Abstract
Cancer is a complex disease that can originate in virtually all tissues of the body, and tumors progress through many different stages during their development. While genetic mutations in the emerging cancer cells drive this disease, it has become increasingly clear that cancer development is strongly influenced by the surrounding microenvironment. Cells of the immune system are critical components of this extrinsic network of cancer regulators, contributing significantly to the microenvironment of most cancers and either promoting or inhibiting the initiation and progression of this disease. Genetically engineered mouse (GEM) mouse models of spontaneous cancer are starting to shape our understanding of how anti-tumor T cells may act to prevent or inhibit cancer progression in some settings and not others. Lessons learned from investigating spontaneous mouse cancer models have important implications for directing clinical efforts that attempt to direct a cancer patient’s immune system to eradicate their disease.
Introduction
Understanding the role of the immune system in human cancer requires the use of animal models that faithfully recapitulate the diversity of interactions between immune cells and the heterogeneous forms of cancer that affect humans. At the same time, these models must allow for hypothesis-driven experimentation, providing reproducible tumor initiation and growth, as well as the capacity to monitor T cells and other cells of the immune system reacting to the tumors. Interest in GEM models of cancer to study anti-tumor immune responses has increased significantly recently, with these models serving as a valuable alternative to the more widely used transplantable and carcinogen induced cancer models. GEM cancer models have led to new biological insights about the importance of tumor antigens, the impact of the tumor type, origin, and underlying genetics in determining immune responses, and the role of immune tolerance versus immunoediting in the process of tumor escape. They have also provided an advanced platform for understanding and improving immunotherapy by revealing aspects of the immune response that can control tumor responsiveness to chemotherapies, targeted therapies, and immunotherapies. The opportunities and limitations of these models compared to alternative cancer models are highlighted in Table 1 and have been recently reviewed [1–3].
Table 1.
A comparison of the different mouse models of cancer
| Features: | Transplanted | Carcinogen- induced | Germline- GEM | Conditional- GEM |
|---|---|---|---|---|
| Cancers modeled: Examples: | All B16 melanoma EL4 lymphoma MC57 fibrosarcoma Lewis Lung carc. TRAMPC prostate |
Limited MCA sarcoma UV fibrosarcoma DMBA+TPA skin carc. |
All RIP-Tag2 pancreatic β-cell hyperplasia PyMT mammary TRAMP (Pro-Tag2) prostate |
All KrasLSL-G12D lung KrasLSL-G12D;PTENf/f ovarian KrasLSL-G12D;p53f/f sarcoma KrasLSL-G12D pancreatic |
| Time to progression: (survival time) | 0–4 weeks (after transplant) | 2–4 months (after induction) | 2–6 months (mouse age) | 2–12 months (after induction) |
| Genetics alterations mimic human cancers? | Unknown/yes | Unknown/yes | Yes (some models) | Yes |
| Histopathology mimics human cancers? | Limited cases | Yes (limited tumor types) | Yes | Yes |
| Tumor initiated by transformation of normal cells? | No | Yes | Yes | Yes |
| Timing of tumor initiation controlled? | Yes | Partially (variable latency & penetrance) | No (empirically defined) | Yes |
| Location of tumor formation controlled? | Yes (orthotopic) | Yes (limited tumor types) | Yes (transgenic); No (tumor suppressor KO) | Yes (limited technology) |
| Tumors in natural microenvironment? | No (maybe orthotopic) | Yes (carcinogens may affect environment) | Yes (oncogenic events not restricted to tumor) | Yes |
| Multifocal disease? | No | Unknown | Yes | Yes |
| Track tumor antigen specific T cell responses? | Yes | No | Yes | Yes |
| Restrict tumor antigen expression to tumors? | Yes | NA | No | Possible |
| Regulated tumor antigen expression? | Possible | No | No | Possible |
Why use spontaneous mouse models of cancer?
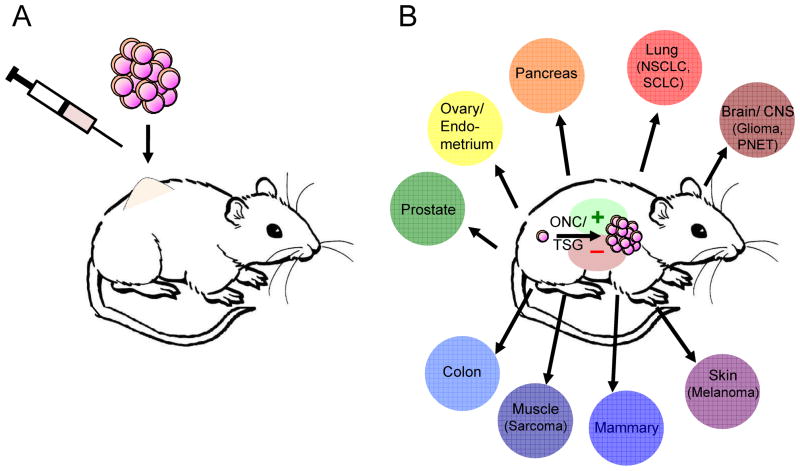
GEM models of cancer represent a diverse collection of genetically modified mice that are predisposed to develop specific types of cancer spontaneously [4]. Such models can be divided into two forms: germline GEM models, which develop cancers in an unregulated (spontaneous) fashion, and conditional GEM models, which provide spatiotemporal control of tumor onset utilizing tissue-specific, ligand-regulated, and/or viral-based technologies [2]. By transforming normal cells in situ with defined genetic events, GEM models can recapitulate the genetic and histopathological characteristics of nearly all forms of human cancer as well as the progression of tumors from the stage of initiation to advanced forms of the disease. In stark contrast, transplant models of cancer involve the introduction of large numbers of fully progressed, relatively homogeneous tumor cell clones into the animal. Furthermore, these often involve ectopic sites (typically subcutaneous) where no equivalent human cancer develops [5]. This form of delivery inevitably leads to massive tumor cell death that can elicit an immune response. In addition, the rapid growth of most transplanted tumors leads to the death of the recipient within a few weeks if untreated. As such, the analysis of the immune response to the tumor, as well as any immune-modulatory therapy, occurs in an acute setting rather than in the context of a more natural course of tumor progression or within an established tumor microenvironment [6,7]. Importantly, critical differences in immunosurveillance and therapeutic responses have been described between equivalent autochthonous and transplanted tumors [8,9], and it is plausible that the number of cells, progressed state, and cellular milieu of transplanted tumors may influence these differences [10–13]. Consequently, interactions between the immune system and cancers in this setting are likely dominated by the transplantation itself, making it difficult to recapitulate the contextual diversity of immune responses that clearly vary in different human cancers (Figure 1). While carcinogen induced cancers can be as good or better than GEM in their capacity to model human cancer, their genetic complexity, considerable variation in progression, and the limited number of cancers that can be modeled are drawbacks. To improve the utility of these models, high doses of carcinogens are often used to increase the penetrance and reduce the latency of tumor formation, potentially generating exceptionally large numbers of carcinogen-induced neoantigens [14,15].
Figure 1. Key differences between transplanted models of cancer and spontaneous or genetically engineered mouse models of cancer affect the immune response to tumors.
A, transplanted tumors introduce large numbers of fully progressed tumor cells into a limited diversity of immune environments (typically subcutaneous inoculation). B, cancers in GEM models originate from single cells that are transformed in situ through the activation of oncogenes (ONC) and inactivation of tumor suppressor genes (TSG) and progress in the unique immune environments of their native tissues. Furthermore, studies using GEM cancers can interrogate the role of defined genetic events that drive each cancer in activating (+) or suppressing (–) immune responses.
Tumor antigens and tracking tumor-reactive T cells
Understanding T cell responses against cancer hinges on our ability to monitor the persistence and function of tumor-reactive T cells. As tumor-reactive T cells may make up only a small fraction of tumor-infiltrating lymphocytes ([8] and unpublished data), it is important that tumor models do not rely on phenotyping bulk CD8 T cell populations. In this regard, transplantable models have advantages, as they are easily modified to express antigens that lead to monitorable anti-tumor immune responses and offer one of the few systems to obtain truly tumor-specific expression of model antigens. Carcinogen-induced cancers are highly immunogenic, harboring tumor-specific antigens (TSAs) that drive anti-tumor T cell responses. However, due to the spontaneous development of these TSAs, it is not possible to track the T cell responses in the setting of primary tumor formation [14]. While GEM models of cancer accurately recapitulate many aspects of human cancer, their stimulation of a tumor-specific immune response is likely to be less pronounced, in part due to the genetic programming of driver mutations in oncogenes and tumor suppressor genes [10,14]. Therefore, GEM cancer models may need to be modified to express antigens to model the anti-tumor T cell responses observed in human cancers [16]. In fact, because GEM cancers remove much of the antigenic complexity that is uncontrolled in transplantable or carcinogen-induced tumors, they may provide the cleanest system to introduce specific antigens and allow for a focused investigation of tumor-specific T cells. One strategy to express model tumor antigens in GEM tumors utilizes tissue-specific promoters to restrict antigens to the same organ, likely modeling tumor-associated antigens (TAAs). However, this approach may fundamentally alter the phenotype of responding T cells because the antigens are also expressed in normal cells of the target tissue and well before tumor formation [17–20]. The recent use of conditional GEMs to study T cell responses to tumors largely circumvent this issue by linking the expression of oncogenes to model antigens, allowing for tumor-specific expression of model antigens [21,22]. Nevertheless, it has proved to be challenging to completely prevent antigen expression in the thymus or other cells prior to tumor formation [21].
Attempts to control antigen expression have ultimately allowed for some functional comparisons of the T cell responses against TSAs versus TAAs [23]. Although T cells responding to germline GEM models of cancer typically become tolerant (often systemically), this may be the result of T cells responding to the more commonly modeled TAAs. In an effort to investigate T cell responses against TSAs expressed from endogenously arising tumors, we developed model systems to either introduce TSAs or overexpress TAAs antigens in autochthonous mouse lung cancers utilizing conditional GEM models [8,21]. These studies, along with others [22,24], indicated that TSAs may be uniquely capable of evoking potent endogenous T cell responses against tumors that delay or prevent cancer development. In another approach, we utilized a conditional GEM model of sarcomagenesis to show that TSA expression was required for the process of immunoediting against endogenous sarcomas [10]. More broadly, it seems tumor immunogenicity results from mutations that generate TSAs (a common characteristic of most cancers) [14], but this may not be an obligatory step in the tumorigenic process. This has important implications for T cell responses against human cancers, as not all cancers may harbor potent TSAs. Nevertheless, targeting TSAs has the benefit of specificity for tumors, reducing the risk of inducing autoimmune reactions, and targeting mutations in tumors that are necessary for driving the disease, precluding tumor escape by antigen loss [24,25]. In the post-genomic era, there is great potential to utilize DNA sequencing to rapidly identify mutations in individual tumors, computationally predict peptides that can best stimulate T cell responses, and vaccinate patients against the unique TSAs in their tumors [26,27].
Tumor type, origin, and genetics affect the T cell response
Over four decades ago, R.T. Prehn speculated that the tissue in which a cancer arises influences how the immune system responds to cancer [28]. However, this issue still has not been adequately addressed experimentally. Instead, discoveries made in a particular model or form of cancer are often interpreted to be broadly applicable to all immune-tumor interactions. While this may have some truth in transplantable cancer models (Figure 1), it does not appear to be the case in human cancers. A tremendous diversity of T cell responses can be observed in different cancers and responses can vary depending on contextual elements of each cancer, such as the originating tissue, the state of immunosurveillance or immunoregulation at that site, the genetics of the developing tumor, or as already described, the nature of the antigens driving the immune response.
Perhaps the best examples of how the contextual elements of cancers can affect the immune response comes from considering the evidence that adaptive immune responses to some tumors can promote tumor progression [29,30]. Mammary and skin cancers are aided by CD4+ T or B cells that promote the activity of innate immune cells that can support tumor development and spread. Interestingly, however, in the context of immunotherapy or chemotherapy, the activity of these adaptive immune cells can be shifted to promote anti-tumor behavior [31,32]. Cytokines are known to have pleitropic activities, and may have opposing roles in different forms of cancer. Cancers of the skin were recently shown to be inhibited by T cell responses that were supported by the presence of thymic stromal lymphopoietin (TSLP), whereas TSLP is known to drive several epithelial cancers by promoting pro-tumorigenic inflammation [33,34]. Tumor necrosis factor (TNF) is a classic example of a cytokine known to promote inflammation and cancer in some settings, while also serving as a critical effector arm for adaptive immune responses against cancer in others [35–37]. Additional cytokines like GM-CSF or IL-10, typically thought to have positive or negative effects on anti-tumor T cell responses against cancers, respectively, were recently shown to have the opposite function in specific cancers [38–40], often via their modulation of immunosuppressive cells that block anti-tumor responses [41,42]. In several cases, the underlying oncogenic drivers of the cancer have been shown to directly regulate cancer cell production of these immune modulators, such as ras-induced GM-CSF or KIT-induced Ido production [38–40,43]. These are intriguing observations given that oncogenes have also been found to activate pathways in tumors that can promote anti-tumor immune responses, including by upregulating stress ligands recognized by NKG2D receptors on NK or T cells [44]. Indeed, the contribution of cytokines to tumor progression can be quite complex and have countervailing roles depending on the type and stage of the disease [45].
Adding to the complexity, recent studies from our lab have shown that T cell responses can diverge dramatically against different cancers even if they have the same TSAs and underlying genetic events. T cell responses against sarcomas of the hind limb induced by expression of oncogenic K-ras and loss of p53 function were fully functional and blocked tumor formation or forced TSA loss. In contrast, in a model of adenocarcinoma of the lung driven by the same mutations and expressing the same TSAs, responding T cells were not fully functional, could not drive TSA loss, and only delayed the malignant progression of the cancers [8,10]. It is tempting to speculate that the different immune environments of the tissues that give rise to these two forms of cancer dictate the divergent anti-tumor immune responses in each context. Because the lung is constantly exposed to irritants, allergens and the antigens of inhaled pathogens, it is likely to harbor sensitive immune-regulatory networks to prevent detrimental immune responses to innocuous encounters, imposing a more stringent threshold for adaptive immune activation against cancer. However, the muscle tissue that gives rise to sarcomas would normally be exposed to antigens solely in the context of a pathogenic infection and, thus, may have fewer regulatory requirements for activating cells of the adaptive immune system to any antigen, including those that arise in developing tumors. These results emphasize the contextual diversity of cancer that can be investigated with different genetic drivers and in different tissues. Importantly, too, therapies targeting particular immune pathways as treatment for cancer must exercise caution and consider the opposing effects such therapies may have on promoting other forms of cancer.
Immune tolerance or immunoediting driven tumor escape? It depends!
Whether cancers progress because of immune tolerance or the evolution of tumor-escape mechanisms has been a topic of great debate. Most likely both mechanisms are relevant in different contexts or at different points in tumor progression, as autoregulatory tolerance mechanisms may provide an alternative route for tumors to progress unedited by dampening functional immune responses. Indeed, an examination of recent studies using spontaneous mouse models of cancer provides evidence that both mechanisms of tumor evasion occur, again likely depending on the contextual elements of each cancer. T cell tolerance may stem from the fact that anti-tumor responses are fundamentally different than responses to acute pathogenic infections, which the immune system has evolved to resist. There are many parallels in the phenotypes of T cells responding to cancer and chronic infections [46]. Immune tolerance to cancer may result from the induction of immune regulatory pathways (potentially co-opted by tumors) that evolved to prevent autoimmune disease in the setting of persistent infectious disease. T cells may be driven to exhaustion or anergy in response to tumors due to chronic antigen presentation at tumor sites [19,47,48]. Vaccination regimens often improve the function of T cells, indicating that natural priming against tumor antigens may be insufficient in many contexts [8,49]. Alternatively, models of pancreatic and breast cancer have shown that tumors actively recruit myeloid-derived suppressor cells (or T regulatory cells) that suppress adaptive immune responses at tumor sites [38,39,42].
Theories of immunoediting postulate that tumors evolve mechanisms to bypass anti-tumor immune responses and, thus, tumors are shaped by these encounters [16]. The remnants of these interactions can be identified by changes in tumor cells that make them less susceptible to immune recognition and destruction (i.e. less immunogenic). Immunoediting by the adaptive immune system has been appreciated for nearly a decade and evidence continues to accumulate for the role of T cells in the elimination of nascent tumors, tumor maintenance in a dormant state (equilibrium phase), and ultimately, tumor escape [10,14,35,45,50]. Broadening the scope of immune cells capable of regulating tumor development, recent studies demonstrated that innate cells of the immune system, particularly NK cells, participate in immunoediting tumors [11,44].
Improving therapy by combining immunotherapy with conventional cancer therapies
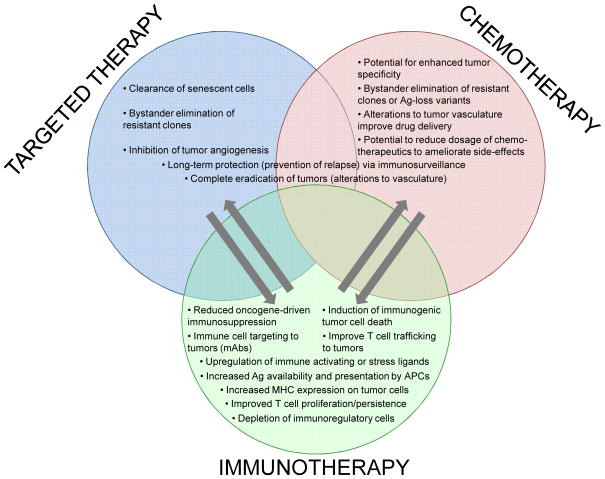
The primary goal of cancer therapy is to induce tumor cell death while sparing normal cells and limiting general toxicity. Immunotherapies utilizing anti-tumor T cells promise tremendous tumor specificity. However, chemotherapies and targeted therapies may also support even better anti-tumor immune responses (Figure 2) [51]. DNA damage responses induced by various chemotherapeutic drugs have been demonstrated to up-regulate stress or danger signals on tumor cells that alert the immune system, stimulating the recruitment and anti-tumor activity of T cells and NK cells or enhancing tumor antigen presentation by dendritic cells [52]. In addition, these therapies can promote anti-tumor immune responses by modulating or depleting immunosuppressive cell populations [51,53,54]. However, it is becoming apparent that the effect of these therapies is connected to the particular immune microenvironments of tumors, once again highlighting the need for experimental models that recapitulate the diversity of tumor immune environments of different human cancers [55,56].
Figure 2. Combining immunotherapy with conventional cancer therapies can enhance treatment efficacy compared to their respective monotherapies.
Arrows indicate potential mechanisms in which one therapy can promote (→) the efficacy of another therapy (also see recent reviews [51,52]). Importantly, while synergism between these therapies is well documented, the mechanisms of action are largely undetermined, especially in the context of different types of cancer and their unique mircoenvironments.
Several elegant proof-of-principle experiments using spontaneous mouse models of cancer have demonstrated that the efficacy of targeted therapies depends on the activity of concomitant anti-tumor T cell responses [43,56,57]. Interestingly, it was shown that complete eradication of tumors targeted with oncogene-specific blockade was only achieved when combined with T cell responses, which more broadly target the tumor tissue, not only destroying tumor cells directly but also the tumor vasculature [56,57]. In the context of chemotherapy, the efficacy of doxorubicin to treat carcinogen-induced sarcomas required CD8+ T cells [58]. However, diverse chemotherapeutics (paclitaxel, doxorubicin) in the treatment of breast cancer were antagonized by the recruitment of innate immune cells to the tumor microenvironment [32,59]. In these settings, blocking the migration of these cells to tumors improved chemotherapeutic responses.
Conclusion
An important next step for cancer immunology should be to embrace the diversity of immune environments that likely shape immune responses to cancers that arise in different tissues. GEM models of cancer provide the means to recapitulate the great diversity of human cancers, preserving the specific contextual elements of different forms of cancer that affect anti-tumor T cell responses. The goal moving forward should be to utilize more models, employing different underlying genetics and tissue origins, as well as developing new strategies to better mimic the T cell response by modulating the nature of the tumor antigens (TSAs versus TAAs) that direct it. In addition to the impressive utilization of these GEM models for testing novel therapies and combinations of therapies, the inclusion of strategies for in vivo imaging and monitoring the dynamic interactions of the cells within the tumor microenvironment are providing an additional layer of mechanistic information about how to improve immunotherapies [47,59,60]. Finally, these studies have revealed that effective immune therapies against cancer not only boost T cell responses to tumors, but also counteract the many regulatory networks that likely restrict the duration of active immune responses. Taking lessons from the study of autoimmune diseases, which represent rare breakdowns in these regulatory networks, may provide important clues to improve immune-based treatments for cancer.
Acknowledgments
We thank A.G. DuPage for editing this manuscript. This work was supported by Grant 1 U54 CA126515-01 from the NIH and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute and the Margaret A. Cunningham Immune Mechanisms in Cancer Research Fellowship Award (M.D.) from the John D. Proctor Foundation. T.J. is a Howard Hughes Investigator and a Daniel K. Ludwig Scholar.
Annotated references
- 1.Dranoff G. Experimental mouse tumour models: what can be learnt about human cancer immunology? Nat Rev Immunol. 2012;12:61–66. doi: 10.1038/nri3129. [DOI] [PubMed] [Google Scholar]
- 2.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 3.Singh M, Murriel CL, Johnson L. Genetically engineered mouse models: closing the gap between preclinical data and trial outcomes. Cancer Res. 2012;72:2695–2700. doi: 10.1158/0008-5472.CAN-11-2786. [DOI] [PubMed] [Google Scholar]
- 4.Zender L, Zuber J, Lowe SW. Snapshot: genetic mouse models of cancer. Cell. 2007;129:838. doi: 10.1016/j.cell.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becher OJ, Holland EC. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66:3355–3358. doi: 10.1158/0008-5472.CAN-05-3827. discussion 3358–3359. [DOI] [PubMed] [Google Scholar]
- 6.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. discussion 3354. [DOI] [PubMed] [Google Scholar]
- 7.Wen FT, Thisted RA, Rowley DA, Schreiber H. A systematic analysis of experimental immunotherapies on tumors differing in size and duration of growth. Oncoimmunology. 2012;1:172–178. doi: 10.4161/onci.1.2.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19:72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Garbe AI, Vermeer B, Gamrekelashvili J, von Wasielewski R, Greten FR, Westendorf AM, Buer J, Schmid RM, Manns MP, Korangy F, et al. Genetically induced pancreatic adenocarcinoma is highly immunogenic and causes spontaneous tumor-specific immune responses. Cancer Res. 2006;66:508–516. doi: 10.1158/0008-5472.CAN-05-2383. These studies [8,9] show that in response to autochthonous cancers of the lung or pancreas T cells acquire tolerance phenotypes. In contrast, in response to transplanted tumors derived from the GEM models, T cell responses are effective and are capable of rejection. [DOI] [PubMed] [Google Scholar]
- 10.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF, Teng MW, et al. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J Exp Med. 2012;209:1869–1882. doi: 10.1084/jem.20112738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikder H, Huso DL, Zhang H, Wang B, Ryu B, Hwang ST, Powell JD, Alani RM. Disruption of Id1 reveals major differences in angiogenesis between transplanted and autochthonous tumors. Cancer Cell. 2003;4:291–299. doi: 10.1016/s1535-6108(03)00245-9. [DOI] [PubMed] [Google Scholar]
- *14.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. This study provides proof-of-principle evidence that DNA sequencing of cancer exomes can be used to identify immunogenic TSAs that can be targeted immunotherapeutically to fight individual cancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 16.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 17.Getnet D, Maris CH, Hipkiss EL, Grosso JF, Harris TJ, Yen HR, Bruno TC, Wada S, Adler A, Georgantas RW, et al. Tumor recognition and self-recognition induce distinct transcriptional profiles in antigen-specific CD4 T cells. J Immunol. 2009;182:4675–4685. doi: 10.4049/jimmunol.0803400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huijbers IJ, Soudja SM, Uyttenhove C, Buferne M, Inderberg-Suso EM, Colau D, Pilotte L, Powis de Tenbossche CG, Chomez P, Brasseur F, et al. Minimal tolerance to a tumor antigen encoded by a cancer-germline gene. J Immunol. 2012;188:111–121. doi: 10.4049/jimmunol.1002612. [DOI] [PubMed] [Google Scholar]
- 19.Bruno TC, Rothwell C, Grosso JF, Getnet D, Yen HR, Durham NM, Netto G, Pardoll DM, Drake CG. Anti-tumor effects of endogenous prostate cancer-specific CD8 T cells in a murine TCR transgenic model. Prostate. 2011;72:514–522. doi: 10.1002/pros.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buschow C, Charo J, Anders K, Loddenkemper C, Jukica A, Alsamah W, Perez C, Willimsky G, Blankenstein T. In vivo imaging of an inducible oncogenic tumor antigen visualizes tumor progression and predicts CTL tolerance. J Immunol. 2010;184:2930–2938. doi: 10.4049/jimmunol.0900893. [DOI] [PubMed] [Google Scholar]
- 21.Cheung AF, Dupage MJ, Dong HK, Chen J, Jacks T. Regulated expression of a tumor-associated antigen reveals multiple levels of T-cell tolerance in a mouse model of lung cancer. Cancer Res. 2008;68:9459–9468. doi: 10.1158/0008-5472.CAN-08-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Homig-Holzel C, Wang Z, Derudder E, Li S, Chakraborty T, et al. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell. 2012;148:739–751. doi: 10.1016/j.cell.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neller MA, Lopez JA, Schmidt CW. Antigens for cancer immunotherapy. Semin Immunol. 2008;20:286–295. doi: 10.1016/j.smim.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Schietinger A, Philip M, Schreiber H. Specificity in cancer immunotherapy. Semin Immunol. 2008;20:276–285. doi: 10.1016/j.smim.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiarle R, Martinengo C, Mastini C, Ambrogio C, D'Escamard V, Forni G, Inghirami G. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat Med. 2008;14:676–680. doi: 10.1038/nm1769. [DOI] [PubMed] [Google Scholar]
- 26.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, Allison JP. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- *27.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, Selmi A, Diken M, Boegel S, Paret C, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 28.Prehn RT. Tumor-specific antigens of putatively nonviral tumors. Cancer Res. 1968;28 :1326–1330. [PubMed] [Google Scholar]
- 29.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreu P, Johansson M, Affara NI, Pucci F, Tan T, Junankar S, Korets L, Lam J, Tawfik D, DeNardo DG, et al. FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell. 2010;17:121–134. doi: 10.1016/j.ccr.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207:637–650. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demehri S, Turkoz A, Manivasagam S, Yockey LJ, Turkoz M, Kopan R. Elevated epidermal thymic stromal lymphopoietin levels establish an antitumor environment in the skin. Cancer Cell. 2012;22:494–505. doi: 10.1016/j.ccr.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Piazza M, Nowell CS, Koch U, Durham AD, Radtke F. Loss of Cutaneous TSLP-Dependent Immune Responses Skews the Balance of Inflammation from Tumor Protective to Tumor Promoting. Cancer Cell. 2012;22:479–493. doi: 10.1016/j.ccr.2012.08.016. [DOI] [PubMed] [Google Scholar]
- **35.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, Fatho M, Lennerz V, Wolfel T, Holzel M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–416. doi: 10.1038/nature11538. This study investigates the use of immunotherapy against a GEM melanoma model using adoptive cell transfer of T cells specific for a melanocyte differentiation antigen (gp100). The study shows that TNF production from the T cells mediates de-differentiation of tumor cells, which blocks the expression of differentiation antigens, such as gp100. In contrast, T cell responses against TSAs are maintained during this de-differentiation process. [DOI] [PubMed] [Google Scholar]
- 36.Sangaletti S, Tripodo C, Ratti C, Piconese S, Porcasi R, Salcedo R, Trinchieri G, Colombo MP, Chiodoni C. Oncogene-driven intrinsic inflammation induces leukocyte production of tumor necrosis factor that critically contributes to mammary carcinogenesis. Cancer Res. 2010;70:7764–7775. doi: 10.1158/0008-5472.CAN-10-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schioppa T, Moore R, Thompson RG, Rosser EC, Kulbe H, Nedospasov S, Mauri C, Coussens LM, Balkwill FR. B regulatory cells and the tumor-promoting actions of TNF-alpha during squamous carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:10662–10667. doi: 10.1073/pnas.1100994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. These studies [38,39] investigated GEM models of pancreatic ductal adenocarcinomas driven by oncogenic Kras and found that ras induced the expression of GM-CSF. This led to the recruitment of myeloid-derived suppressor cells that act to support tumor development by blocking anti-tumor CD8+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumm JB, Emmerich J, Zhang X, Chan I, Wu L, Mauze S, Blaisdell S, Basham B, Dai J, Grein J, et al. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell. 2012;20:781–796. doi: 10.1016/j.ccr.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Tanikawa T, Wilke CM, Kryczek I, Chen GY, Kao J, Nunez G, Zou W. Interleukin-10 ablation promotes tumor development, growth, and metastasis. Cancer Res. 2012;72 :420–429. doi: 10.1158/0008-5472.CAN-10-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer C, Sevko A, Ramacher M, Bazhin AV, Falk CS, Osen W, Borrello I, Kato M, Schadendorf D, Baniyash M, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc Natl Acad Sci U S A. 2011;108:17111–17116. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Balachandran VP, Cavnar MJ, Zeng S, Bamboat ZM, Ocuin LM, Obaid H, Sorenson EC, Popow R, Ariyan C, Rossi F, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med. 2011;17 :1094–1100. doi: 10.1038/nm.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NM, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dougan M, Li D, Neuberg D, Mihm M, Googe P, Wong KK, Dranoff G. A dual role for the immune response in a mouse model of inflammation-associated lung cancer. J Clin Invest. 2011;121:2436–2446. doi: 10.1172/JCI44796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim PS, Ahmed R. Features of responding T cells in cancer and chronic infection. Curr Opin Immunol. 2010;22:223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012;21 :402–417. doi: 10.1016/j.ccr.2012.01.008. This study utilized intravital imaging to monitor tumor antigen presentation by antigen presenting cells (APCs) at the tumor site in a GEM model of mammary cancer. The use of in vivo imaging techniques provides a valuable perspective for understanding how T cell responses to cancer are regulated and the authors show that the interactions of T cells with APCs in the tumor microenvironment promotes tolerance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donkor MK, Sarkar A, Savage PA, Franklin RA, Johnson LK, Jungbluth AA, Allison JP, Li MO. T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-beta1 cytokine. Immunity. 2011;35:123–134. doi: 10.1016/j.immuni.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran Thang NN, Derouazi M, Philippin G, Arcidiaco S, Di Berardino-Besson W, Masson F, Hoepner S, Riccadonna C, Burkhardt K, Guha A, et al. Immune infiltration of spontaneous mouse astrocytomas is dominated by immunosuppressive cells from early stages of tumor development. Cancer Res. 2010;70:4829–4839. doi: 10.1158/0008-5472.CAN-09-3074. [DOI] [PubMed] [Google Scholar]
- *50.Eyles J, Puaux AL, Wang X, Toh B, Prakash C, Hong M, Tan TG, Zheng L, Ong LC, Jin Y, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120:2030–2039. doi: 10.1172/JCI42002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12:237–251. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Galluzzi L, Senovilla L, Zitvogel L, Kroemer G. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov. 2012;11:215–233. doi: 10.1038/nrd3626. [DOI] [PubMed] [Google Scholar]
- 53.Le DT, Jaffee EM. Regulatory T-cell modulation using cyclophosphamide in vaccine approaches: a current perspective. Cancer Res. 2012;72:3439–3444. doi: 10.1158/0008-5472.CAN-11-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rech AJ, Mick R, Martin S, Recio A, Aqui NA, Powell DJ, Jr, Colligon TA, Trosko JA, Leinbach LI, Pletcher CH, et al. CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci Transl Med. 2012;4:134ra162. doi: 10.1126/scitranslmed.3003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gough MJ, Crittenden MR. Immune system plays an important role in the success and failure of conventional cancer therapy. Immunotherapy. 2012;4:125–128. doi: 10.2217/imt.11.157. [DOI] [PubMed] [Google Scholar]
- **56.Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. doi: 10.1016/j.ccr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **57.Anders K, Buschow C, Herrmann A, Milojkovic A, Loddenkemper C, Kammertoens T, Daniel P, Yu H, Charo J, Blankenstein T. Oncogene-targeting T cells reject large tumors while oncogene inactivation selects escape variants in mouse models of cancer. Cancer Cell. 2012;20:755–767. doi: 10.1016/j.ccr.2011.10.019. These studies [56,57] demonstrate that the efficacy of targeted therapies that inactivate the driving oncogene in cancers are dependent on intact T cell responses. T cells aid in tumor destruction by targeting the vasculature within the tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattarollo SR, Loi S, Duret H, Ma Y, Zitvogel L, Smyth MJ. Pivotal role of innate and adaptive immunity in anthracycline chemotherapy of established tumors. Cancer Res. 2011;71:4809–4820. doi: 10.1158/0008-5472.CAN-11-0753. [DOI] [PubMed] [Google Scholar]
- *59.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21 :488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]