Abstract
This study describes the clinical, histopathological, and virological characterization of teat papillomatosis from Brazilian dairy cattle herds. Four types of bovine papillomavirus were identified (BPV6, 7, 9, and 10); one of these (BPV7) is being detected for the first time in Brazilian cattle.
Keywords: papilloma, BPV, genotyping
Papillomaviruses (PVs) are small (52–55 nm), non-enveloped, double-stranded DNA oncoviruses that replicate in the nucleus of squamous epithelial cells and can induce warts in the skin and mucosal epithelia of most higher vertebrate species. Some specific viral types have the potential to cause malignant progression in papillomatous lesions of animals and humans (Antonsson et al., 2002; de Villiers et al., 2004). In cattle, bovine papillomavirus (BPV) is the etiological agent of cutaneous and teat papillomatosis, cancers of the upper gastrointestinal tract and urinary bladder (Campo 2002; Borzacchiello et al., 2003; Wosiacki et al., 2006).
BPV produces characteristic gross lesions that are either exophytic (proliferating outwards) or endophytic (inverted) and are composed of a hyperplastic epithelium supported by a discrete dermal tissue containing dilated capillaries (Ginn et al., 2007; Hargis et al., 2012).
Currently, the genomes of thirteen BPVs (BPV1 to 13) have been fully characterized. They are classified into four genera based on their genome relatedness and biological properties (de Villiers et al., 2004). These genera are Deltapapillomavirus (BPV1, 2, and 13), Xipapillomavirus (BPV3, 4, 6, 9, 10, 11, and 12), Epsilonpapillomavirus (BPV5 and 8), and a yet unnamed PV genus (BPV7) (Bernard et al., 2010; Hatama et al., 2011; Zhu et al., 2012; Lunardi et al., 2013). Moreover, the occurrence of numerous additional viral types has been proposed based on partial nucleotide sequence analysis of the major capsid protein L1, obtained from both benign cutaneous lesions and swab samples of healthy skin from cattle (Antonsson and Hansson 2002; Maeda et al., 2007; Ogawa et al., 2007; Claus et al., 2008; Claus et al., 2009c).
Teat papillomatosis has been reported in dairy herds worldwide as a cattle health problem resulting in economic losses (Campo 2003). While milking can become difficult in markedly affected individuals, ulceration and rupture of established cutaneous lesions might predispose dairy cattle to mastitis and distortion of the milk ducts. Additionally, the maintenance of affected cows with alteration in mammary gland shape and/or even of herds with a high number of affected animals may prevent economic profits in the dairy industry (Campo 2002; Borzacchiello et al., 2008).
Although papilloma lesions found in teats and udders can be caused by diverse BPV types, the BPV6 has often been identified in this anatomical location. Therefore, the association of BPV1, 3, 5, 7, 8, 9, 10, 11, and other types still to be characterized, have been confirmed in mammary glands of cattle (Campo et al., 1981; Jarrett et al., 1984; Ogawa et al., 2004; Claus et al., 2007; Maeda et al., 2007; Claus et al., 2008).
Despite the high number of cattle herds affected by cutaneous papillomatosis in several geographical regions of Brazil, studies investigating BPV diversity associated with different clinical outcomes are sporadic (Stocco dos Santos et al., 1998; Freitas et al., 2003; Wosiacki et al., 2005; Wosiacki et al., 2006; Claus et al., 2008; Claus et al., 2009b; Claus et al., 2009c; Lunardi et al., 2010).
The aim of this report is to describe the identification of BPVs 6, 7, 9, and 10 in association with cases of teat papillomatosis from two dairy cattle herds of Brazil. This study represents the first detection of the BPV7 in Brazilian cattle.
Seven exophytic teat papillomas were individually collected from three dairy cows belonging to two different cattle herds from the Northern region of Parana state in Southern Brazil. These lesions were observed on different parts of the teat and coded for identification. A portion of each lesion was fixed in 10% buffered formalin solution and routinely processed for histopathological evaluation. Fragments from each papilloma specimen were grounded in phosphate-buffered saline solution (PBSpH 7.2) and suspensions (10–20% w/v) were centrifuged for 15 min at 3,000 × g at 4 °C. Aliquots (250 μL) from supernatant were treated with lysis buffer (10 mM Tris; 1 mM EDTA; 0.5% Nonidet P40; 1% SDS; and 0.2 mg/mL proteinase K) (Invitrogen, Life Technologies, Carlsbad, CA, USA). After homogenization, the samples were incubated at 56 °C for 30 min. For DNA extraction, a combination of the phenol/chloroform/isoamyl alcohol and silica/guanidine iso-thiocyanate methods was performed as previously described (Alfieri et al., 2006). DNA was eluted in 50 μL of ultrapure sterile water and kept at −20 °C until used. Aliquots of ultrapure sterile water were included as negative controls in the DNA extraction procedures.
The PCR assay was carried out by using the primer pair FAP59 (forward: 5′-TAACWGTIGGICAYCCWTATT-3′) and FAP64 (reverse: 5′-CCWATATCWVHCATITCICCATC-3′) (Forslund et al., 1999), with modifications (Claus et al., 2007). Aliquots from the PCR products were analyzed by electrophoresis in a 2% agarose gel in TBE buffer, pH 8.4 (89 mM Tris; 89 mM boric acid; 2 mM EDTA) at constant voltage (90 V) for approximately 45 min, stained with ethidium bromide (0.5 μg/mL), and visualized under UV light.
PCR bands suggestive of PV amplification were excised from the agarose gel and purified by using illustra GFX PCR DNA and Gel Band Purification kit (GE Healthcare, Little Chalfont, UK). Direct sequencing was then performed in the 3500 Genetic Analyzer (Applied Biosystems, Carlsbad, USA), with the FAP59 and FAP64 primers. The obtained sequences were examined with the PHRED software for quality analysis of chromatogram readings. The sequences were accepted if the base quality was equal to or higher than 20. Consensus sequences were determined by using the CAP3 software, and the sequence identity was verified with all sequences deposited in the GenBank by using the BLAST software. The guidelines of the Papillomavirus Nomenclature Committee 1995 (14th International Papillomavirus Conference, Quebec City, Quebec, Canada) were followed to identify PV types (de Villiers et al., 2004).
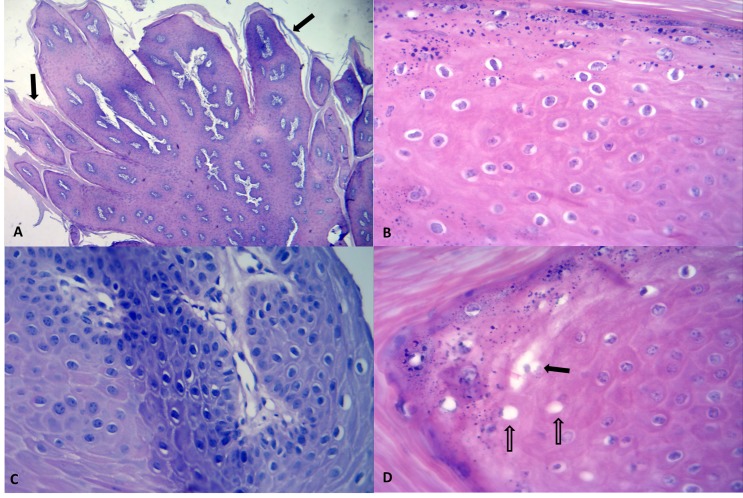
Grossly, all cutaneous lesions were exophytic and were histologically classified as benign squamous cell neoplasms (cutaneous papilloma) irrespective of the type of virus isolated. Microscopically, all tumorous growths demonstrated similar histological features; being characterized by varying degrees of hyperkeratosis or parakeratosis with elongated digital-like proliferation of the squamous epithelium (Figure 1A). In all tumors fragments evaluated, most keratinocytes within the stratum spinosum demonstrated clear perinuclear halo, some having pyknotic nucleus (characterized as koilocytes) others revealed discrete ballooning degeneration; in some areas, two or more adjacent degenerated cells fused to produce microvesciles (Figure 1B–D). Further, there was acanthosis, reduced mitotic index (1–2 per 40× Obj.), foci of apoptosis of squamous epithelium, and severe accumulations of irregular keratohyalin granules within cells of the stratum granulosum. However, characteristic basophilic intranuclear viral inclusion bodies were not observed.
Figure 1.
Histopathological features of teat papillomas; skin, mammary gland, cow. There is outward digital-like proliferation of the squamous epithelium with hyperkeratosis (arrows) of the stratum corneum (A). Observe that the nucleuses of most keratinocytes are surrounded by a clear halo (B) and the severely condensed basophilic-staining (pyknotic) nucleus (C) of the koilocytes. There are several degenerated keratinocytes (open arrows) and observe some that adjacent swollen cells are coalesced into a larger microvescile (closed arrow) (D). (A, Hematoxylin and eosin, 4 × Obj.; B–D, Hematoxylin and eosin, 40 × Obj.).
Amplicons of the expected length, approximately 480 bp, were obtained from each of the seven teat papilloma DNA samples, via the FAP PCR assay. Negative controls were not amplified.
Through direct sequencing of the FAP products, BPV6, 7, 9, and 10 were identified in the examined papilloma specimens. Two cows with more than one teat papillomas evaluated were coinfected by different BPV types (BPV7 and 10, and BPV6 and 9, respectively). The BPV strains identified were designated as BPV6/BR-UEL, BPV7/BR-UEL, BPV9/BR-UEL, and BPV10/BR-UEL (GenBank accession numbers: HM245430, HM245431, HM245433, and HM245432, respectively).
The gross characteristics of the cutaneous lesions as well as the PV type classification and GenBank accession numbers of the obtained sequences are indicated in Table 1. However, an association between the macroscopic characteristics and the viral type present in the cutaneous teat lesions was not observed in our analysis.
Table 1.
Lesion characteristics and classification of BPV strains identified in teat lesions from dairy cattle herds.
| Herd | Animal | Lesion | Macroscopic characteristics of true papillomas | Nucleotide sequence | |
|---|---|---|---|---|---|
|
|
|||||
| BPV classification | GenBank accession number* | ||||
| 1 | A | A1 | Filiform | 7 | HM245431 |
| B | B1 | Cauliflower | 7 | - | |
| B2 | Cauliflower | 10 | HM245432 | ||
| B3 | Cauliflower | 10 | - | ||
| 2 | C | C1 | Cauliflower | 6 | HM245430 |
| C2 | Flat and round | 6 | - | ||
| C3 | Flat and round | 9 | HM245433 | ||
A single representative of each BPV type had its sequence deposited at the GenBank database.
BPV types 7, 9, and 10, in addition to BPV type 6, were identified as causative agents of warts in dairy cow teats. The histopathological findings of this study are consistent with those lesions associated with BPV (Ginn et al., 2007; Hargis and Ginn 2012). Although viral inclusions were not observed, the characteristic cytopathic effects induced by this virus (Ginn et al., 2007; Hargis and Ginn 2012) was identified in all tissue fragments evaluated.
The current taxonomic classification system for the Papillomaviridae family has recently proposed the introduction of the taxonomic terms “species” and “genus” for this viral family, establishing the relationship between phylogenetic assemblages and biological and pathological properties shared by viruses grouped together (de Villiers et al., 2004). The three Xipapillomavirus representatives (BPV6, 9, and 10) identified in our study were found determining true papillomas in teat skin. These histological features are in agreement with the biological properties described for this specific genus (de Villiers et al., 2004). Regarding the histological characteristics of BPV7-induced cutaneous lesions, this viral type also gave rise to true epithelial papilloma at teat skin. The histological analyses of cutaneous lesions determined by BPV7 were lacking in the study that established its phylogenetic position and characterized its genome (Ogawa et al., 2007). Therefore, based on our findings, it is likely that future isolates that are to be grouped with BPV7 in its yet unnamed genus should also be able to induce true cutaneous papillomas in cattle.
The identification of four different viral types in teat warts herein evaluated demonstrates the high diversity in BPV types involved in this specific condition. Although BPV6 is the viral type that is classically associated with teat papilloma worldwide, our findings of diverse BPV types are in agreement with the results obtained by other studies investigating viral diversity in Japanese cattle herds affected by teat papillomatosis. Such investigations have confirmed the high BPV diversity that occurs in benign teat lesions and healthy teat skin, in a commensal form (Campo 2002; Ogawa et al., 2004; Maeda et al., 2007). To the best of the authors’ knowledge, this study represents the first identification of the BPV type 7 in cattle from South America. The identification of this BPV type in a geographical region not related to the location where it was first isolated, Japan, supports the hypothesis that this viral type is widespread and affect dairy cattle herds worldwide (Ogawa et al., 2007; Hatama et al., 2008). Previous studies involving the identification of BPV types 9 and 10 have documented that these viral types are easily detected from naturally occurring teat papilloma but not from healthy teat skin (Ogawa et al., 2004; Maeda et al., 2007; Hatama et al., 2008). This observation correlates well with our findings that confirm the specific association of these viral types with teat papillomatosis. More recently, the tumorigenic potential of BPV9 was tested through experimental infection of the teat skin of heifers, resulting in the development of papillomatosis (Hatama et al., 2009). Conversely, BPV7 has been detected mainly from swab samples of healthy teat skin, and besides the identification herein reported, it was identified in only a few teat lesions from dairy cattle in Japan (Ogawa et al., 2004).
The identification of BPV7 in teat papillomas from Brazilian cattle coupled with data obtained in a recent study involving genotyping of viral strains associated with cutaneous warts from other anatomical sites from cattle from the northeastern region, clearly indicates that almost all fully characterized BPV types (BPV1 to 11) are circulating in Brazilian herds (Carvalho et al., 2012). Besides the high diversity verified in affected herds, the presence of multiple viral types in different lesions evaluated from a single individual was demonstrated in this study. Previously, other investigations showed that the occurrence of coinfections in a single lesion, adding up two to six viral types, is a common field event (Claus et al., 2009a; Schmitt et al., 2010; Carvalho et al., 2012). Surprisingly, we found that BPV6 and 7 produced lesions with different gross appearances. Whether the differences observed are associated with the presence of undetected viral types establishing coinfections is yet to be clarified.
The limited number of recent studies aiming to identify BPV types associated with lesions of the mammary gland of cattle has reported a considerable diversity of the viral types involved. Therefore, the viral type (BPV6) that has been classically associated with this anatomical location is not the only causative agent of these lesions in affected herds.
Detailed epidemiological investigations conducted in different geographical areas are required to define the most prevalent BPV types associated with this disease. Since the host immunity against PV infections is type-specific, the availability of such information would allow the elaboration of a multivalent immunogen based on antigens from the viral types capable of causing cutaneous papillomas. It is noteworthy to remember that currently there are no commercially available alternatives for the effective treatment or prevention of teat papillomatosis.
Acknowledgments
The authors thank the Brazilian Institutes CNPq, CAPES, FINEP, and Fundaçao Araucaria (FAP/PR) for financial support. Alfieri, A.A., Headley, S.A., and Alfieri, A.F. are recipients of CNPq fellowships.
References
- Alfieri AA, Parazzi ME, Takiuchi E, Medici KC, Alfieri AF. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998–2002. Trop Anim Health Prod. 2006;38:521–526. doi: 10.1007/s11250-006-4349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Hansson BG. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H-U, Burk RD, Chen Z, van Doorslaer K, Hausen Hz, de Villiers E-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzacchiello G, Iovane G, Marcante ML, Poggiali F, Roperto F, Roperto S, Venuti A. Presence of bovine papillomavirus type 2 DNA and expression of the viral oncoprotein E5 in naturally occurring urinary bladder tumours in cows. J Gen Virol. 2003;84:2921–2926. doi: 10.1099/vir.0.19412-0. [DOI] [PubMed] [Google Scholar]
- Borzacchiello G, Roperto F. Bovine papillomaviruses, papillomas and cancer in cattle. Veterinary Rresearch. 2008;39:45. doi: 10.1051/vetres:2008022. [DOI] [PubMed] [Google Scholar]
- Campo MS. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Campo MS. Papillomavirus and disease in humans and animals. Vet Comp Oncol. 2003;1:3–14. doi: 10.1046/j.1476-5829.2003.00001.x. [DOI] [PubMed] [Google Scholar]
- Campo MS, Moar MH, Laird HM, Jarrett WF. Molecular heterogeneity and lesion site specificity of cutaneous bovine papillomaviruses. Virology. 1981;113:323–335. doi: 10.1016/0042-6822(81)90159-8. [DOI] [PubMed] [Google Scholar]
- Carvalho CC, Batista MV, Silva MA, Balbino VQ, Freitas AC. Detection of bovine papillomavirus types, co-infection and a putative new BPV11 subtype in cattle. Transbound Emerg Dis. 2012;59:441–447. doi: 10.1111/j.1865-1682.2011.01296.x. [DOI] [PubMed] [Google Scholar]
- Claus MP, Lunardi M, Alfieri AA, Otonel RAA, Ferracin LM, Fungaro MHP, Alfieri AF. A bovine teat papilloma specimen harboring Deltapapillomavirus (BPV-1) and Xipapillomavirus (BPV-6) representatives. Braz Arch Biol Technol. 2009a;52:87–91. [Google Scholar]
- Claus MP, Lunardi M, Alfieri AA, Otonel RAA, Sartori D, Fungaro MHP, Alfieri AF. Multiple bovine papillomavirus infections associated with cutaneous papillomatosis in brazilian cattle herds. Braz Arch Biol Technol. 2009b;52:93–98. [Google Scholar]
- Claus MP, Lunardi M, Alfieri AF, Ferracin LM, Fungaro MH, Alfieri AA. Identification of unreported putative new bovine papillomavirus types in Brazilian cattle herds. Vet Microbiol. 2008;132:396–401. doi: 10.1016/j.vetmic.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Claus MP, Lunardi M, Alfieri AF, Sartori D, Fungaro MHP, Alfieri AA. Identification of the recently described new type of bovine papillomavirus (BPV-8) in a Brazilian beef cattle herd. Pesq Vet Bras. 2009c;29:25–28. [Google Scholar]
- Claus MP, Vivian D, Lunardi M, Alfieri AF, Alfieri AA. Phylogenetic analysis of bovine papillomavirus associated with skin warts in cattle herds from the state of Parana. Pesq Vet Bras. 2007;27:314–318. [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80(Pt 9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Freitas ACd, Carvalho Cd, Brunner O, Birgel-Junior EH, Dellalibera AMMP, Benesi FJ, Gregory L, Beçak W, Santos RdCSd. Viral DNA sequences in peripheral blood and vertical transmission of the virus: a discussion about BPV-1. Braz J Microbiol. 2003;34:76–78. [Google Scholar]
- Ginn PE, Mansell JEKL, Rakich PM. Skin and appendages. In: Maxie MG, editor. Jubb, Kennedy, and Palmer’s Pathology of Domestic Animals. Vol. 1. Saunders/Elsevier; Philadelphia: 2007. pp. 748–751. [Google Scholar]
- Hargis AM, Ginn PE. The integument. In: Zachary JF, McGavin MD, editors. Pathologic Basis of Veterinary Disease. Elsevier/Mosby; St. Louis: 2012. pp. 1025–1027. [Google Scholar]
- Hatama S, Ishihara R, Ueda Y, Kanno T, Uchida I. Detection of a novel bovine papillomavirus type 11 (BPV-11) using xipapillomavirus consensus polymerase chain reaction primers. Arch Virol. 2011;156:1281–1285. doi: 10.1007/s00705-011-0970-7. [DOI] [PubMed] [Google Scholar]
- Hatama S, Nishida T, Kadota K, Uchida I, Kanno T. Bovine papillomavirus type 9 induces epithelial papillomas on the teat skin of heifers. Vet Microbiol. 2009;136:347–351. doi: 10.1016/j.vetmic.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Hatama S, Nobumoto K, Kanno T. Genomic and phylogenetic analysis of two novel bovine papillomaviruses, BPV-9 and BPV-10. J Gen Virol. 2008;89:158–163. doi: 10.1099/vir.0.83334-0. [DOI] [PubMed] [Google Scholar]
- Jarrett WFH, Campo MS, Oweil BW, Laird HM, Coggins LW. A novel bovine papillomavirus (BPV-6) causing true epithelial papillomas of the mammary gland skin: A member of a proposed new BPV subgroup. Virology. 1984;136:255–264. doi: 10.1016/0042-6822(84)90162-4. [DOI] [PubMed] [Google Scholar]
- Lunardi M, Alfieri AA, Otonel RA, de Alcantara BK, Rodrigues WB, de Miranda AB, Alfieri AF. Genetic characterization of a novel bovine papillomavirus member of the Deltapapillomavirus genus. Vet Microbiol. 2013;162:207–213. doi: 10.1016/j.vetmic.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Lunardi M, Claus MP, Alfieri AA, Fungaro MHP, Alfieri AF. Phylogenetic position of an uncharacterized Brazilian strain of bovine papillomavirus in the genus Xipapillomavirus based on sequencing of the L1 open reading frame. Genet Mol Biol. 2010;33:745–749. doi: 10.1590/S1415-47572010005000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Shibahara T, Wada Y, Kadota K, Kanno T, Uchida I, Hatama S. An outbreak of teat papillomatosis in cattle caused by bovine papilloma virus (BPV) type 6 and unclassified BPVs. Vet Microbiol. 2007;121:242–248. doi: 10.1016/j.vetmic.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Tomita Y, Okada M, Shinozaki K, Kubonoya H, Kaiho I, Shirasawa H. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. Journal of general virology. 2004;85:2191–2197. doi: 10.1099/vir.0.80086-0. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Tomita Y, Okada M, Shirasawa H. Complete genome and phylogenetic position of bovine papillomavirus type 7. J Gen Virol. 2007;88:1934–1938. doi: 10.1099/vir.0.82794-0. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Fiedler V, Muller M. Prevalence of BPV genotypes in a German cowshed determined by a novel multiplex BPV genotyping assay. J Virol Methods. 2010;170:67–72. doi: 10.1016/j.jviromet.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Stocco dos Santos RC, Lindsey CJ, Ferraz OP, Pinto JR, Mirandola RS, Benesi FJ, Birgel EH, Pereira CA, Becak W. Bovine papillomavirus transmission and chromosomal aberrations: an experimental model. J Gen Virol. 1998;79(Pt 9):2127–2135. doi: 10.1099/0022-1317-79-9-2127. [DOI] [PubMed] [Google Scholar]
- Wosiacki SR, Barreiro MA, Alfieri AF, Alfieri AA. Semi-nested PCR for detection and typing of bovine Papillomavirus type 2 in urinary bladder and whole blood from cattle with enzootic haematuria. J Virol Methods. 2005;126:215–219. doi: 10.1016/j.jviromet.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Wosiacki SR, Claus MP, Alfieri AF, Alfieri AA. Bovine papillomavirus type 2 detection in the urinary bladder of cattle with chronic enzootic haematuria. Mem Inst Oswaldo Cruz. 2006;101:635–638. doi: 10.1590/s0074-02762006000600009. [DOI] [PubMed] [Google Scholar]
- Zhu W, Dong J, Shimizu E, Hatama S, Kadota K, Goto Y, Haga T. Characterization of novel bovine papillomavirus type 12 (BPV-12) causing epithelial papilloma. Arch Virol. 2012;157:85–91. doi: 10.1007/s00705-011-1140-7. [DOI] [PubMed] [Google Scholar]