Abstract
Purpose
Typically, apolipoproteins are individually measured in blood by immunoassay. In this report, we describe the development of a multiplexed selected reaction monitoring (SRM) based assay for a panel of apolipoproteins and its application to a clinical cohort of samples derived from acute stroke patients.
Experimental Design
An SRM assay for a panel of nine apolipoproteins was developed on a triple quadrupole mass spectrometer. Quantitative data for each apolipoprotein were analyzed to determine expression ratio and receiver operating characteristic (ROC) values for ischemic versus hemorrhagic stroke.
Results
The optimized SRM assay was used to interrogate a small cohort of well-characterized plasma samples obtained from patients with acute ischemic and hemorrhagic strokes. The ROC analyses demonstrated good classification power for several single apolipoproteins, most notably apoC-III and apoC-I. When a novel multi-marker ROC algorithm was applied, the ischemic versus hemorrhagic groups were best differentiated by a combination of apoC-III and apoA-I with an area under the curve (AUC) value of 0.92.
Conclusions and clinical relevance
This proof-of-concept study provides interesting and provocative data for distinguishing ischemic versus hemorrhage within first week of symptom onset. However, the observations are based on one cohort of patient samples and further confirmation will be required.
Keywords: Biomarker, Cerebrovascular, Hemorrhagic, Ischemic, Mass spectrometry
1 Introduction
A stroke is the rapidly developing loss of brain function due to disturbance of blood supply to the brain. Strokes can be classified into two major types: ischemic and hemorrhagic [1]. Ischemic strokes occur due to lack of blood flow arising from the formation of a clot or other obstruction, whereas hemorrhagic strokes are caused by rupture of a blood vessel or an abnormal vascular structure. The majority of all strokes are of ischemic (90%) origin [2–4].
Proper medical treatment of a stroke victim relies on accurate and rapid differentiation between ischemic and hemorrhagic stroke. Not only do ischemic and hemorrhagic stroke have completely divergent therapeutic options, the treatment itself can convert ischemic stroke to hemorrhage. For example, ischemic stroke can be treated by the administration of tissue plasminogen activator (tPA) or other thrombolytic agent designed to dissolve clots, thereby restoring blood flow to the brain. Potentially, not only will a hemorrhagic stroke patient die if given blood-thinning medications, after the administration of tPA and other clot-dissolving medication, ischemic stroke convert to lethal hemorrhage within the first week due to reperfusion injury [4–6]. Clinically, it is therefore crucial to monitor and distinguish ischemia versus hemorrhage within the first week of symptom onset to prevent adverse outcome. In current practice, diagnosis of hemorrhage versus ischemia is performed by computerized tomography (CT) or magnetic resonance imaging (MRI) scans. While imaging technology is sensitive, it is time consuming, difficult to implement in extremely sick and immobile patient, and also requires the availability of expensive equipment, highly skilled radiological expertise to interpret the results, and repeat radiation exposure. Against this background, there remains a need for a reliable, relatively inexpensive method for differentiating between ischemic and hemorrhagic stroke in patients—potentially a point-of-care (POC) assay that can be performed on a daily basis within the first week of stroke onset—to help triage a patient’s diagnostic and therapeutic algorithm.
Over the past several years, a body of literature has accumulated on the clinical usefulness of a biomarker-based diagnostic test for acute stroke [7,8]. To date, most biomarkers associated with stroke and proposed as diagnostics in the emergency room for acute stroke are blood-borne proteins of tissue injury such as CRP, MMP9, D-dimer, S100β, and B-type natriuric peptide [7,9]. There is also copious evidence that the relative levels of plasma lipoproteins can be significant risk factors for stroke [10–12]. Genetic polymorphisms of the apoA-I/apoC-III gene cluster may indicate increased risk for atherosclerotic brain infarctions [13]. Several years ago, apoC-I and apoC-III were identified as potential plasma markers to distinguish between ischemic and hemorrhagic stroke in a surface-enhanced laser desorption/ionization (SELDI) based study with confirmation by enzyme-linked immunosorbent assay (ELISA) [14]. To date, this is the only report of the potential application of apolipoprotein levels for diagnosis and/or classification of stroke.
Typically, apolipoproteins are individually measured in blood by immunoassay. The availability of a multiplexed assay that could simultaneously and quantitatively measure a panel of apolipoproteins would be an extremely useful clinical research tool [15]. Mass spectrometry-based selective reaction monitoring (SRM) is rapidly becoming a preferred technology for the development of quantitative protein or peptide assays for clinical research because it delivers high sensitivity, selectivity, throughput, and accurate quantification [16,17]. In this report, we describe the development of a multiplexed SRM-based assay for a comprehensive panel of apolipoproteins. We subsequently used this assay to interrogate a heterogeneous cohort of clinical samples obtained from stroke patients in an effort to discover whether apolipoprotein ratios could distinguish between normal (nonstroke), ischemic, and hemorrhagic stroke patient samples.
2 Materials and methods
2.1 Clinical serum samples
A total of 111 plasma samples were obtained from ischemic and hemorrhagic stroke patients, and healthy controls with similar risk factors from our tertiary medical center. Strokes are defined as an appropriate clinical syndrome, with MRI or CT findings consistent with ischemic or hemorrhagic origin, and without evidence of structural disease or co-morbidities that may bias analysis. Rigorous exclusion criteria were applied to ensure homogeneity within each group. Ischemic strokes are all arterial and embolic in etiology and those due to small artery disease (lacunes), vasculitis, endocarditis, and venous infarction were excluded. Hemorrhagic strokes due to trauma, coagulapathy, and structural disease (such as malignancy) were excluded. All patients with active infection, pregnancy, which may alter protein signaling profiles, and those with absence of baseline CT or MRI needed to confirm clinical stroke syndromes were excluded. Controls were recruited from subjects with similar baseline risk factors, especially lipid profiles to match the study population. Subject demographics (N = 54 ischemic stroke, N = 26 hemorrhagic, N = 31 control) are listed in Table 1. Patients were consecutively, prospectively enrolled post ischemic or hemorrhagic stroke in accordance with the approval of institutional IRB.
Table 1.
Patient clinical characteristics and sample collection
| Characteristic | Ischemic (N = 54) |
Hemorrhagic (N = 26) |
Normal (N = 31) |
|---|---|---|---|
| Age | 50 | 51 | 44 |
| (mean and range) |
(26–76) | (18–82) | (22–72) |
| Gender | 57% M | 50% | 48% |
| Race | 93% | 90% | 94% |
| Caucasian | Caucasian | Caucasian | |
| Hyperlipidemia | 38% | 34% | 30% |
| Hypertension | 38% | 35% | 30% |
| Diabetes | 15% | 12% | 10% |
| Time from onset of stroke symptom |
1–8 days | 1–7 days | N/A |
Venous plasma was obtained within about 1 week of stroke onset in EDTA-coated tubes, processed, aliquoted, and frozen within 30 min of collection at —80°C. Standard operating procedures were strictly observed for all samples. All personnel were trained with SOP to process samples in the same fashion as follows: blood was obtained from venous puncture and immediately processed (within 5 min) to obtain plasma by centrifugation at 3400 rpm for 15 min at 20°C (to avoid platelet activation), removing the plasma supernatant without disturbing the clot, aliquoted immediately, and frozen at −80°C within 30 min of venipuncture to ensure minimal protein degradation.
Frozen samples were carefully transported to be processed at the same time, in random order, by investigators blinded to the clinical data to avoid bias and batch variations.
2.2 Trypsin digestion, reduction/alkylation, and desalting
Serum samples (25 µL) were thawed on ice and processed as previously described [18]. Experiments using a concatenated, heavy labeled synthetic protein standard demonstrated consistent digestion with an average digestion efficiency of 64% [19]. Digested samples were desalted with HyperSep™− 96 C18 solid phase extraction media (Thermo Fisher Scientific, San Jose, CA, USA).
2.3 High resolution LC-MS/MS
High-resolution LC-MS/MS analysis was carried out on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) as previously described [18]. All samples were analyzed by high-resolution LC-MS/MS before SRM assay development.
2.4 LC-MS/MS data analysis and protein identification
LC-MS/MS data analysis, protein identification, and peak-list generation were performed using the SIEVE v.1.2.1 (Thermo Fisher Scientific) algorithm [20] incorporating the SEQUEST® v.28 search engine and Percolator™ [21], as previously described [18]. Individual MS/MS spectra with ion assignments for single-peptide identifications are provided in File 1 in Supporting Information.
2.5 SRM assays
SRM assays were developed on a TSQ Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific), as previously described [17,18]. There were three technical replicates per sample, i.e. each sample was digested once and analyzed three times in the SRM assay.
2.5.1 Calibration curve generation
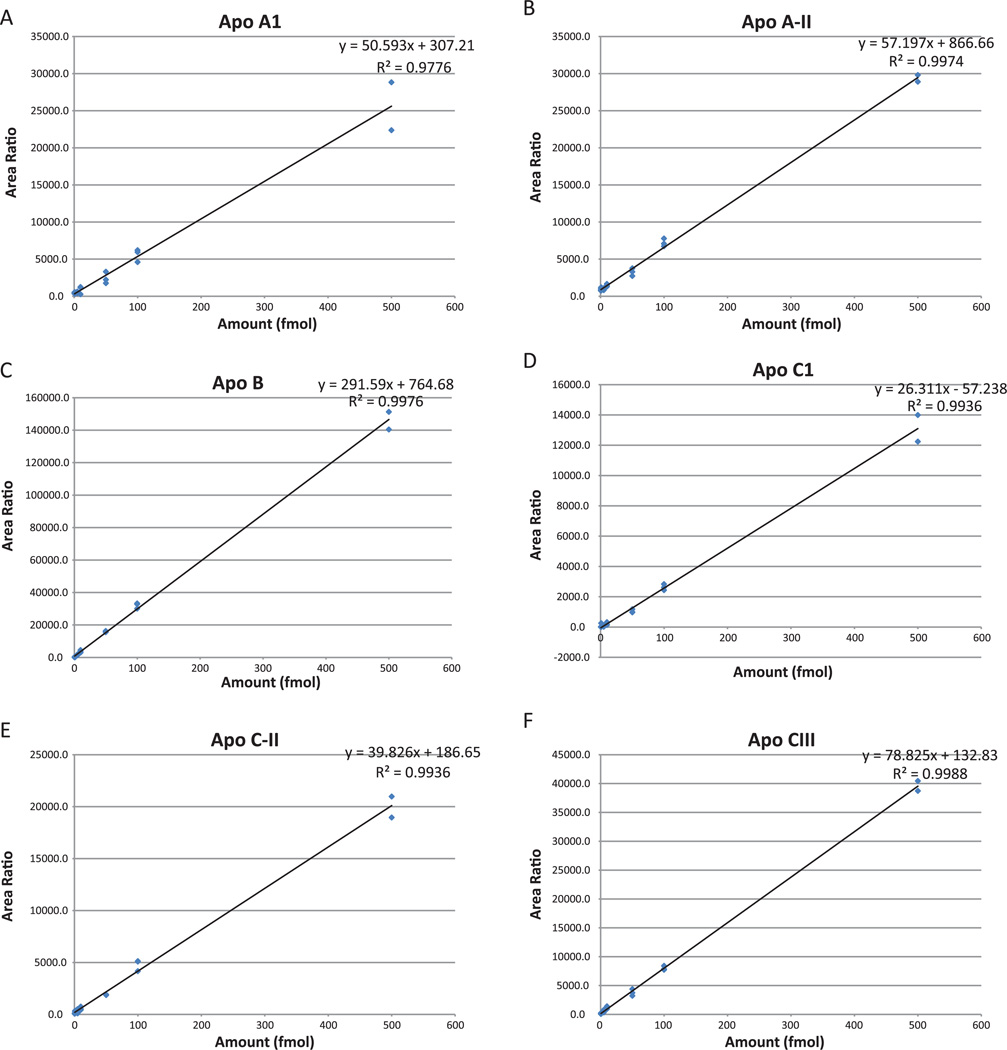
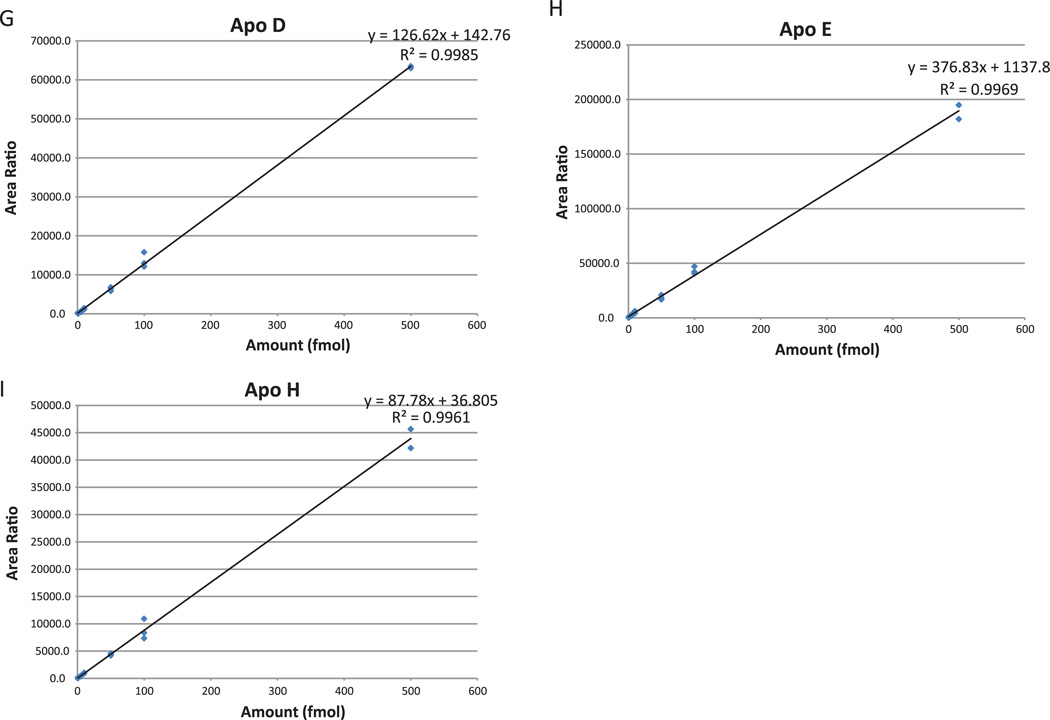
Seven-point calibration curves (0.5 fmol, 1 fmol, 5 fmol, 10 fmol, 50 fmol, 100 fmol, 500 fmol, Fig. 2A–I) were created with a pool (30 µg) of all samples as a background matrix. Since we did not know the biological variability and therefore the range of endogenous amounts of apolipoproteins in the serum samples, we used a broad range for the calibration curve. Each point on the calibration curve (and every sample analyzed) included 100 fmol of heavy labeled peptides. We used reverse curves to generate calibration curves, as we did not have matrix free of the endogenous analyte. Full details of this type of curve have been presented previously [22]. In addition, all samples were dissolved in a solution of 200 µg/mL of glucagon in 97% water, 3% ACN, and 0.2% formic acid to minimize binding to plastic surfaces [17]. Finally, the calibration curve replicate points were run adjacently. Since each calibration point contained the same amount of light peptide (endogenous), variance in the heavy peptide (or endogenous peptide) amounts was used to calculate the run-to-run variance. Also, heavy peptides were spiked into each clinical sample and the low run-to-run variance added confidence to the analysis.
Figure 2.
Pinpoint-generated calibration curves for apolipoprotein heavy peptides in a background of 30 µg raw serum digest. The 8-point curves measured concentrations from 250 attomoles to 500 femtomoles on column, in triplicate. LLODs were estimated at 250–500 attomoles and LOQs were calculated between 1 and 5 femtomoles for each peptide. The linear correlation coefficients ranged from 0.93 to 0.99 and the CVs for points above the LOQ ranged from 0 to 20%. (A) apoA-I; (B) apoA-II; (C) apoB; (D) apoC-I; (E) apoC-II; (F) apoC-III; (G) apoD; (H) apoE; (I) apoH.
2.5.2 Choice of proteins, peptides, and transitions
Pinpoint software (Thermo Fisher Scientific) was used for targeted protein quantification [17,18]. Our initial choice of proteins was based on two criteria:
A review of the literature suggested apolipoproteins that were likely to be involved in cardiovascular disease; this is discussed in the Introduction and Discussion section.
Examination of high-resolution LC-MS/MS discovery data indicated which apolipoprotein peptides were clearly detected and provided robust signal in samples that had not been subjected to any complex sample preparation such as albumin depletion or fractionation. This was crucial in order to provide the robustness and rigorous quantification needed in a clinical research assay. For the workflow described in this report, the peptides identified in the LC-MS/MS discovery MS data were imported directly into Pinpoint and transitions were chosen based upon the predominant fragments observed in the discovery data (>5 transitions per peptide). We subsequently chose one proteotypic peptide and several transitions per target protein in order to simplify the assay (Table 2 and Supporting Information Table S1). As described in a previous publication [18], the decision to limit the number of peptides from each protein to one was based on two factors: (a) multiple peptides might quantify different isoforms of the same protein thereby producing conflicting results and our goal was to monitor the most common isoform (and hence the most abundant peptide) and (b) the choice of a nonredundant peptide sequence and its characterization using a synthetic standard with multiple, co-eluting transitions ensured the verification of peptide identity (eliminating interferences) and thus its validity as a surrogate for the target protein. Peptides were identified by co-eluting light and heavy labeled transitions in the chromatographic separation. For additional verification and elimination of interferences, the transition ratios were confirmed using discovery spectra. Time alignment and relative quantification of the transitions were performed with Pinpoint. All clinical samples were assayed in triplicate. The raw quantitative data for all sample measurements are given in Supporting Information Table S2.
Table 2.
Apolipoproteins and targeted peptide sequences
| Protein ID | Peptide sequence | AA |
|---|---|---|
| Apolipoprotein A-I | LLDNWDSVTSTFSK | 70–83 |
| Apolipoprotein A-II | SKEQLTPLIK | 68–77 |
| Apolipoprotein B | TGISPLALIK | 220–229 |
| Apolipoprotein C-I | LKEFGNTLEDK | 37–47 |
| Apolipoprotein C-II | ESLSSYWESAK | 42–52 |
| Apolipoprotein C-III | GWVTDGFSSLK | 61–71 |
| Apolipoprotein D | NILTSNNIDVK | 143–153 |
| Apolipoprotein E | FWDYLR | 51–56 |
| Apolipoprotein H | ATVVYQGER | 252–260 |
2.5.3 t-SRM (scheduling of SRMs)
Transitions were scheduled based on the retention time observed in the discovery experiments. Over the course of multiple iterative serum runs, the retention times varied by less than 10 s or < 2% of the gradient length.
2.5.4 Light and heavy labeled peptides
Light and heavy versions (97% purity) of each target peptide were synthesized (Thermo Fisher Scientific). Heavy peptides had identical sequences to the light peptides, but the C-terminal Lysine or Arginine was fully labeled (>98.5%) with 13C or 15N (16), see Supporting Information Table S1 for peptide sequences.
2.6 Receiver operating characteristic (ROC) analysis
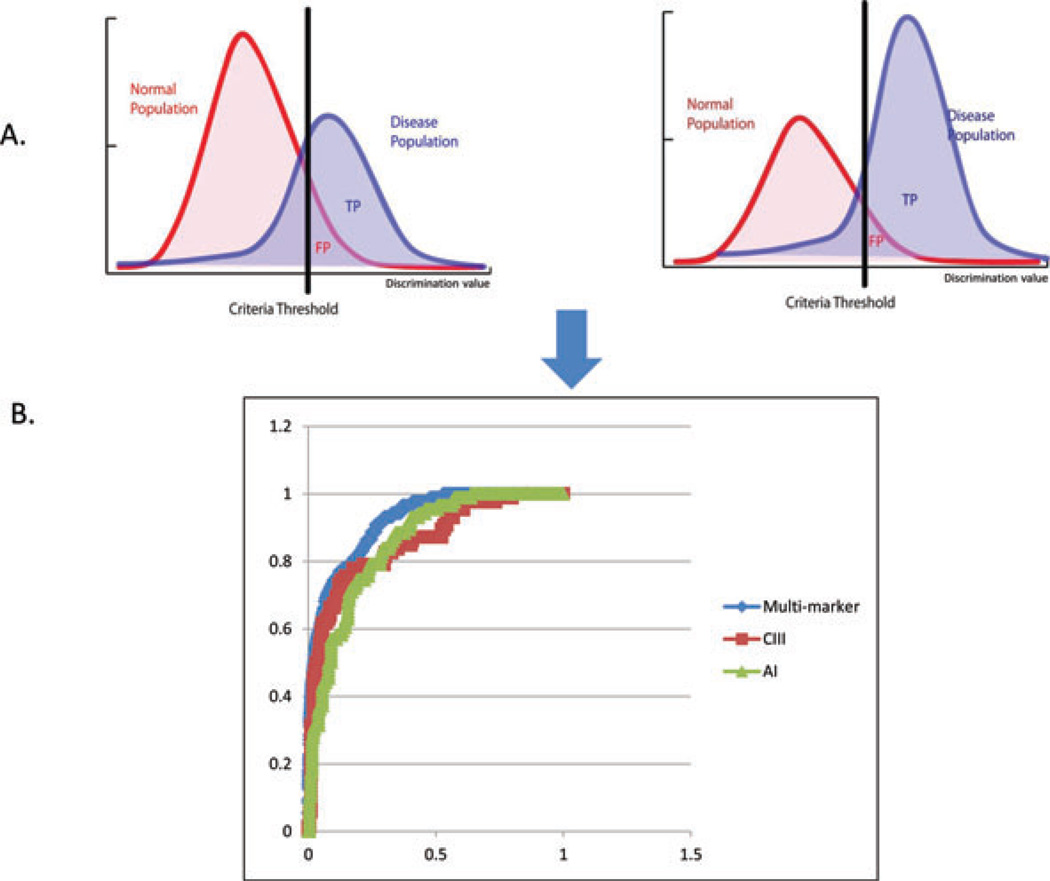
Traditional ROC analysis is a useful technique for quantifying the capacity of a single marker to discriminate between two classes of measurements by plotting the true-positive (TP) rate (sensitivity) versus the false-positive (FP) rate (1 specificity). However, in a biological system there may be multiple marker candidates that work in tandem (with their own individual discriminating capability), therefore, the overall discriminating capability may be improved if multiple markers (instead of a single marker) are used. To address this, we developed a novel method where the TP and FP are derived by the optimal combined probability for each marker threshold. The combined probabilities (TP and FP) are calculated for all possible thresholds for each marker within a set of markers. The maximum TP for each FP is used to build the optimized multi-marker ROC curve (Fig. 3). Although the multi-marker algorithm is theoretically applicable to an indefinite number of markers, we found that there was no significant improvement when we combined more than two markers in this dataset. The multi-marker algorithm is freely downloadable from the URL http://www.vastsci.com/rocstation/. File 2 in Supporting Information is an example and tutorial using data from this study.
Figure 3.
(A) Graphical representation of the multi-marker ROC algorithm. The different thresholds are varied for the two different marker populations. The highest TP rate for a given FP rate generates the final ROC curve. (B) Overlaid ROC plots of the single markers and marker pair with the highest AUC for differentiating ischemic from hemorrhagic samples (see Table 3).
The overall discriminating power of the marker can be summarized by a single number corresponding to the integrated area under the curve (AUC). AUC can range from 1 (maximum discriminating power) to 0.5 (ambiguous). To address uncertainty in ROC curves and AUC values, measurement uncertainties are propagated through the ROC calculation where extreme values of the ROC curve ares considered.
ROC curves are typically used to perform cross-validation where there is a learning algorithm involved. For example, when performing a fivefold cross-validation it is typical to compute a separate ROC curve for each of the fivefolds using the hypothesis and then build a mean ROC curve to validate the algorithm. In this case, we have skipped this step, and used the ROC curves to automatically validate the usefulness of the individual markers themselves. The area-under-the-ROC curve has been used extensively to understand sensitivity and specificity of learning algorithms and is statistically robust because it can take biological variation into account.
2.7 Differential expression analysis
For a particular protein, group differential expression ratio (e.g. between normal and ischemic) was calculated within Pinpoint by the following formula:
Group differential expression ratios can only be calculated for single markers (not multiple markers).
3 Results
3.1 SRM assays
3.1.1 Assay development
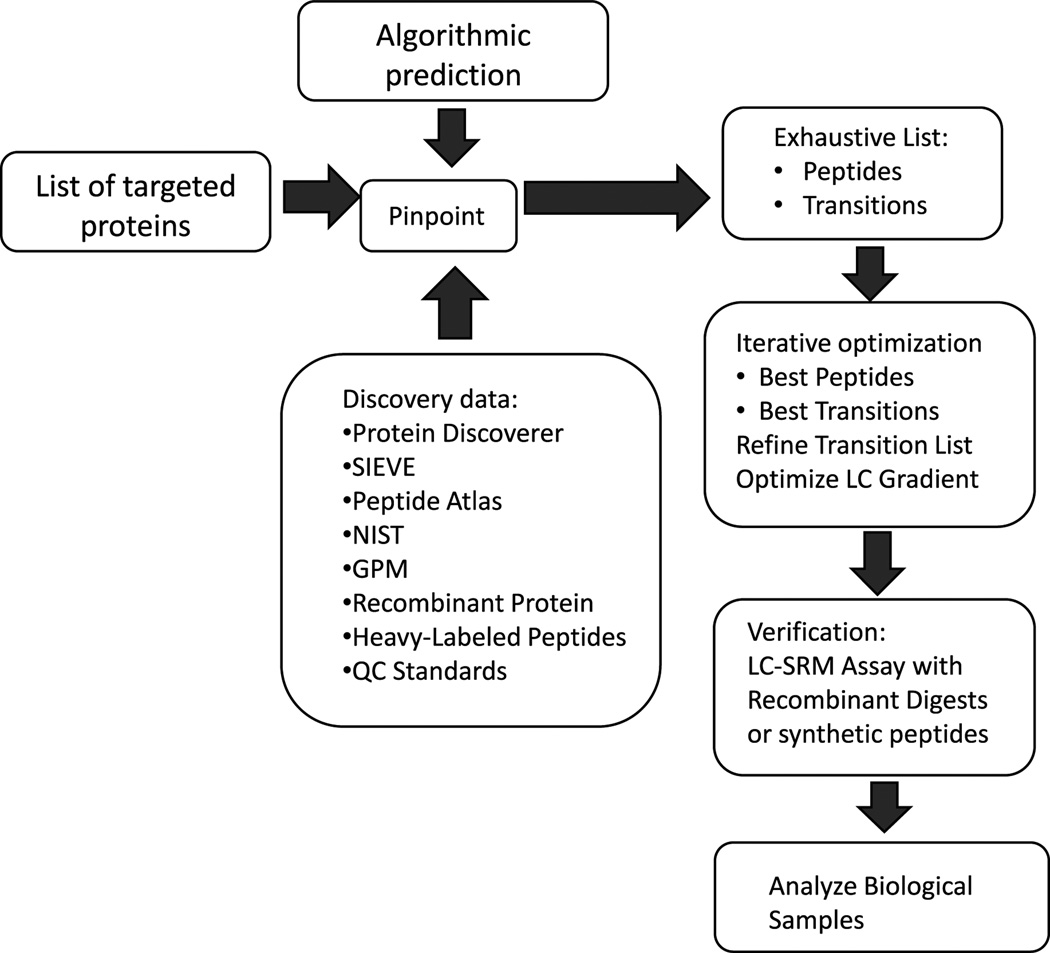
Figure 1 illustrates the workflow we applied for the development of the multiplexed apolipoprotein SRM assay. Incorporation of previously obtained high-resolution LC-MS/MS data (Data 1 in Supporting Information) combined with algorithmic prediction (Pinpoint) facilitated the creation of an initial list of peptides and transitions for targeting. Because the apolipoproteins we were targeting (Table 1) are highly abundant in serum, we bypassed optimization using recombinant proteins and instead tested the method directly on control serum samples. After iterative optimization, the best single (highest intensity and least interference) peptide and several fragment transitions were chosen for each protein. As discussed in the methods and in previous publications [18], we chose to measure only one peptide per protein in order to simplify the multiplexed assay and its subsequent analysis (Table 1). In this case, nonredundant peptide sequences were chosen and characterized with synthetic standards. Multiple co-eluting transitions ensured peptide identity and eliminated interferences, thus confirming the target peptide as a valid surrogate for the target protein.
Figure 1.
Workflow for automated development and optimization of SRM assays.
The complete list of targeted proteins, peptides, and 85 transitions is given in Table 1 and Supporting Information Table S1. Each LC iteration was approximately 1 h in duration and the entire assay optimization process was completed within 1 day. Corresponding heavy labeled peptides were then synthesized and used for final LC optimization and as internal quantitative standards.
3.1.2 Assay sensitivity and precision
Eight point calibration curves using heavy labeled peptides in a background of serum matrix were constructed to test assay sensitivity and precision (Fig. 2). The lower limit of detection (LLOD) for all nine peptides ranged from 250 to 500 attomoles on column and the limit of quantitation (LOQ) ranged from 1 to 5 femtomoles on column. All peptides demonstrated linear behavior (correlation coefficients ranged from 0.93 to 0.99) in the range between 250 attomoles and 500 femtomoles on column and the coefficients of variation (CVs) for points above the LOQ ranged from 0 to 20%.
3.2 ROC analyses and differential expression ratios
The optimized SRM assay was used to interrogate the clinical samples. Quantitative data were obtained for all samples and then analyzed in Pinpoint to determine differential expression group ratios and classification power (determined by ROC analyses) associated with the different apolipoprotein peptides. The raw quantitative data for all sample measurements are given in Supporting Information Table S2. In this study, we used descriptive statistics throughout for error estimates. ROC calculations were performed with proper propagation of measurement error through final results. The dataset was not large enough for more classical modeling analysis such as logistic regression.
Three different groups were created (Table 3): (i) is-chemic versus hemorrhagic, (ii) ischemic versus normal, and (iii) hemorrhagic versus normal. ROC curves with AUC close to 1 have high selectivity and sensitivity; whereas, curves with areas close to 0.5 correspond to cases where the putative marker effectively cannot distinguish the two classes. The ROC analyses (Fig. 3, Table 3) demonstrated good classification power for several single apolipoproteins, most notably apoC-III and apoC-I. ApoC-III differentiated ischemic from hemorrhagic and ischemic from normal stroke samples with an AUC of 0.85 and 0.91, respectively, and a differential expression ratio of 0.5 for both (see Supporting Information Table S3 for complete ROC classification data). The hemorrhagic versus normal groups were best (single marker) differentiated by apoC-I, with an AUC of 0.95 and a differential expression ratio of 2.5. When the multiple marker algorithm was applied to these group analyses, the AUC values were increased. The ischemic versus hemorrhagic groups were differentiated best by a combination of apoC-III and apoA-I with an AUC value of 0.92. The ischemic and normal groups were best differentiated by multiple markers apoC-III and apoC-I with AUC value increased to 0.93. And finally, the multiple markers that best differentiated the hemorrhagic versus normal groups were apoC-I and apoA-II with a resulting AUC value of 0.98. Differential expression group ratios cannot be calculated for multiple markers.
Table 3.
ROC AUC values for the top discriminating markers
| Group comparison | Top marker(s) | AUC | Differential expression group ratioa) |
t-test p-value |
|---|---|---|---|---|
| Ischemic hemorrhagic | ||||
| Single | ApoC-III | 0.85 +0.077 −0.154 | 0.5 ± 0.32 | 1.56 × 10−6 |
| Multiple | ApoC-III, ApoA-I | 0.92 +0.058 −0.182 | NA | |
| Ischemic versus normal | ||||
| Single | ApoC-III | 0.91 +0.078 −0.401 | 0.5 ± 0.29 | 4.78 × 10−7 |
| Multiple | ApoC-III, ApoC-I | 0.93 +0.057 −0.199 | NA | |
| Hemorrhagic versus normal | ||||
| Single | ApoC- I | 0.95 +0.043 −0.379 | 2.5 ± 1.82 | 4.46 × 10−9 |
| Multiple | ApoC-I, ApoA-II | 0.98 +0.022 −0.376 | NA | |
Differential expression ratios cannot be calculated for multiple markers.
4 Discussion
The goals of this study were twofold: (i) To test the hypothesis that the levels of blood apolipoproteins can potentially be useful for practical discrimination of ischemic and hemorrhagic strokes within 1 week of stroke onset when the risk of hemorrhagic transformation causing adverse outcome is highest and (ii) To apply single and novel multi-marker ROC algorithms early in the discovery process to speed the evaluation of putative biomarker data acquired with targeted SRM assays.
4.1 Apolipoproteins as putative biomarker candidates for cerebrovascular disease
The relative levels of various apoliproteins have been identified as important in a variety of cardiovascular and cerebrovascular pathologies including heart disease, stroke, Alzheimer’s disease, diabetes, and metabolic syndrome [11–13, 15, 23–28]. Several studies suggest that apolipoproteins are better markers than traditional serum lipid measurements for prediction of cardiovascular disease risk in the general population [26–28]. In addition, most patients have multiple risk factors that interact and potentiate disease phenotypes, for example heart disease, cerebrovascular disease, and obesity are risk factors for each other and influence each other greatly. Therefore, an assay measuring a panel of apolipoproteins could be potentially useful for individualized risk assessment, monitoring therapeutic efficacy of medical therapy, or detection of acute conditions. Although apolipoproteins are typically measured (indirectly) by conventional immunoassays, they can suffer from serious limitations including interfering antibodies, poor interplatform concordance, cross-reactivity, and high-dose hook effects [26]. We were interested in the application of highly quantitative LC-MS/MS for the direct measurement of apolipoproteins in blood to discover if they had potential application for the differentiation of acute stroke subtypes.
4.2 ROC curves coupled with multiplexed SRM assays to expedite proteomic discovery?
Typical mass spectrometry-based proteomic discovery experiments generate long lists of putative biomarkers for diseases and pathologies. Verification of these markers in multiplexed assays poses a statistical challenge because relative fold-change or expression ratios are typically used as discriminates despite the fact that the actual ratio or fold-change does not necessarily relate to the ability of the marker to distinguish normal from disease cases. Expression ratios are calculated as the mean across different groups and can be highly skewed by outliers. In addition, ratios cannot be calculated for panels of markers.
ROC curves are commonly used in clinical study reports as a concise visual representation describing the outcome of a specific test measurement on a patient population. Traditional ROC curves used to calculate the sensitivity and specificity of a diagnostic or predictive assay are based on single markers. The ability to combine quantitative information from several markers could potentially improve the accuracy of existing tests and facilitate the development of new tests. However, panels of markers are not commonly used in part because multi-marker ROC curves are computationally very expensive. In this report, we describe a novel algorithm for efficient computation of multi-marker ROC curves and apply it to multiplexed SRM analysis in order to rapidly identify significant markers for differentiating types of strokes. Application of this algorithm to quickly evaluate the usefulness of a list of putative biomarkers in a targeted discovery approach can significantly expedite the search and verification process. In the current study, the entire process of assay development, sample testing, and data analysis was completed in less than a week as opposed to traditional shotgun proteomics discovery experiments that can take months to complete. The analysis of proteomic data with this approach allows a fast, objective, and quantitative method to rank the “usefulness” of a putative biomarker.
4.3 Practical considerations
The multiplexed and quantitative SRM assay described herein can simultaneously monitor nine different apolipoproteins, does not require complicated sample preparation and is applicable to standard LC fast-flow configurations coupled to triple quadrupole mass spectrometers typically found in many clinical research laboratories [17]. High-abundance proteins present in biological samples at concentrations of 1 µg/mL or higher have been successfully assayed previously by SRM in complex matrices such as tissues and plasma [29–31]. However, the published methods require sample preparation such as albumin depletion and nanoflow LC, difficult to deploy in a clinical setting due to low throughput, lack of robustness, need for operator expertise, and economic considerations.
The data generated using this assay and communicated in this report present preliminary evidence (based on a single clinical cohort) that the plasma levels of specific apolipoproteins, apoC-III, apoC-I, apoAI, and apoA-II clearly discriminated (with high sensitivity and specificity) ischemic, hemorrhagic, and normal patient sample groups from each other. These conclusions are based on two different types of measurements, differential expression ratios and single and multi-marker ROC analyses. The differential expression data demonstrated significant underexpression of apoC-III in ischemic stroke samples versus normal and hemorrhagic stroke patient samples. Conversely, the hemorrhagic stroke samples, (including subarachnoid and intraventricular), demonstrated an overexpression of apoC-I versus normal or ischemic patient samples. These data are interesting and provocative but the observations are again based on one cohort of patient samples and further confirmation will require blinded studies with many additional patient samples. It is important to also note that a copious body of literature on apoC-III and its potential as a cardiovascular risk factor is rapidly accumulating [32–39].
4.4 Limitations and future opportunities
This is a small proof-of-concept study where we attempt to match patients with similar risk factors to limit confounders, but larger studies are imperative to further investigate these findings. We designed this study to find markers from the most important time window to monitor ischemia versus hemorrhage as the peak time to distinguish tPA-related and other anticoagulant-related hemorrhage are within the first week from symptom onset. During this time, patient with ischemic stroke given thrombolytics or anticoagulants often has to undergo repeat CT to rule out hemorrhage. While we obtained good results when testing the panel’s ability to discriminate (in order to reduce repeat exposure with CT monitoring), during the first week, we do not have the power to distinguish these changes within the first few hours which may have implication for initial tPA administration and this certainly warrant future study. However, our panel seems to be stable over the first week and is not affected by fasting state, and therefore may be robust enough to discriminate stroke subtype both in the acute and chronic period. Further studies with blood from earlier and more chronic time window will be one of our important future goals to test the stability of this panel. The importance of protein micro-heterogeneity has increasingly become a focus in proteomics [40,41]. Future next-generation assays for protein analytes will certainly require rational design incorporating clinically important variants. Recently, top-down analysis of apolipoproteins by mass spectrometry revealed quantitative differences in glycosylated isoforms of apoC-III isolated from human high density lipoprotein (HDL) 3 [42]. Fortunately, intelligent design of SRM assays allows the incorporation of variant-specific peptides and provides the opportunity to update and refine existing assays as new molecular information is uncovered. Future plans in the author’s laboratory include the characterization of apoC-I and apoC-III clinical variants and their incorporation into functional SRM assays.
Supplementary Material
Clinical Relevance.
Medical treatment of stroke victims currently relies on the application of imaging technology to accurately and rapidly differentiate between ischemic and hemorrhagic stroke. Not only do ischemic and hemorrhagic stroke have completely divergent therapeutic options, the treatment itself can convert ischemic stroke to hemorrhage. For example, ischemic stroke can be treated by the administration of tissue plasminogen activator (tPA) or other thrombolytic agent designed to dissolve clots, thereby restoring blood flow to the brain. Potentially, not only will a hemorrhagic stroke patient die if given blood-thinning medications, after the administration of tPA and other clot-dissolving medication, ischemic stroke can convert to lethal hemorrhage within the first week due to reperfusion injury [6]. Clinically, it is therefore crucial to monitor and distinguish ischemia versus hemorrhage within the first week of symptom onset to prevent adverse outcome. Imaging technology is highly sensitive but not only requires the availability of expensive equipment and skilled radiological expertise to interpret the results, but also has adverse effects of radiation exposure. There is a need for a rapid, reliable, and relatively inexpensive method for differentiating between ischemic and hemorrhagic stroke to help triage a patient’s diagnostic and therapeutic algorithm. This report presents preliminary evidence that the plasma levels of specific apolipoproteins, apoC-III, apoC-I, apoAI, and apoA-II measured by a multiplexed, mass spectrometric selective reaction monitoring (SRM) assay discriminated, with high sensitivity and specificity, ischemic, hemorrhagic, and normal patient acute stroke samples from each other. Apolipoproteins are typically measured by immunoassays but these suffer from serious limitations including interfering antibodies, poor interplatform concordance, cross-reactivity, and high-dose hook effects. The multiplexed, SRM-based assay described herein may have clinical application for classification of stroke subtype.
Abbreviations
- apo
apolipoprotein
- CT
computerized tomography
- FP
false positive
- FPR
false-positive rate
- LLOD
lower limit of detection
- MRI
magnetic resonance imaging
- ROC
receiver operating characteristic
- SRM
selective reaction monitoring
- TP
true positive
- tPA
tissue plasminogen activator
Footnotes
Colour Online: See the article online to view Fig. 2 in colour.
References
- 1.Fisher M. The challenge of mixed cerebrovascular disease. Ann. N.Y. Acad. Sci. 2010;207:18–22. doi: 10.1111/j.1749-6632.2010.05758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elizabeth G, Nabel MD. Incidence and Prevalence: 2006 Chart Book on Cardiovascular and Lung Diseases. Bethesda, MD: National Heart, Lung, and Blood Institute; 2006. [Google Scholar]
- 3.B. Kase CS, Caplan LR. Intracerebral Hemorrhage. Boston: Butterworth-Heinemann; 1996. [Google Scholar]
- 4.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, et al. Executive summary: heart disease and stroke statistics–2010 update: a report from the American Heart Association. Circulation. 2010;121:948–954. doi: 10.1161/CIRCULATIONAHA.109.192666. [DOI] [PubMed] [Google Scholar]
- 5.Study Group. Tissue plasminogen activator for acute is-chemic stroke. The National Institute of Neurological Disorders and Stroke rt-PA Stroke Study Group. N. Engl. J. Med. 1995;333:1581–1587. doi: 10.1056/NEJM199512143332401. [DOI] [PubMed] [Google Scholar]
- 6.Ning MM, Wang X, Lo EH. Reperfusion injury after stroke: neurovascular proteases and the blood-brain barrier. Handb. Clin. Neurol. 2008;92:117–136. doi: 10.1016/S0072-9752(08)01906-4. [DOI] [PubMed] [Google Scholar]
- 7.Laskowitz DT, Kasner SE, Saver J, Remmel KS, et al. Clinical usefulness of a biomarker-based diagnostic test for acute stroke: the biomarker rapid assessment in ischemic injury (BRAIN) study. Stroke. 2009;40:77–85. doi: 10.1161/STROKEAHA.108.516377. [DOI] [PubMed] [Google Scholar]
- 8.Glickman SW, Phillips S, Anstrom KJ, Laskowitz DT, et al. Discriminative capacity of biomarkers for acute stroke in the emergency department. J. Emerg. Med. 2010;41:333–339. doi: 10.1016/j.jemermed.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Foerch C, Montaner J, Furie KL, Ning MM, et al. Searching for oracles? Blood biomarkers in acute stroke. Neurology. 2009;73:393–399. doi: 10.1212/WNL.0b013e3181b05ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benn M. Apolipoprotein B levels, apoB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis. 2009;206:17–30. doi: 10.1016/j.atherosclerosis.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Davidsson P, Hulthe J, Fagerberg B, Carmejo G. Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2010;302:156–63. doi: 10.1161/ATVBAHA.108.179317. [DOI] [PubMed] [Google Scholar]
- 12.Milan J, Pinto X, Munoz A, Zuniga M, et al. Lipoprotein ratios; physiological significance and clinical usefulness in cardiovascular prevention. Vasc. Health Risk Manag. 2009;5:757–65. [PMC free article] [PubMed] [Google Scholar]
- 13.Groenendijk M, Cantor RM, de Bruin TW, Dallinga-Thie GM. The apoAI-CIII-AIV gene cluster. Atherosclerosis. 2001;157:1–11. doi: 10.1016/s0021-9150(01)00539-1. [DOI] [PubMed] [Google Scholar]
- 14.Allard L, Lescuyer P, Burgess J, Leung KY, et al. ApoC-I and ApoC-III as potential plasmatic markers to distinguish between ischemic and hemorrhagic stroke. Proteomics. 2004;4:2242–2251. doi: 10.1002/pmic.200300809. [DOI] [PubMed] [Google Scholar]
- 15.Davidson MH. Apolipoprotein measurements; is more widespread use clinically indicated? Clin. Cardiol. 2009;32:482–486. doi: 10.1002/clc.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008;222:1–14. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash A, Rezai T, Krastins B, Sarracino D, et al. Platform for establishing interlaboratory reproducibility of selected reaction monitoring-based mass spectrometry pep-tide assays. J. Proteome. Res. 2010;9:6678–6688. doi: 10.1021/pr100821m. [DOI] [PubMed] [Google Scholar]
- 18.Lopez MF, Kuppusamy R, Sarracino D, Prakash A, et al. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester trisomy 21 maternal serum. J. Proteome. Res. 2011;10:133–142. doi: 10.1021/pr100153j. [DOI] [PubMed] [Google Scholar]
- 19.Krastins B, Prakash A, Peterman S, Sarracino D, et al. ASMS Poster Presentation. Denver, CO: 2010. Development of a synthetic protein quality control (QC) standard for the assessment of sample proteolysis reproducibility. [Google Scholar]
- 20.Sutton J, Richmond T, Shi X, Athanas M, et al. Performance characteristics of an FT MS-based workflow for label-free differential MS analysis of human plasma: standards, reproducibility, targeted feature investigation, and application to a model of controlled myocardial infarction. Proteomics Clin. Appl. 2008;2:862–881. doi: 10.1002/prca.200780057. [DOI] [PubMed] [Google Scholar]
- 21.Ka¨ll L, Canterbury J, Weston J, Stafford Noble W, et al. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 22.Campbell J, Rezai T, Prakash A, Krastins B, et al. Evaluation of absolute peptide quantitation strategies using selected reaction monitoring. Proteomics. 2011;11:1148–1152. doi: 10.1002/pmic.201000511. [DOI] [PubMed] [Google Scholar]
- 23.Mihic M. Modi, P. Metabolic syndrome-risk factors for atherosclerosis and diabetes. Curr. Diabetes Rev. 2008;4:122–128. doi: 10.2174/157339908784220750. [DOI] [PubMed] [Google Scholar]
- 24.Duron E, Hanon O. Vascular risk factors, cognitive decline, and dementia. Vasc. Health Risk Manag. 2008;4:363–381. doi: 10.2147/vhrm.s1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sniderman AD, Faraj M. Apolipoprotein B, apolipoprotein A-I, insulin resistance and the metabolic syndrome. Curr. Opin. Lipidol. 2007;6:633–637. doi: 10.1097/MOL.0b013e3282f0dd33. [DOI] [PubMed] [Google Scholar]
- 26.Agger SA, Marney LC, Hoofnagle AN. Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography-multiple-reaction-monitoring mass spectrometry. Clin. Chem. 2010;56:1804–1813. doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holme I, Aastveit AH, Hammar N, Jungner I, et al. Lipoprotein components and risk of congestive heart failure in 84,740 men and women in the Apolipoprotein MOrtality RISk study (AMORIS) Eur. J. Heart Fail. 2009;11:1036–1042. doi: 10.1093/eurjhf/hfp129. [DOI] [PubMed] [Google Scholar]
- 28.Lamarche B, Moorjani S, Lupien PJ, Cantin B, et al. Apolipoprotein A-I and B levels and the risk of ischemic heart disease during a five-year follow-up of men in the Quebec cardiovascular study. Circulation. 1996;94:273–278. doi: 10.1161/01.cir.94.3.273. [DOI] [PubMed] [Google Scholar]
- 29.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Addona TA, Abbatiello SE, Schilling B, Skates SJ, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring based measurements of proteins in plasma. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzyk MA, Smith D, Yang J, Cross TJ, et al. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooi EM, Barrett PH, Can DC, Watta GF. Apolipoprotein C-III: understanding an emerging cardiovascular risk factor. Clin. Sci. (Lond) 2008;114:611–624. doi: 10.1042/CS20070308. [DOI] [PubMed] [Google Scholar]
- 33.Chan DC, Chen MM, Ooi EM, Watts GF. An ABC of apolipoprotein C-III; a clinically useful new cardiovascular risk factor? Int. J. Clin. Pract. 2008;62:799–809. doi: 10.1111/j.1742-1241.2007.01678.x. [DOI] [PubMed] [Google Scholar]
- 34.Stankovik S, Majkic-Singh N. Genetic aspects of is-chemic stroke: coagulation, homocysteine, and lipoprotein metabolism as potential risk factors. Crit. Rev. Clin. Lab. Sci. 2010;47:72–123. doi: 10.3109/10408361003791520. [DOI] [PubMed] [Google Scholar]
- 35.Kawakami A, Yoshida M. Apolipoprotein CIII links dyslipi-demia with atherosclerosis. J. Atheroscler. Thromb. 2009;16:6–11. doi: 10.5551/jat.e607. [DOI] [PubMed] [Google Scholar]
- 36.Van Dijk KW, Rensen PC, Voshol PJ, Havekes LM. The role and mode of actionofapolipoproteins CIII and AV: synergistic actors in triglyceride metabolism? Curr. Opin. Lipidol. 2004;15:239–246. doi: 10.1097/00041433-200406000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Shachter NS. Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Curr. Opin. Lipodol. 2001;12:297–304. doi: 10.1097/00041433-200106000-00009. [DOI] [PubMed] [Google Scholar]
- 38.Vaisar T, Mayer P, Nilsson E, Zhao XQ, et al. HDL in humans with cardiovascular disease exhibits a proteomic signature. Clin. Chim. Acta. 2010;411:972–979. doi: 10.1016/j.cca.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng C, Khoo C, Furtado J, Sacks FM. Apolipoprotein c-III and the metabolic basis for hypertriglyceridemia and the dense low-density lipoprotein phenotype. Circulation. 2010;121:1722–1734. doi: 10.1161/CIRCULATIONAHA.109.875807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borges CR, Rehder DS, Jarvis JW, Schaab MR, et al. Full-length characterization of proteins in human populations. Clin. Chem. 2010;56:202–211. doi: 10.1373/clinchem.2009.134858. [DOI] [PubMed] [Google Scholar]
- 41.Lopez MF, Rezai T, Sarracino DA, Prakash A, et al. Selected reaction monitoring-mass spectrometric immunoas-say responsive to parathyroid hormone and related variants. Clin. Chem. 2010;56:281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 42.Mazur MT, Cardasis HL, Spellman DS, Liaw A, et al. Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. USA. 2010;107:7728–7733. doi: 10.1073/pnas.0910776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.