Abstract
Among current in vitro methods for identification of pathogenic Listeria monocytogenes (L. monocytogenes) rely on growth in culture media, followed by isolation, and biochemical and serological identification. Now PCR (Polymerase Chain Reaction) has been used for the rapid, sensitive and specific detection of pathogenic L. monocytogenes. The pathogenicity of the organism is highly correlated with haemolytic factor known as listeriolysin O (LLO). A total of 400 samples from meat and 250 samples from raw milk and their products were collected from various local dairy farms, dairy units and butcheries in Bareilly, India. Pure isolates of L. monocytogenes obtained after enrichment in Buffered Listeria enrichment broth (BLEB) followed by plating onto Listeria oxford agar. The DNA extracted from pure isolates and used for the detection of bacterial pathogen. The oligonucleotide primer pairs (F: CGGAGGTTCCGCAAAAGATG; R: CCTCCAGAGTGATCGATGTT) complementary to the nucleotide sequence of the hlyA gene selected for detection of L. monocytogenes using polymerase chain reaction (PCR). PCR products of 234 bp generated with DNA from all of L. monocytogenes isolates. The highest occurrence of haemolytic L. monocytogenes isolates from various meat samples was in raw chicken (6.0%), followed by fish meat (4.0%), and then beef (2.5%). Among various milk and milk products, curd (2.0%) showed the highest prevalence, followed by raw milk (1.3%). The cytotoxic effects of haemolytic L. monocytogenes isolates were screened on vero cell lines. The cell lines with cell free culture supernatant (CFCS) examined at 1 min, 10 min, 30 min, and 60 min. The significant changes in vero cells were observed at 30 min with both 30 μL and 50 μL of volume. We conclude that application of PCR approaches can provide critical information on distribution of haemolytic strains of L. monocytogenes in food processing environments. Vero cell cytotoxicity assay (in vitro) resulted positive in twenty four strong haemolysin producing L. monocytogenes isolates. The vero cytotoxicity assay could be suggested as a further step towards an alternative assay for detection of haemolytic strains of L. monocytogenes.
Keywords: L. monocytogenes, haemolysis, PCR, vero cytotoxicity
Introduction
Food safety has emerged as an important global issue with international trade and public health implications (Sudershan et al., 2009). L. monocytogenes associated with outbreaks have been reported from a wide variety of food types (Swaminathan and Gerner-Smidt, 2007) while the bacterium has been isolated from meat, milk and milk products in all over the world (Ryser and Marth, 1991; Jardat et al., 2002; El-Malek et al., 2010; Ikeh et al., 2010). However, the published information on the status of L. monocytogenes from these foods is very scattered and unsystematic, both in the veterinary and public health sectors.
Listeria monocytogenes is a gram-positive rod-shaped bacterium and has emerged as an important foodborne pathogen known to be the causative agent of listeriosis. Listeriosis is associated with the highest case fatality rate of 30% approximately, unlike infection with other common foodborne pathogens, such as Salmonella, which rarely results in fatalities (Gouws and Liedmann, 2005). Immunocompromised patients, neonates and pregnant women are more susceptible to listeriosis. However, the disease can also develop in seemingly healthy individuals (Parihar et al., 2008). The role of haemolysin called listeriolysin (LLO) in pathogenicity has been well documented. The hlyA gene based detection for L. monocytogenes has been frequently adopted by various investigators (Barbuddhe et al., 2000; Aznar and Alarcon, 2002; Dumen et al., 2008). The haemolytic factor from the virulent strain EGD has been purified and characterized as a 60 kDa thiol-activated exotoxin designated listeriolysin O, which is a polypeptide of 529 amino acids, sharing strong homologies with SLO and pneumolysin (Mangaud et al., 1988; Karger and Ghasemi, 2009).
Thus, proficient and dependable techniques for the isolation and identification of L. monocytogenes in food samples are very important to facilitate clinical and epidemiological studies. These include cultural methods and rapid methods (based on the antibodies and nucleic acids) can be categorised broadly into immunological (for example, latex agglutination test, ELISA) and nucleic acid (polymerase chain reaction - PCR) based methods (Adzitey and Huda, 2010). 1). L monocytogenes can be also identified by mouse pathogenicity test, Anton’s test and toxicity of chicken embryos, and these assays require whole animal availability (Furrer et al., 1991).
Current microbiological methods of identification rely on growth in culture media, followed by isolation, biochemical and serological identification. Moreover, these methods are laborious and time consuming, requiring a minimum of five days to recognize pathogen (Khan et al., 2011) and about ten days to identify L. monocytogenes by confirmatory tests (Anon, 1996). On the otherside, immediate attention should be taken in case of contamination to ensure the safety of food products, especially in the case of those food matrices having short shelf-lives, such as meat or dairy products. In the past years, advancements in molecular methods have resulted in the development of rapid methods that reduce analysis time and offer great sensitivity and specificity in the detection of pathogens. Among these, PCR has been increasingly used for the rapid, sensitive and specific detection of foodborne pathogens (Amagliani et al., 2004). The PCR used in this study, for the detection of L. monocytogenes, has been previously described and this has been adopted by various investigators for detecting haemolysin gene among L. monocytogenes (Furrer et al., 1991). It has also been found to be applicable in food sources, like meat and milk sources (Jaradat et al., 2002; Aznar and Elizaquivel, 2008). The cell line methods (tissue culture method) have been used to identify pathogenic L. monocytogenes from non-pathogenic L. monocytogenes earlier. However, very few literatures are available on such activity, and those available are very scattered and from clinical sources (Farber and Speirs, 1987; Kathariou and Pine, 1991; Starvu et al., 2011).
Keeping things in mind, the published information on status of L. monocytogenes in meat and milk sources and methods available for the detection of pathogenic L. monocytogenes, we investigated the prevalence of L. monocytogenes in various samples of meat, milk and milk products from local butcheries, dairy farms and confectioneries in Bareilly, India, and examined in vitro LLO based pathogenicity of L. monocytogenes by conventional, molecular and tissue culture method.
Materials and Methods
Sample collection and transportation
A total of 650 commercial food samples (400 from raw meat and 250 from raw milk and milk products) were randomly collected from butcheries (8), dairy farms (2) and dairy/confectionery shops (3) at various locations in the Bareilly city, Uttar Pradesh, India. The locations of sample collection are shown in Table 1. The sampling was done in winter (early December 2006), summer (end of May 2007) and fall (early October 2007) seasons. Sampling visits were made on every Monday for entire month. All the samples were transported in ice packs containers within an hour to the laboratory.
Table 1.
Samples collected in this work.
| Source | No. of samples | Locations | Time of collection (month/year) |
|---|---|---|---|
| Milk and milk products | |||
| (i) Raw milk | 50 | Local dairy farm Partapur Bareilly India | Dec 2006 |
| (ii) Cheese | 20 | Prince sweets &confectionary Partapur Bareilly India | Dec 2006 |
| (iii) Curd | 20 | Prince sweets &confectionary Partapur Bareilly India | Dec 2006 |
| Raw Meat | |||
| (i) Raw chicken | 50 | Hanif chicken shop Peer Bahora Bareilly India | Dec 2006 |
| (ii) Beef | 20 | Sameer meat shop Peer Bahora Bareilly India | Dec 2006 |
| (iii) Fish meat | 50 | Open air fish stall Peer Bahora Bareilly | Dec 2006 |
| Milk and milk products | |||
| (i) Raw milk | 50 | Krishna milk bar D.D.Puram Bareilly India | May 2007 |
| (ii) Cheese | 20 | Krishna milk bar D.D.Puram Bareilly India | May 2007 |
| (iii) Curd | 20 | Krishna milk bar D.D.Puram Bareilly India | May 2007 |
| Raw Meat | |||
| (i) Raw chicken | 50 | Chicken shop Badi Bihar market Bareilly India. | May 2007 |
| (ii) Beef | 30 | Meat shop Badi Bihar Bareilly India | May 2007 |
| (iii) Fish meat | 50 | Open air fish stall Badi Bihar Bareilly India | May 2007 |
| Milk and milk products | |||
| (i) Raw milk | 50 | Indian Veterinary Research Institute (IVRI) Izatnagar Bareilly India | Oct 2007 |
| (ii) Cheese | 10 | Kamal dairy & confectionary Rajender nagar Bareilly India | Oct 2007 |
| (iii) Curd | 10 | Kamal dairy &confectionary Rajender nagar Bareilly India | Oct 2007 |
| Raw Meat | |||
| (i) Raw chicken | 50 | Rehmania meat shop Dela Peer Bareilly India | Oct 2007 |
| (ii) Fish meat | 100 | Open air fish stall Dela Peer Bareilly India | Oct 2007 |
| Total | 650 | ||
Isolation of L. monocytogenes
L. monocytogenes from different food samples were isolated as per USFDA/BAM/CFSAN method, described by Hitchins (1998). Briefly, 25 g of raw meat, cheese and curd, while 25 mL of milk sample, was taken and homogenized in 225 mL of Buffered Listeria Enrichment Broth (BLEB), incubated at 30 °C for 4 h, and added selective agent [0.5% (w/v) acriflavin and nalidixic acid and 1.0% (w/v) cycloheximide, Hi-media, Mumbai, India]. Selective enrichment for isolation was continued at 30 °C for 48 h. Next, a loopful of each sample was streaked on Listeria Oxford agar (Hi-media, Mumbai, India) and incubated at 37 °C for 20 h. Brown-green coloured colonies with a black halo with varying radii were examined for their size, colour, consistency, shape and microscopic examination after Gram staining. Those colonies showing typical characteristics of Listeria were selected and purified by repeated streaking, given an isolate number and maintained at 4 °C. The isolates were subjected to biochemical assays (utilization of rhamnose, mannitol, catalase, motility at 25 °C and 37 °C and NO3 reduction) as recommended in USFDA/BAM/CFSAN described by Hitchins (1998). The standard methods described by Cruickshank et al. (1975) were adopted for their identification as L. monocytogenes.
CAMP assay
The CAMP (Christie, Atkins, Munch-Peterson) assay was performed according to standard procedure as described by Hitchins (1998). Briefly, streak out the L. monocytogenes isolates along with Staphylococcus aureus and Rhodococcus equi on 5% sheep blood agar plates (Merck, India) and observing zones of haemolysis after 24–48 h incubation at 35–37 °C.
Haemolysis on sheep blood agar
The characteristic beta (β) haemolysis was tested with the standard procedure as described by Hitchins (1998). Briefly, L. monocytogenes isolates were streaked on 5% sheep blood agar plates (Merck, India) and observing complete clear zones of haemolysis after 24–48 h incubation at 35–37 °C.
Detection of L. monocytogenes isolates based on haemolysin (hlyA) gene by PCR
The DNA extraction method was adopted as described earlier by Sambrook and Russel (2001) from BHI overnight culture of typical bacterial colonies on Listeria Oxford agar. Briefly, 10 mL of overnight BHI broth culture of L. monocytogenes centrifuged at 6000 rpm for 15 min at 4 °C. The pellet washed twice with TRIS-EDTA (TE) buffer and finally suspended in 567 μL of TE buffer (10 mM Tris, 1 mM EDTA) by gentle vortexing. To the above suspension, 30 μL of 10% SDS and 3 μL of 20 μg/mL proteinase-K (100 μg/mL proteinase-K in 0.5% SDS) added and mixed thoroughly by inversion and incubated at 37 °C for 1 h in a water bath. To this, 100 μL of 5 M NaCl added and mixed thoroughly followed by addition of 80 μL of CTAB/NaCl (10% CTAB dissolved in 0.7 M NaCl), after proper mixing and incubation at 35 °C for 10 min in a water bath. An equal volume (0.7–0.8 μL) of phenol; chloroform; isoamyl alcohol (25:24:1) was added, mixed thoroughly and centrifuged at 13,000 rpm for 20 min. A thick white layer of protein and polysaccharide was formed at the interface of the two layers. The upper aqueous phase was transferred to a fresh microcentrifuge tube. Equal volume of phenol; chloroform; isoamyl alcohol was again added to the aqueous supernatant, re-extracted by thorough mixing and spun at 13,000 rpm for 10–15 min. The supernatant was transferred to a fresh microcentrifuge tube. The procedure was repeated once again. To the final supernatant, 0.6 volumes of isopropanol added at room temperature and centrifuged at 13,000 rpm for 5 min then rinsed with 400 μL of 70% ethanol. The DNA was harvested by spinning at 13,000 rpm for 5 min. The pellet was dried and resuspended in 50 μL sterile triple glass distilled water, added RNase (10 μg/mL) and incubated for 20 min at room temperature.
The primers (synthesized by Genuine Chemical Corporation, India) for the virulence associated gene hlyA were used as described earlier (Furrer et al., 1991). The amplification of hlyA gene (234 bp) by PCR was performed as follows: 2.5 μL of 10X PCR buffer (20 mM Tris HCl, pH 8.0 at 25 °C, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol, 0.5% tween 20 and 0.5% Nonidet-P40); 1.5 mM MgCl2; 2.5 mM of dNTPs mix; 1.0 μL of both forward and reverse primers (15 pmol); 1 U of Taq DNA polymerase enzyme; and 2 μL of DNA as template. Nuclease free water was added to obtain a final volume of 25 μL. PCR conditions included an initial denaturation step at 95 °C for 5 min, followed by 40 subsequent cycles consisting of heat denaturation at 95 °C for 30 s, annealing at 54 °C for 1 min, and extension at 72 °C for 1 min. A final extension was performed at 72 °C for 5 min. PCR products were separated in 1.5% agarose gel electrophoresis, stained by ethidium bromide (Sigma-Aldrich, USA), and visualized under UV light.
Detection of L. monocytogenes isolates based on vero cell cytotoxicity assay
The cell free culture supernatant (CFCS) was prepared as described by Farber and Speirs (1987). Briefly, each L. monocytogenes isolate was inoculated in 5 mL of Tryptic Soy Broth (TSB) (Himedia, Mumbai, India) and incubated at 35 °C for 24 h. About 2 mL of broth culture was re-inoculated into 20 mL of TSB and incubated at 35 °C for 16 h in a low speed shaking water bath. The bacterial cells pelleted by centrifugation (10,000 × g) at 4 °C for 30 min, and the resultant supernatant was filtered through 0.22 μm pore size membrane filter (Nalgene, India). The sterility of CFCS was checked by streaking on Tryptic Soy Agar (TSA) plates (Hi-media, Mumbai, India), incubated at 35 °C for 24 h. The sterile CFCS were then stored at −20 °C.
The vero cell lines were propagated with a standard method described by Konowalchuk et al. (1978). Glasgow minimum essential medium (G-MEM) (Gibco, Mumbai, India) of pH 7.4, containing sodium bicarbonate solution (2.7%), was supplemented with New Born Calf Serum (NBCS) (10% for growth and 20% for maintenance), used for propagation and maintenance of the cell lines. The trypsin-versene solution and Glasgow minimum essential medium (G-MEM) were sterilized by Seitz filtration, and incubated overnight at 37 °C, to check sterility. The cell lines were maintained by regular sub-culturing.
The toxin assay was performed in 6-well microtiter tissue culture plates (SPL life sciences, Korea) as the standard method described by Konowalchuk et al. (1978). Vero cell culture in each well was seeded and allowed to grow as confluent monolayers for 24–48 h. At the same time media was discarded, monolayers were washed twice with Hank’s Balanced Salt Solution (HBSS) (Difco, Haryana, India). Volumes of 30 μL and 50 μL of CFCS were inoculated into each well in duplicate, and 400 μL of the medium supplemented with New Born Calf Serum (NBCS) was then added into each well. The microtiter plates were then incubated at 35 °C under 5% CO2 atmosphere and examined daily by inverted microscope (Olympus, Japan) up to 3 days, for changes in morphological characteristics of vero cell lines.
Results
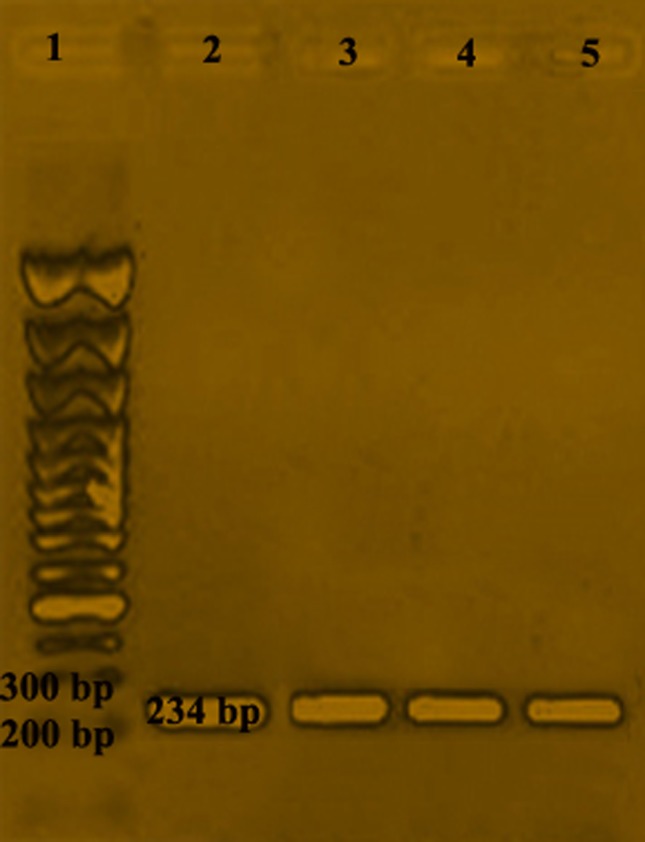
A total of 650 samples (400 from raw meat and 250 from raw milk and milk products) collected from various locations were examined by cultural and biochemical methods. The haemolysis assay on sheep blood agar plate was performed to determine the haemolysis. A clear and complete zone of haemolysis around the streak line of the L. monocytogenes isolate was observed (Figure 1). All isolates showed positive reactions (Table 3). A total of 25 L. monocytogenes isolates were obtained. Overall, 9 samples (6.0%) from raw chicken, 8 samples (4.0%) from fish meat, and 5 samples (2.5%) from beef were found to be positive for L. monocytogenes. Among various milk and milk products samples, 1 sample from curd and 2 samples from raw milk were found to be positive for L. monocytogenes. However, none of the samples found positive from cheese for the presence of L. monocytogenes. Further, the presence of a haemolytic factor in all isolates including reference strain was also confirmed by using a PCR assay (Table 2) producing 234 bp products after amplification of hlyA gene (Figure 2).
Figure 1.
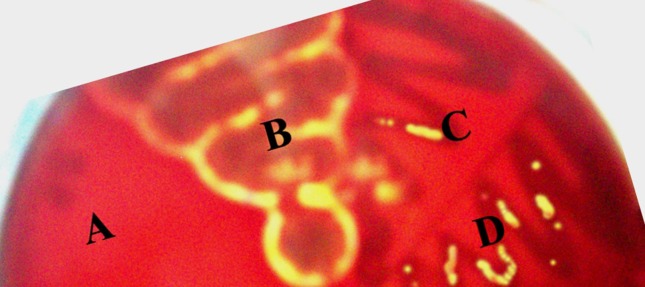
Haemolysis on sheep blood agar; (A) L. inoccua NCTC 11288, (B) L. monocytogenes MTCC 657, (C) FLM-44 (L. monocytogenes isolate from fish meat) (D) FLM-147 (L. monocytogenes isolate from fish meat).
Table 3.
Number of various food samples found positive for L. monocytogenes by cultural and PCR method.
| Type of samples | No. of samples | No. of positive samples by cultural method | No. of positive samples by PCR |
|---|---|---|---|
| Cheese | 50 | 00 | 00 |
| Curd | 50 | 01 | 01 |
| Raw milk | 150 | 02 | 02 |
| Beef | 50 | 05 | 05 |
| Fish meat | 200 | 08 | 08 |
| Raw chicken | 150 | 09 | 09 |
| Total | 650 | 25 | 25 |
Table 2.
Detection of L. monocytogenes isolates by cultural, PCR and Vero cytotoxic assay.
| Isolate No. | Source | Haemolysis on blood agar | Haemolysin (hlyA) gene PCR assay | Vero cytotoxicity |
|---|---|---|---|---|
| CULM-19 | Curd | + | + | + |
| MLM-31 | Raw milk | + | + | + |
| MLM-47 | Raw milk | + | + | + |
| BLM-3 | Beef | + | + | + |
| BLM-17 | Beef | + | + | + |
| BLM-20 | Beef | + | + | + |
| BLM-32 | Beef | + | + | + |
| BLM-35 | Beef | + | + | + |
| FLM- 13 | Fish meat | + | + | + |
| FLM- 21 | Fish meat | + | + | + |
| FLM- 44 | Fish meat | +a | + | − |
| FLM- 45 | Fish meat | + | + | + |
| FLM- 73 | Fish meat | + | + | + |
| FLM-147 | Fish meat | +a | + | − |
| FLM-148 | Fish meat | + | + | + |
| FLM-173 | Fish meat | + | + | + |
| CLM- 2 | Raw chicken | + | + | + |
| CLM- 5 | Raw chicken | + | + | + |
| CLM- 6 | Raw chicken | + | + | + |
| CLM- 19 | Raw chicken | + | + | + |
| CLM- 24 | Raw chicken | + | + | + |
| CLM- 43 | Raw chicken | + | + | + |
| CLM-111 | Raw chicken | + | + | + |
| CLM-116 | Raw chicken | + | + | + |
| CLM-129 | Raw chicken | + | + | + |
| MTCC-657 | Reference strain | + | + | + |
+: weak haemolysis.
Figure 2.
hlyA gene based detection of L. monocytogenes using PCR; (Lane 1)-100 bp DNA ladder, (Lane 2)-MTCC 657, (Lane 3)-CULM 19 (L. monocytogenes isolate from curd), (Lane 4)-MLM 47 (L. monocytogenes isolate from milk), (Lane 5)-BLM 3 (L. monocytogenes isolate from beef).
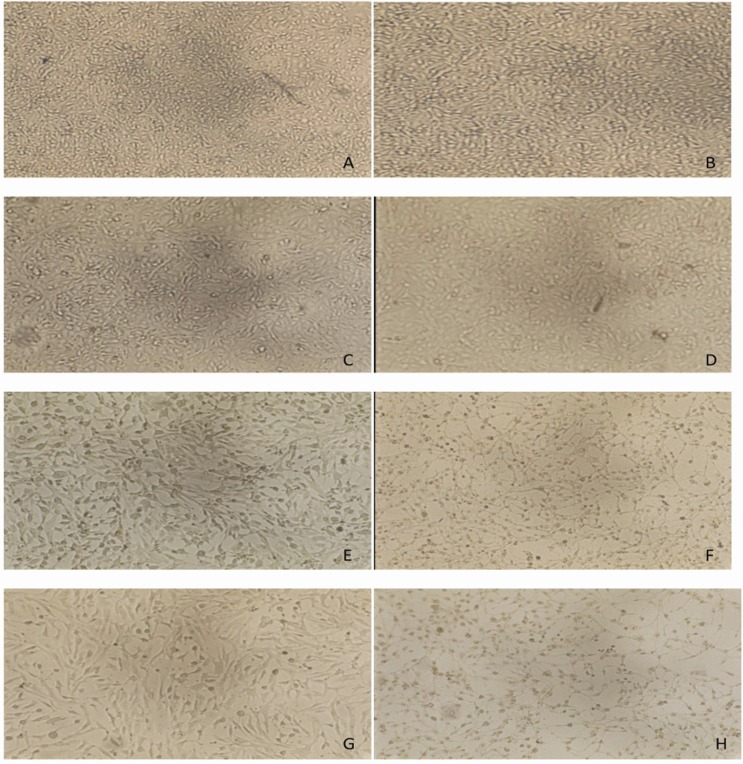
To know the effect of pathogenic L. monocytogenes isolates on continuous cell lines, the cytotoxicity assay was performed on vero cell lines. The microtitre plates were examined for morphological changes in cell lines. After incubation with CFCS from L. monocytogenes, the results were examined at 1 min, 10 min, 30 min, and 60 min. At 10 min, vero cells started to change morphologically and changes were clearer with the 50 μL of CFCS (Figure 3C and 3D). The significant changes in vero cells were observed at 30 min with both 30 μL and 50 μL of volume. The vero cells changed from spindle (characteristic of normal vero cells) to round and shrivelled cells (Figure 3E and 3F), and changes continued with further incubation (Figure 3G and 3H). All twenty five isolates and the reference strain of L. monocytogenes identified by cultural and PCR methods were found to be positive in vero cell cytotoxicity assay, except two strains from fish meat source (FLM 44 and FLM 147) (Tables 2 and 3).
Figure 3.
Cytopathic effect of CFCS from L. monocytogenes on Vero cells (100× magnification); (A) at 1 min (30 μL), (B) at 1 min (50 μL), (C) at 10 min (30 μL), (D) at 10 min (50 μL), (E) at 30 min (30 μL), (F) at 30 min (50 μL), (G) at 60 min (30 μL), (H) at 60 min (50 μL).
Discussion
Various meat and milk products are frequently consumed in all over the world, including India, and they can act as transmission vehicles for L. monocytogenes. Several reports revealed L. monocytogenes contamination in beef (Bohaychuk et al., 2006; Stavru et al., 2011), chicken (Genigeorgis et al., 1989; Bailey et al., 1990; Mahmood et al., 2003; Paszkowska et al., 2005), fish (Furrer et al., 1991; Kargar and Ghasemi, 2009), milk and milk products (Barbuddhe et al., 2000; Jaradat et al., 2002; Sudershan et al., 2009; Saikia and Joshi, 2010) in the present investigation. Among all the samples from all sources, the higher incidence of L. monocytogenes was noticed with meat samples. In developing countries like India, the possible reason behind this may be the use of chopping boards, mincing machines, cleaning clothes and carcasses, which are frequently used in butcheries and kept for an indefinite period of time. Moreover, methods of sterilization or disinfection with low efficacy are adopted to keep these equipment and utensils sanitized (Nayak et al., 2010). Our investigation provided an important insight about the L. monocytogenes contamination in various meat, milk and milk products in Bareilly, India, and other states where consumers are regularly supplied with contaminated food. It was observed that there is a need to implement HACCP in the unorganised sector, as it accounts for a large amount of food produced and processed in India.
The positive CAMP and haemolysis assay and the presence of hlyA gene by PCR in all isolates suggest that these virulent strains may have the potential to invade host cells and cause listeriosis (Ryser and Marth, 2007). Due to no confirmation of the pathogenic nature of the L. monocytogenes isolates, the vero cell cytotoxicity assay was performed. According to assay results, all twenty five L. monocytogenes isolates, including the reference strain, showed pathogenic effect on vero cell lines, except two strains from fish meat (FLM 44 and FLM 147), that showed weak haemolytic pattern. These two fish meat isolates were also confirmed by PCR for the exact presence of a haemolytic factor, and they were positive for the presence of LLO. However, these two strains exhibited no cytotoxicity in vero cell assay. L. monocytogenes is a stronger producer of haemolysin and pathogenic for humans and mice. It has also been described that the ability of L. monocytogenes to produce haemolysin is strongly correlated with its virulence. However, some strains produce weak haemolysis reactions, that can appear to be negative during culture base biochemical assay (Farber and Speirs, 1987). Thus, through the phenotypic interpretation, it is difficult to determine haemolysis of L. monocytogenes strains (USFDA/BAM/CFSAN, 1998). In our knowledge, there are very few literatures available on cytotoxic effect of L. monocytogenes on vero cells, especially from food sources (Farber and Speirs, 1987). The vero cell cytotoxicity assay described in this work could be suggested as an alternative or adjunct assay for haemolytic L. monocytogenes strains identification. The test is easy to perform, and a positive reaction can occur within 10 min of addition of undiluted bacterial culture filtrates. However, weak haemolytic strains are matter for further investigation.
Acknowledgement
We express our thanks to the Director, IVRI and Head, Department of Biochemistry and Department of Animal Biotechnology, IVRI, for providing facilities, Vero cells and their constant encouragement.
References
- Adzitey F, Huda N. Listeria monocytogenes in foods: incidences and possible control measures. Afr J Microbiol Res. 2010;4:2848–2855. [Google Scholar]
- Amagliani G, Brandi G, Omiccioli E, Casiere A, Bruce IJ, Magnani M. Direct detection of Listeria monocytogenes from milk by magnetic based DNA isolation and PCR. Food Microbiol. 2004;21:597–603. [Google Scholar]
- Anonymous. Rapporti ISTISAN ISSN 1123–3117, 96/35. Amaglianni G, Giammarini C, Omiccioli E, Brandi G, Magnani M (2007) Detection of Listeria monocytogenes using a commercial PCR kit and different DNA extraction methods. Food Cont. 1996;18:1137–1142. [Google Scholar]
- Aznar R, Alarcon B. On the specificity of PCR detection of Listeria monocytogenes in food: a comparison of published primers. Syst Appl Microbiol. 2002;25:109–119. doi: 10.1078/0723-2020-00079. [DOI] [PubMed] [Google Scholar]
- Aznar R, Elizaquivel P. Reliability of Listeria monocytogenes identification by specific PCR assessed by phenotypic and genotypic techniques. Food Anal Meth. 2008;1:243–251. [Google Scholar]
- Bailey JS, Fletcher DL, Cox NA. Listeria monocytogenes colonization of broiler chickens. Poultry Sci. 1990;69:457–461. doi: 10.3382/ps.0690457. [DOI] [PubMed] [Google Scholar]
- Barbuddhe SB, Malik SVS, Bhilegaonkar KN, Kumar P, Gupta LK. Isolation of Listeria monocytogenes and anti-listeriolysin O detection in sheep and goats. Small Rumi Res. 2000;38:151–155. [Google Scholar]
- Bohaychuk VM, Gensler GE, King RK, Manninen KI, Sorensen O, Wu JT, Stiles ME, Mcmullen LM. Occurrence of pathogens in raw and ready-to-eat meat and poultry products collected from the retail market place in Edmonton, Alberta, Canad. J Food Prot. 2006;69:2176–2182. doi: 10.4315/0362-028x-69.9.2176. [DOI] [PubMed] [Google Scholar]
- Cruickshank R, Duguid JP, Marmion BP, Swain RHA. Medical microbiology- the practice of medical microbiology. Churchill Livingstone; Edinberg, London and New York: 1975. [Google Scholar]
- Dumen E, Baca AU, Dumen E. Comparative detection of Listeria monocytogenes in raw milk by microbiological method and PCR. Medycyna Wet. 2008;1:59–63. [Google Scholar]
- El-Malek AMA, Ali SFH, Hassanein R, Mohamed MA, Elsayh KI. Occurrence of Listeria species in meat, chicken products and human stools in Assiut city, Egypt with PCR use for rapid identification of Listeria monocytogenes. Vet. World. 2010;3:353–359. [Google Scholar]
- Farber JM, Speirs JI. Potential use of continuous cell lines to distinguish between pathogenic and non pathogenic Listeria spp. J Clin Microbiol. 1987;25:1463–1466. doi: 10.1128/jcm.25.8.1463-1466.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs JA, Surendran PK. Incidence of Listeria in tropical fish and fishery products. Lett Appl Microbiol. 1989;9:49–51. [Google Scholar]
- Furrer B, Candrian V, Hofelein C, Luthy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- Genigeorgis CA, Dutulescu D, Garayzabel JF. Prevalence of Listeria spp. in turkey meat at supermarket and slaughter house level. J Food Prot. 1989;52:618–24. doi: 10.4315/0362-028X-52.9.618. [DOI] [PubMed] [Google Scholar]
- Gouws PA, Liedemann I. Evaluation of Diagnostic PCR for the Detection of Listeria monocytogenes in Food Products. Food Tech Biotech. 2005;43:201–205. [Google Scholar]
- Hitchins AD. Detection and enumeration of Listeria monocytogenes in foods. [Accessed 6 March 2008];Bacteriological Analytical Manual Online. 1998 Available at: http://www.cfsan.fda.gov/~ebam/bam-10.html.
- Ikeh MAC, Obi SKC, Ezeasor DN, Ezeonu IM, Moneke AN. Incidence and pathogenicity profile of Listeria sp. isolated from food and environmental samples in Nsukka, Nigeria. Afr J Biotechnol. 2010;9:4776–4782. [Google Scholar]
- Jaradat ZW, Schutze GE, Bhunia AK. Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int J Food Microbiol. 2002;76:1–10. doi: 10.1016/s0168-1605(02)00050-8. [DOI] [PubMed] [Google Scholar]
- Kargar M, Ghasemi A. Role of Listeria monocytogenes hlyA gene isolated from fresh cheese in human habitual abortion in Marvdasht. Iran J Clin Infect Dis. 2009;4:214–218. [Google Scholar]
- Karunasagar I, Segar K, Karunasagar I, Goebel W. Incidence of Listeria spp. in tropical sea foods. XI International symposium on problems of listeriosis; Copenhagen Denmark. 1992. Abstract 155. [Google Scholar]
- Kathariou S, Pine L. The type strain(s) of Listeria monocytogenes: A source of continuing difficulties. Int J Syst Bacteriol. 1991;41:328–330. doi: 10.1099/00207713-41-2-328. [DOI] [PubMed] [Google Scholar]
- Khan JA, Rathore RS, Ahmad I, Khan S. Molecular Strategies: Detection of Foodborne Bacterial Pathogens. In: Ahmad I, Ahmad F, Pitchel J, editors. Microbes and Microbial Technology. Springer; New York: 2011. pp. 189–206. [Google Scholar]
- Konowalchuk J, Dickie N, Stavric S, Speirs J. Properties of an Escherichia coli cytototoxin. Infect Immun. 1978;20:575–577. doi: 10.1128/iai.20.2.575-577.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood MS, Ahmed AN, Hussain I. Prevalence of Listeria monocytogenes in poultry meat, poultry meat products and other related inanimates at Faisalabad. Pak J Nut. 2003;2:346–349. [Google Scholar]
- Mengaud J, Vicente MF, Chenevert J, Pereira JM, Geoffroy C, Gicquel-Sanzey B, Baquero F, Perez-Diaz JC, Cossart P. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of L. monocytogenes. Infect Immun. 1988;56:766–772. doi: 10.1128/iai.56.4.766-772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak JB, Brahmbhatt MN, Savalia CV, Bhong CD, Roy A, Kalyani IH, Parmar BC. Detection and characterization of Listeria species from buffalo meat. Buff Bull. 2010;29:83–94. [Google Scholar]
- Parihar VS, Barbuddhe SB, Danielsson-Tham ML, Tham W. Isolation and characterization of Listeria species from tropical seafoods. Food Cont. 2008;19:566–569. [Google Scholar]
- Paszkowska KK, Bania J, Bystron J, Molenda J, Czerw M. Occurrence of Listeria spp. in raw poultry meat and poultry meat products. Bull Vet Inst Pul. 2005;49:219–222. [Google Scholar]
- Ryser ET, Marth EH. Listeria, Listeriosis and Food Safety. Marcel Dekker; New York (Basel): 1991. [Google Scholar]
- Ryser ET, Marth EH. Listeria, Listeriosis, and Food Safety. Taylor and Francis; Boca Raton, FL: 2007. [Google Scholar]
- Saikia P, Joshi SR. Retail market poultry meats of NorthEast India- A microbiological survey for pathogenic contaminants. Res J Microbiol. 2010;5:36–43. [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; New York: 2001. [Google Scholar]
- Stavru F, Frederic B, Anna S, Daniel R, Pascale C. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. PNAS. 2011 doi: 10.1073/pnas.1100126108. www.pnas.org/cgi/doi/10.1073/pnas.1100126108. [DOI] [PMC free article] [PubMed]
- Sudershan RV, Rao P, Polasa K. Food safety research in India: a review. Asian J Food Ag Ind. 2009;2:412–433. [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- USFDA/BAM/CFSAN. Bacteriological analytical manual online. 1998 Available at: www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm.