Abstract
The present study describes integron mediated multiple antibiotic resistance in extended-spectrum β-lactamase producing clinical isolates of Klebsiella pneumoniae. One hundred and four clinical isolates of K. pneumoniae from two Iranian hospitals were screened for extended-spectrum β-lactamase production and susceptibility of the extended-spectrum β-lactamase producing isolates was determined to 17 antibiotics by disc diffusion. Presence of integron classes 1, 2 and 3 was detected by PCR and integrase specific primers. Isolates harboring class 1 integron were then screened for variable regions using PCR. Fifty isolates (48%) produced extended-spectrum β-lactamases among which, 22 (44%) harbored class 1, 3 (6%) carried class 2 and none contained class 3 integons. Integron carriage was significantly associated with higher rates of multiple antibiotic resistance in extended-spectrum β-lactamase producing clinical isolates of K. pneumoniae. Integron harboring isolates were more resistant to aztreonam (51.3%), ceftazidime (42.6%), cefotaxime (43.3%), cefepime (24.6%), kanamycin (43.2%), tobramycin (30.7%), norfloxcacin (32%) and spectinomycin (25.6%) compared to the organisms without integrons. On the other hand, resistance to nitrofurantoin and streptomycin was significantly higher among the integron negative isolates. PCR amplification of class1 integron variable regions revealed 9 different sized DNA fragments and isolates with similar profiles for class 1 integron variable regions showed the same antibiotic resistance phenotypes.
Keywords: Klebsiella pneumoniae, extended-spectrum β-lactamase, multiple antibiotic resistance, integron
Introduction
Klebsiella pneumoniae is an opportunistic pathogen responsible for up to 10% of all nosocomial infections and one of the leading causes of urinary, respiratory and blood infections particularly in immuno-compromised patients (Brisse et al., 2006; Podschun and Ullmann, 1998). Emergence and spread of multidrug resistant K. pneumoniae, specifically the extended spectrum β-lactamase (ESBL) producing strains, is often responsible for the failure of antibiotic treatment in hospital settings (Paterson and Bonomo, 2005). Mobile genetic elements such as plasmids and transposons are reported to carry genetic units named integrons which contain genes for site-specific recombination, and are capable of capturing and mobilizing gene cassettes (Hall and Collis, 1995; Stokes and Hall, 1989). These can be subdivided into two main groups, the superintegrons and the antibiotic resistance integrons (ARIs) (Cambray et al., 2010; Fluit and Schmitz, 2004). Dissemination of antibiotic resistance genes among bacteria can occur by mobile genetic elements containing the ARIs (Cambray et al., 2010; Hall, 1997; Hall and Collis, 1995). Among different classes of ARIs, classes 1, 2, and 3 have been historically implicated with multiple antibiotic resistance (MAR) phenotypes and are defined based on their respective integrase (intI) genes (Cambray et al., 2010; Hall, 1997). Class 1 integrons are the most widespread and have been frequently found in ESBL producing clinical isolates of Enterobacteriaceae including K. pneumoniae (Jones et al., 2003; Machado et al., 2007; Rao et al., 2006; Yao et al., 2007). Class 2 integrons occur less frequently in ESBL producing E. coli and K. pneumoniae and finally, class 3 integrons are rarely found in ESBL producing K. pneumoniae (Bhattacharjee et al., 2010; Correia et al., 2003; Machado et al., 2005). We investigated the antimicrobial susceptibility and prevalence of class 1, 2 and 3 integrons in ESBL producing clinical isolates of K. pneumoniae from Iranian patients. The variable regions of class 1 integrons were also detected. Finally, the association between integron carriage and MAR phenotypes were determined.
Materials and Methods
Bacterial isolates
One hundred and four clinical isolates of K. pneumoniae were collected from hospitalized patients in two teaching hospitals in Tehran (Imam Hussein and Labbafinejad) from March 2008 to March 2009. The majority of the isolates were from urine (n = 90; 86.5%) followed by wound (n = 4; 3.84%), trachea (n = 4; 3.84%), sputum (n = 2; 1.92%), blood (n = 2; 1.92) and other unknown sources (n = 2; 1.92%). The ATCC standard strains K. pneumoniae (ATCC 10031) and E. coli (ATCC 25922) were used as susceptibility controls. Klebsiella pneumoniae (ATCC 1029) was kindly provided by Dr. Rajabnia (Babol University, Medical School) for use as positive control for class 1 integron in PCR tests.
ESBL production
The isolates were initially screened for ESBL production by the double disc synergy test (DDST) using cefotaxime (30 μg), ceftazidime (30 μg) and cefepime (30 μg) placed 20 mm center to center from an amoxicillin/clavulanic acid disk (20/10 μg) (Himedia, India). ESBL production was detected when synergy was observed between the inhibition zones of cephalosporins and amoxicillin/clavulanic acid and was further confirmed by the phenotypic confirmatory test (PCT) using ceftazidime and cefotaxime alone or in combination with clavulanic acid (CLSI, 2011; Drieux et al., 2008).
Antimicrobial susceptibility
In vitro antimicrobial susceptibility of ESBL producing isolates was determined to 17 antibiotics according to the CLSI criteria (CLSI, 2011). The antimicrobial discs (Himedia, India) were; cefepime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), imipenem (10 μg), gentamicin (10 μg), tobramycin (10 μg), aztreonam (30 μg), amikacin (30 μg), nitrofurantoin (300 μg), streptomycin (10 μg), spectinomycin (100 μg), norfloxacin (10 μg), kanamycin (30 μg), ampicilin/sulbactam (10/10 μg), colistin (10 μg), co-amoxiclav (20/10 μg), piperacilin (100 μg) and pipercilin/tazobactam (100/10 μg).
Detection of class 1, 2 and 3 integrons by PCR
Isolates were grown overnight (18 h) in Luria Bertani (LB) broth (Difco, Detroit, MI, USA) at 37 °C and genomic DNA was extracted using High Pure PCR template Prep kit for Genomic DNA extraction (Roche Diagnostics, Mannheim, Germany). PCR amplification of classes 1, 2 and 3 integrase genes was performed in 25 μL reaction mixtures containing 30 ng DNA template, 0.4 mM of each dNTP, 150 μM MgCl2, 0.2 U Super Taq DNA polymerase (CinnaGen, Tehran, Iran) and 1 pM of each primer (FazaBiotech, Tehran, Iran) as follows: Int1F; CCTCCCGCACGATGATC, Int1R; TCCACGCATCGT CAGGC, Int2F; TTATTGCTGGGATTAGGC, Int2R; ACGGCTACCCTCTGTTATC, Int3F; AGTGGGTGGC GAATGAGTG, Int3R; TGTTCTTGTATCGGCAGGTG) (Goldstein et al., 2001). Amplifications were performed in a Bioer TC25/HH Thermal Cycler (Bioer Technology Co. Ltd, Hangzhou, China) using the following program: initial denaturation at 95 °C for 5 min followed by 35 cycles of 1 min at 94 °C, 1 min at 60 °C and 1 min at 72 °C with a final extension at 72 °C for 10 min. For the class 1 integron carrying isolates, PCR amplification of variable regions was performed in 20 μL reaction mixtures containing 30 ng DNA template, 20 μM of each dNTP, 150 μM MgCl2, 0.2 U Super Taq DNA polymerase and 1 pM of each primer (5′-CS; GGCATCCAAGCAGCAAG, 3′-CS AAGCAGA CTTGACCTGA) (Levesque et al., 1995). Amplification of the integron variable regions was carried out with initial denaturation of 4 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C with a final extension of 10 min at 72 °C. The amplified PCR products were resolved by electrophoresis in 1% agarose gels and visualized after staining with ethidium bromide.
Statistical analysis
To compare the antibiotic resistance profiles between integron positive and integron negative isolates, non-parametric analysis was performed using the two-tailed Mann-Whitney U test allowing for continuous variable and non-normal distribution.
Results
Among the 104 K. pneumoniae clinical isolates, 50 (48%) were confirmed to produce ESBL. The antibiotic susceptibility results showed that all ESBL producing isolates were resistant to piperacillin. Resistance rate to other antibiotics were: amoxiclav (96%), aztreonam, kanamycin and cefotaxime (68%), tobramycin (66%), ceftazidime (64%), ampicillin/sulbactam (60%), spectinomycin (56%), cefepime (52%), norfloxacin (48%), gentamicin (36%), nitrofurantoin (24%), amikacin (22%), piperacillin/tazobactam (20%), colistin (14%) and streptomycin (10%). All isolates were susceptible to imipenem (Table 1).
Table 1.
Association between antimicrobial susceptibility and integron carriage in 50 extended-spectrum β-lactamase producing clinical isolates of Klebsiella pneumoniae.
| Antibiotic | Integron positive isolates (n= 23) | Integron negative isolates (n=27) | Level of significance* | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| % R | % I | % S | % R | % I | % S | ||
| Kanamycin | 91.4 | 4.3 | 4.3 | 48.2 | 40.7 | 11.1 | ** |
| Tobramycin | 82.6 | 0 | 17.4 | 51.9 | 0 | 48.1 | * |
| Gentamicin | 47.8 | 13 | 39.2 | 25.9 | 11.1 | 63 | NS |
| Norfloxacin | 65.3 | 4.3 | 30.4 | 33.3 | 22.3 | 44.4 | NS |
| Aztreonam | 95.7 | 4.3 | 0 | 44.4 | 11.2 | 44.4 | *** |
| Cefotaxime | 91.4 | 4.3 | 4.3 | 48.1 | 3.8 | 48.1 | *** |
| Ceftazidime | 87 | 8.7 | 4.3 | 44.4 | 22.3 | 33.3 | ** |
| Cefepime | 65.3 | 4.3 | 30.4 | 40.7 | 0 | 59.3 | NS |
| Spectinomycin | 69.6 | 26.1 | 4.3 | 44.4 | 40.7 | 14.9 | NS |
| Piperacillin | 100 | 0 | 0 | 100 | 0 | 0 | NS |
| Piperacillin/Tazobactam | 26.1 | 34.8 | 39.1 | 14.8 | 29.6 | 55.6 | NS |
| Streptomycin | 0 | 30.4 | 69.6 | 18.5 | 48.2 | 33.3 | ** |
| Ampicilin/sulbactam | 73.9 | 13.1 | 13 | 48.2 | 18.5 | 33.3 | NS |
| Colistin | 13 | 0 | 87 | 14.8 | 0 | 85.2 | NS |
| Amikacin | 17.4 | 34.8 | 47.8 | 25.9 | 3.7 | 70.4 | NS |
| Nitrofurantoin | 8.7 | 26.1 | 65.2 | 37 | 26 | 37 | * |
| Imipenem | 0 | 0 | 100 | 0 | 0 | 100 | NS |
| Amoxyclav | 100 | 0 | 0 | 92.6 | 0 | 7.4 | NS |
The significance level was determined using the two-tailed Mann-Whitney U test.
5%;
1% and
0.1% level.
NS, non-significant; R, resistant; I, intermediately resistant; S, sensitive.
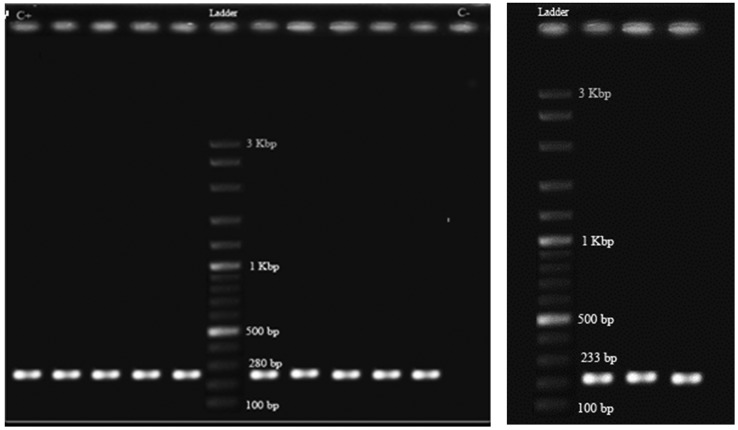
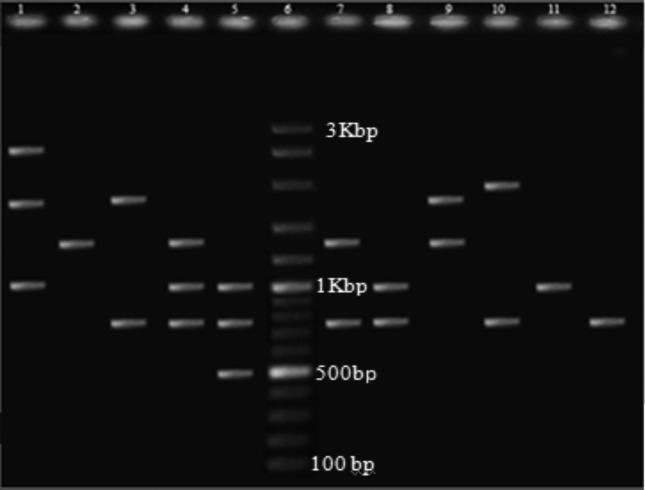
PCR amplifications for classes 1 and 2 integrase genes showed that 22 (44%) of the ESBL producing K. pneumoniae isolates harbored class 1 (Figure 1A) and 3 (6%) carried class 2 integrons (Figure 1B), two of which had both classes. None of the isolates carried class 3 integron. Class 1 integron harboring isolates possessed one to three DNA sequences manifested as nine different sized amplicons in the range of 0.5 to 3 kb (Figure 2). The integron harboring isolates were significantly more resistant to aminoglycosides including kanamycin (43.2%) and tobramycin (30.7%). The same trend was observed for other antibiotics where integron carrying isolates were more resistant to cefotaxime (43.3%), ceftazidime (42.6%), aztreonam (51.3%), norfloxcacin (32%), cefepime (24.6%) and spectinomycin (25.6%) (Table 1). On the other hand, resistance to streptomycin and nitrofurantoin was significantly higher in integron negative isolates (p ≤ 0.05) (Table 1). The association between integron carriage and antibiotic resistance was most significant for aztreonam (p = 0.000) followed by cefotaxime (p ≤ 0.001), kanamycin, ceftazidime and streptomycin (p ≤ 0.01) and finally tobramycin (p ≤ 0.05) (Table 1). Comparison of MAR phenotype (resistance to 6 or more antibiotics) in the two groups of isolates showed that integron harboring strains had a much higher rate of MAR (95.65%) compared to integron negative isolates (48.15%) (p = 0.000). There was no significant association between the type of specimen with ESBL production, antibiotic susceptibility or integron carriage. However, isolates with similar profiles for class 1 integron variable regions had the same antibiotic resistance phenotypes (Table 2).
Figure 1.
PCR amplification product of class 1 (A) and class 2 (B) integrase genes in a number of ESBL producing Klebsiella pneumoniae clinical isolates. Ladder, DNA molecular weight marker (100 bp, Fermentas); C+, positive control for class 1 integron; C−, negative control (no DNA template).
Figure 2.
PCR amplification of class 1 integron variable regions in ESBL producing Klebsiella pneumoniae isolates (Lanes 1–5 and 7–12). Lane 6, DNA molecular weight marker (100 bp, Fermentas).
Table 2.
Distribution of variable region fragments sizes among class1 integron carrying ESBL producing K. pneumoniae clinical isolates.
| No. of isolates | No. of bands | Band size (bp) | Antibiotic resistance profiles |
|---|---|---|---|
| 2 | 1 | 750 | ATM, CAZ, AMC, CTX, KM, CPM, NOR, AS, PIP, TN |
| 1 | 1 | 1000 | CTX, PIP |
| 1 | 1 | 1300 | ATM, CAZ, AMC, CTX, KM, AK, NF, SP, NOR, AS, CL, PIP, TN |
| 8 | 2 | 750–2000 | ATM, CAZ, AMC, CTX, KM, CPM, SP, PIP |
| 4 | 2 | 750–1000 | ATM, CAZ, AMC, KM, CPM, SP, PIP |
| 1 | 2 | 1300–1800 | ATM, CAZ, AMC, CTX, SM, SP, NOR, AS, PIP, TN |
| 1 | 2 | 750–1800 | ATM, AMC, CTX, KM, GM, CPM, NOR, SP, AS, TN |
| 1 | 2 | 750–2500 | ATM, CAZ, AMC, CTX, GM, KM, CPM, PZT, SM, SP, NOR, AS, PIP, TN |
| 1 | 3 | 500–750–1000 | CAZ, CTX, KM, PIP, A/S |
| 1 | 3 | 750–1000–1300 | ATM, CAZ, AMC, CTX, GM, KM, CPM, SM, SP, NOR, AS, PIP, TN |
| 1 | 3 | 250–1000–1700 | CAZ, AMC, CTX, KM, CPM, AK, PZT, SM, SP, NOR, AS, CL, PIP, TN |
Amikacin (AK), Gentamicin (GM), Kanamycin (KM), Imipenem (IPM), Aztreonam (ATM), Amoxyclav (AMC). Cefotaxime (CTX), Ceftazidime (CAZ), Cefepime (CPM), Piperacillin (PRL), Piperacillin/Tazobactam (PZT), Ampicilin/sulbactam (AS), Streptomycin (SM), Spectinomycin (SP), Tobramycin (TN), Colistin (CL), Nitrofurantoin (NF), Norfloxacin (NOR).
Discussion
ESBL producing K. pneumoniae have been reported to harbor higher rates of class 1 integrons compared to non-ESBL strains (Bhattacharjee et al., 2010; Goldstein et al., 2001; Jones et al., 2003; Machado et al., 2007; Martinez-Freijo et al., 1998; Rao et al., 2006; White et al., 2001; Yao et al., 2007). In the current study, 44% of the ESBL positive isolates carried class 1 integrons. We found a significant association between integron carriage and higher rates of resistance to β-lactams and aminoglycosides. Other reports have shown that resistance gene cassettes to all aminoglycosides are carried by class 1 integrons (Cambray et al., 2010; Fluit and Schmitz, 2004).
The size range of the amplified fragments for class 1 integron variable regions indicates presence of one or more gene cassettes, providing a possible explanation for various MAR phenotypes, or more than one integron in an isolate. It may also be concluded that these isolates either contain more than one class1 integron with a variety of integrated gene cassettes or integrons which have undergone possible rearrangements (Levesque et al., 1995). Identical antibiotic resistance phenotypes in the isolates carrying similar sized fragments in variable regions could mean that amplicons of the same size carry the same resistance gene cassettes (El-Najjar et al., 2010).
According to our results, 6% of the ESBL positive isolates harbored class 2 integrons similar to the report by Bhattacharjee et al. (Bhattacharjee et al., 2010). Several studies have reported restricted distribution of class 2 integrons among the family Enterobacteriaceae including Escherichia coli, Salmonella, Enterobacter cloacae and also in Acinetobacter baumannii (Goldstein et al., 2001; Ramirez et al., 2010). To our knowledge, there is only one other report describing class 2 integron in ESBL producing K. pneumoniae (Bhattacharjee et al., 2010).
We did not find any significant concordance between the presence of the ARIs and a specific MAR phenotype. However, in the integron positive isolates, the frequency of resistance to seven antibiotics from five classes was significantly higher than integron negative strains (p < 0.05). In addition, integron positive isolates were significantly more likely to show MAR phenotypes than integron negative isolates (p = 0.000). It could be concluded that ARI carriage may at least be partially responsible for not only higher rates of resistance to several antibiotics, but also higher prevalence of MAR phenotypes in K. pneumoniae isolates (Martinez-Freijo et al., 1998; Yao et al., 2007).
Interestingly, there was an inverse association between resistance to streptomycin and nitrofurantoin with integron carriage, where integron negative isolates were more resistant to these antibiotics. No relationship was found between resistance to amikacin and colistin with integron carriage. Martinez-Freijo and colleagues also found no significant association between presence of integrons and amikacin resistance (Martinez-Freijo et al., 1998). Similar observations have been made by White and colleagues regarding amikacin and nitrofurantoin (White et al., 2001). These results suggest that resistance to nitrofurantoin and amikacin are independent of resistance gene cassettes carried by ARIs.
Irrespective of whether or not resistance genes are contained within the integrons, the results presented here indicate wide dissemination of ARIs in ESBL producing K. pneumoniae and correlation of integron carriage with increased drug resistance rates as well as multidrug resistance. Presence of integron associated resistance in the community has been shown to be widespread and can contribute to multidrug resistance in hospital settings (Leverstein-van Hall et al., 2002). It has been suggested that screening for ARIs could complement existing antibiotic resistance surveillance programs by providing information on resistance profiles as well as dissemination of resistance between commensal and infectious strains (Jones et al., 2003).
Acknowledgments
The financial support of this research was provided by the Shahid Beheshti University Research Council.
Footnotes
Disclosure Statement
No competing financial interests exist.
References
- Bhattacharjee A, Sen MR, Prakash P, Gaur A, Anupurba S, Nath G. Observation on integron carriage among clinical isolates of Klebsiella pneumoniae producing extended spectrum β-lactamases. Ind J Med Microbiol. 2010;28:207–210. doi: 10.4103/0255-0857.66472. [DOI] [PubMed] [Google Scholar]
- Brisse S, Grimont F, Grimont PAD. The genus Klebsiella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: a handbook on the biology of bacteria. Vol. 6. Berlin: Springer-Verlag; 2006. pp. 159–196. [Google Scholar]
- Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. 1. Vol. 30. Clinical and Laboratory Standards Institute; Wayne, PA: 2011. M100-S21. [Google Scholar]
- Correia M, Boavida F, Grosso F, Salgado MJ, Lito LM. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2003;47:2838–2843. doi: 10.1128/AAC.47.9.2838-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14:90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- El-Najjar NG, Farah MJ, Hashwa FA, Tokajian ST. Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. Lett Appl Microbiol. 2010;51:456–461. doi: 10.1111/j.1472-765X.2010.02926.x. [DOI] [PubMed] [Google Scholar]
- Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, Grady M, Liebert C, Summers AO, White DG, Maurer JJ. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother. 2001;45:723–726. doi: 10.1128/AAC.45.3.723-726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- Hall RM. Mobile gene cassettes and integrons: moving antibiotic resistance genes in Gram-negative bacteria. CIBA Found Symp. 1997;207:192–205. doi: 10.1002/9780470515358.ch12. [DOI] [PubMed] [Google Scholar]
- Jones LA, Mclver CJ, Rawlinson WD, White PA. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intel. 2003;27:103–110. [PubMed] [Google Scholar]
- Leverstein-van Hall MA, Paauw A, Box ATA, Blok HEM, Verhoef J, Fluit AC. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J Clin Microbiol. 2002;40:3038–3040. doi: 10.1128/JCM.40.8.3038-3040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque C, Piche L, Larose C, Roy pH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado E, Ferreira J, Novais A, Peixe L, Cantón R, Baquero F, Coque TM. Preservation of integron types among Enterobacteriaceae producing extended-spectrum β-lactamases in a Spanish hospital over a 15-year period (1988 to 2003) Antimicrob Agents Chemother. 2007;51:2201–2204. doi: 10.1128/AAC.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado E, Cantón R, Baquero F, Galán JC, Rollán A, Peixe L, Coque TM. Integron content of extended spectrum β-lactamase producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother. 2005;49:1823–1829. doi: 10.1128/AAC.49.5.1823-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Freijo P, Fluit AC, Schmitz FJ, Grek VSC, Verhoef J, Jones ME. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, Pineiro S, Centron D. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother. 2010;54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AN, Barlow M, Clark LA, Boring JR, Tenover FC, McGowan JE. Class 1 integrons in resistant Escherichia coli and Klebsiella spp., US Hospitals. Emerg Infect Dis. 2006;12:1011–1014. doi: 10.3201/eid1206.051596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: Integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Quian Y, Chen S, Wang P, Huang Y. Incidence of extended spectrum β-lactamases and characterization of integrons in extended spectrum β-lactamase producing Klebsiella pneumoniae isolated in Shantou, China. Acta Biochim Biophys Sin. 2007;39:527–532. doi: 10.1111/j.1745-7270.2007.00304.x. [DOI] [PubMed] [Google Scholar]