2 s test pulses to voltages ranging from -120 to +50 mV in 10 mV increments from a holding potential of -100 mV, followed by a 0.5 s voltage pulse to -90 mV. For experiments examining an interaction between the PAS-cap and CNBHD (with mutations in residues R7-R8 and E627), the mEAG1 currents were elicited by a series of 50 ms test pulses to voltages ranging from -140 mV to +200 mV in 10 mV increments, from a holding potential of -100 mV, following by a 200 ms voltage pulse to -100 mV. Currents were not leak subtracted. Data were acquired with Pulse software (HEKA Elektronik) and analyzed with Igor (WaveMetrics, Inc).
To measure the conductance-voltage relationships for all electrophysiology experiments, peak tail current amplitudes at -90 or -100 mV, were normalized to the largest peak conductance amplitude. Plots of the normalized conductance versus the test voltage were fit with a Boltzmann function:
Where V represents the test voltage (mV), V0.5 is the midpoint activation voltage (mV), and s is the slope of the relation (mV).
Supplementary Material
Acknowledgments
We thank S. Cunnington and S. Camp for technical assistance. We thank the beamline staff at the Advanced Light Source (ALS). We also thank G. Flynn and J. Bankston, M. Puljung and S. Gordon for helpful discussions and critical reading of the manuscript. This work was supported by the National Institutes of Health (NIH) grant R01 EY010329 (W.N.Z.), The International Human Frontier Science Program Organization (HFSPO) long-term postdoctoral fellowship LT-001025/2011 (Y.H.), and NIH grant F32 HL095241 (A.E.C.). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Author contributions: Y.H. and W.N.Z. conceived the experiments. Y.H. performed the biochemical and crystallographic experiments and data analysis. A.E.C. conceived the PAS-cap related experiments and performed all the electrophysiology experiments and data analysis. Y.H. and W.N.Z. wrote the manuscript.
Author information: Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 4LLO. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Warmke J, Drysdale R, Ganetzky B. A distinct potassium channel polypeptide encoded by the Drosophila eag locus. Science. 1991;252:1560–2. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- 2.Ganetzky B, Robertson GA, Wilson GF, Trudeau MC, Titus SA. The eag family of K+ channels in Drosophila and mammals. Ann N Y Acad Sci. 1999;868:356–69. doi: 10.1111/j.1749-6632.1999.tb11297.x. [DOI] [PubMed] [Google Scholar]
- 3.Bauer CK, Schwarz JR. Physiology of EAG K+ channels. J Membr Biol. 2001;182:1–15. doi: 10.1007/s00232-001-0031-3. [DOI] [PubMed] [Google Scholar]
- 4.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–9. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Deletion of the potassium channel Kv12.2 causes hippocampal hyperexcitability and epilepsy. Nat Neurosci. 2010;13:1056–8. doi: 10.1038/nn.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huffaker SJ, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15:509–18. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho J. Ether a go-go potassium channels and cancer. Cancer Lett. 2006;233:1–9. doi: 10.1016/j.canlet.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Morais Cabral JH, et al. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–55. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 9.Gustina AS, Trudeau MC. HERG potassium channel regulation by the N-terminal eag domain. Cell Signal. 2012;24:1592–8. doi: 10.1016/j.cellsig.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, et al. NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker. Biochem Biophys Res Commun. 2010;403:126–32. doi: 10.1016/j.bbrc.2010.10.132. [DOI] [PubMed] [Google Scholar]
- 11.Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. Structure of the carboxy-terminal region of a KCNH channel. Nature. 2012;481:530–3. doi: 10.1038/nature10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques-Carvalho MJ, et al. Structural, biochemical, and functional characterization of the cyclic nucleotide binding homology domain from the mouse EAG1 potassium channel. J Mol Biol. 2012;423:34–46. doi: 10.1016/j.jmb.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Brelidze TI, Carlson AE, Zagotta WN. Absence of direct cyclic nucleotide modulation of mEAG1 and hERG1 channels revealed with fluorescence and electrophysiological methods. J Biol Chem. 2009;284:27989–97. doi: 10.1074/jbc.M109.016337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens L, Ju M, Wray D. Roles of surface residues of intracellular domains of heag potassium channels. Eur Biophys J. 2009;38:523–32. doi: 10.1007/s00249-009-0402-8. [DOI] [PubMed] [Google Scholar]
- 15.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Zou A, Splawski I, Keating MT, Sanguinetti MC. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J Biol Chem. 1999;274:10113–8. doi: 10.1074/jbc.274.15.10113. [DOI] [PubMed] [Google Scholar]
- 17.Splawski I, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–85. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Trudeau MC, Zappia AM, Robertson GA. Regulation of deactivation by an amino terminal domain in human ether-a-go-go-related gene potassium channels. J Gen Physiol. 1998;112:637–47. doi: 10.1085/jgp.112.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muskett FW, et al. Mechanistic insight into human ether-a-go-go-related gene (hERG) K+ channel deactivation gating from the solution structure of the EAG domain. J Biol Chem. 2011;286:6184–91. doi: 10.1074/jbc.M110.199364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brelidze TI, Gianulis EC, Dimaio F, Trudeau MC, Zagotta WN. Structure of the C-terminal region of an ERG channel and functional implications. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adaixo R, Harley CA, Castro-Rodrigues AF, Morais-Cabral JH. Structural Properties of PAS Domains from the KCNH Potassium Channels. PLoS One. 2013;8:e59265. doi: 10.1371/journal.pone.0059265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng CA, et al. The N-terminal tail of hERG contains an amphipathic alpha-helix that regulates channel deactivation. PLoS One. 2011;6:e16191. doi: 10.1371/journal.pone.0016191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano C, et al. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–80. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 24.Cherubini A, et al. Human ether-a-go-go-related gene 1 channels are physically linked to beta1 integrins and modulate adhesion-dependent signaling. Mol Biol Cell. 2005;16:2972–83. doi: 10.1091/mbc.E04-10-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun XX, Hodge JJ, Zhou Y, Nguyen M, Griffith LC. The eag potassium channel binds and locally activates calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2004;279:10206–14. doi: 10.1074/jbc.M310728200. [DOI] [PubMed] [Google Scholar]
- 26.Ziechner U, et al. Inhibition of human ether a go-go potassium channels by Ca2+/calmodulin binding to the cytosolic N- and C-termini. FEBS J. 2006;273:1074–86. doi: 10.1111/j.1742-4658.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Myers CD, Robertson GA. Dynamic control of deactivation gating by a soluble amino-terminal domain in HERG K(+) channels. J Gen Physiol. 2000;115:749–58. doi: 10.1085/jgp.115.6.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–17. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Zagotta WN, et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–5. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
Methods references
- 1.Brelidze TI, Carlson AE, Sankaran B, Zagotta WN. Structure of the carboxy-terminal region of a KCNH channel. Nature. 2012;481:530–3. doi: 10.1038/nature10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods. 2009;6:532–7. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–81. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat Protoc. 2011;6:365–87. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 6.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 9.Zagotta WN, Hoshi T, Aldrich RW. Gating of single Shaker potassium channels in Drosophila muscle and in Xenopus oocytes injected with Shaker mRNA. Proc Natl Acad Sci U S A. 1989;86:7243–7. doi: 10.1073/pnas.86.18.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.