Abstract
The mangrove ecosystem is an unexplored source for biotechnological applications. In this unique environment, endemic bacteria have the ability to thrive in the harsh environmental conditions (salinity and anaerobiosis), and act in the degradation of organic matter, promoting nutrient cycles. Thus, this study aimed to assess the cellulolytic activities of bacterial groups present in the sediment from a mangrove located in Ilha do Cardoso (SP, Brazil). To optimize the isolation of cellulolytic bacteria, enrichments in two types of culture media (tryptone broth and minimum salt medium), both supplemented with 5% NaCl and 1% of cellulose, were performed. Tests conducted with the obtained colonies showed a higher occurrence of endoglycolytic activity (33 isolates) than exoglycolytic (19 isolates), and the degradation activity was shown to be modulated by the presence of NaCl. The isolated bacteria were clustered by BOX-PCR and further classified on the basis of partial 16S rRNA sequences as Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, Firmicutes or Bacteroidetes. Therefore, this study highlights the importance of studies focusing on the endemic species found in mangroves to exploit them as novel biotechnological tools for the degradation of cellulose.
Keywords: cellulose; endo-1,4-β-D-glucanase; exo-1,4-β-D-glucanase; salinity
Introduction
Mangrove ecosystems are geographically widely distributed, covering approximately 60 to 75% of the shoreline in tropical and sub-tropical rainforests. These unique ecosystems are most abundant in Brazil, Indonesia and Australia, and in Latin America, mangroves cover approximately 400,000 hectares (Hoguin et al., 2001). On the southeastern Brazilian shore, the state of São Paulo has an area of 125 km2 covered by mangroves, which are distributed from Ubatuba (north coast) to the city of Cananéia (south coast) (Said and Pietro, 2004).
These ecosystems reveal significant biological diversity of fish, crustaceans, mollusks, birds, reptiles and mammals (Lankau and Strauss, 2007). This diversity requires a high availability of nutrients at the beginning of the food web (Dias et al., 2009), what is provided by microbes that are adapted to the variability of salinity and the low availability of oxygen within the mangrove sediment (Lane et al., 1985; Taketani et al., 2010). Such microorganisms constitute the major route for nutrient cycling by aiding in the decomposition of organic matter and performing mineral transformations (Flores-Mireles et al., 2007). Thus, one can conclude that the microbial communities form the nutritional basis of this ecosystem (Hoguin et al., 2001).
In addition to the processes of photosynthesis performed by mangrove plants, another source of carbon in this environment is the degradation of organic matter, mainly achieved by the senescence of plants and the releases of plant roots. Cellulolytic microorganisms can perform the degradation of cellulose-based compounds, resulting in the generation of simple-sugar derivatives in these sediments (Hanson and Hanson, 1996; Tengerdy and Szakacs, 2003). From a biotechnological perspective, the unique environmental conditions present in mangroves may serve as a selective pressure for the generation of unique cellulolytic organisms that are able to degrade cellulose in harsh environments (Wilson, 2009). The literature alludes to the usefulness of mangrove species, like bacteria, to produce esterase, lipases and other enzymes (Dias et al., 2009).
The cellulases are the second largest group of carbohydrases that have been commercially exploited (Angelo, 2004), mainly due to the high specificity and efficiency of degradation. The conversion of cellulose into simple forms of carbon may be useful for the production of bioenergy (Bisaria and Ghose, 1981; Schallmey et al., 2004). In general, the degradation of cellulose occurs by the induction of three types of enzymes: endo-1,4-β-D-glucanase (endocellulase), exo-1,4-β-D-glucanase (exocellulase) and β-glucosidase (cellubiose hydrolase) (Eveleigh, 1981; Hamada et al., 1999). Enzymes called endoglucanases or endocellulases represent a group of cellulases active in the cleavage of random regions within cellulose, producing oligosaccha-rides with variable sizes. The exocellulases or exoglucanases are also known as cellodextrinases and act on the terminals of oligosaccharide chains generated by endocellulases, releasing glucose or cellobiose. Thus, cellulases are capable of breaking the glycosidic bonds of cellulose microfibrils, resulting in the release of oligosaccharides and improving the digestibility of cellulose (Dillon, 2004).
Thus, the discovery of bacteria that produce enzymes with cellulolytic activity in environments with unique features, such as mangroves, is of great interest to broaden the genetic base on which biotechnological processes can be explored (Akhtar et al., 2008; Thauer and Shima, 2006). The present work aimed to screen the cellulolytic activity of endo- and exoglucanases in bacteria isolated from mangrove sediment by the enrichment of samples collected in a preserved area at the Ilha do Cardoso (Cananéia, Brazil).
Materials and Methods
Sample analysis
The samples used in this study were derived from sediments collected in a mangrove located at the Ilha do Cardoso in the city of Cananéia (SP, Brazil). The samples were collected from the superficial sediment (0–10 cm), aseptically stored in sterilized plastic bags and transported to the laboratory at 4 ºC. A total of five sub-samples were collected in the environment. These samples were later homogenized, resulting in one composite sample.
Enrichment methodology
Enrichments were performed in triplicate using aliquots of five grams of sediment. Flasks containing 50 mL of two types of culture media were used for sediment enrichment: Trypic Soy Broth (TSB) at 5% of the recommended concentration (1.5 g.L−1) and MM (6.0 g.L−1 NaNO3, 1.5 g.L−1 KH2PO4, 0.5 g.L−1 KCl, 0.5 g.L−1 MgSO4, 0.01 g.L−1 FeSO4 and 0.01 g.L−1 ZnSO4). These flasks were also supplemented with NaCl (5%) and cellulose (1%) (Cellulose Powder, Sigma) or glucose (1% in control tubes) as the carbon sources. In total, 12 flasks were used for enrichments (2 medium × 2 treatments × 3 replicates), which were incubated at 28ºC with shaking at 150 rpm. The samples were then transferred to new culture medium (10% v/v) at periods of 3, 6, 12 and 24 days after the initiation of the enrichment and incubated at the same conditions. This methodology is based on the enrichment of bacterial populations that are able to utilize cellulose as a carbon source.
Isolation of bacteria from the enrichments
During the transfer of enrichments at days 12 and 24 after setup, aliquots were used for bacterial isolation. Serial dilutions (10−1 to 10−5) from the aliquots were used to inoculate Petri dishes containing the same media used in the enrichment but supplemented with 1.5% (w/v) of agar. The plates were incubated at the same temperature at which the enrichments were performed. After 7 days of incubation, the development of bacterial colonies was recorded. Additionally, colonies from each treatment were randomly selected, comprising a total of 132 isolates. The number of 33 isolates from each enrichment was selected to allow the BOX-PCR analysis in one gel lane, and also to represent roughly half of the colonies obtained from each enrichment plating. These isolates were grown in liquid media and preserved in a 20% glycerol solution at −80 ºC.
Evaluation of cellulolytic activities of isolates
Determination of the capacity of the isolates in degrading cellulose was indirectly inferred by the use of two marker molecules: the degradation of carboxymethyl cellulose (CMC), indicating the endoglycolytic activity, and the degradation of Avicel, providing evidence regarding the exoglycolytic activity. To determine the production of endoglucanases, the isolates were grown on medium containing CMC (0.5 g.L−1 NaNO3, 1.0 g.L−1 K2HPO4, 0.5 g.L−1 MgSO4.7H2O, 0.01 g.L−1 FeSO4, 1.0 g.L−1 yeast extract, 10.0 g.L−1 CMC, and 15.0 g.L−1 agar), and incubated at 28 ºC for 72 h. After bacterial growth was detected, 5 mL of 1% iodine solution was added to each plate, and the presence of a stainless halo around the colony indicated production of the enzyme (Kasana et al., 2008). A semi-quantitative approach was taken using the values obtained by the ratio of the halo size (enzyme activity) and diameter of the bacterial colony, resulting in what is named here as the enzymatic index (Lima et al., 2005).
Similarly, the exoglycolytic activity was estimated by the cultivation of isolates in plates containing MM medium supplemented with microcrystalline cellulose-avicel (6.0 g.L−1 NaNO3, 1.5 g.L−1 KH2PO4, 0.5 g.L−1 KCl, 0.5 g.L−1 MgSO4.7H2O, traces of FeSO4, ZnSO4, 5.0 g.L−1 Avicel and 15.0 g.L−1 agar). In this case, the observation of halos was not possible, and the presence of the exoglucanases was shown by the development of bacterial colonies. To rule out the possibility that the bacteria were growing by using a residual carbon source from the previous culture media, isolates were subjected to three replications to guarantee the use of avicel as a carbon source.
Additionally, a test was performed to evaluate the response of the isolates to salinity shifting. The test was based on the endoglycolytic activity (CMC) using the same technique described above except that media lacking or containing 5% NaCl (w/v) was also compared. The induction or repression of endoglycolytic activity was considered to have occurred when the enzymatic index was doubled or diminished by half of that observed in the medium free of NaCl, respectively.
Identification of isolates
Identification of the isolates was based on partial sequencing of the 16S rDNA gene. DNA was extracted by the commonly used bead beating methodology and purified with phenol:chloroform. Briefly, cells were suspended in 500 μL of TE buffer, and lysis was promoted by adding 0.1 g of glass beads (0.1 mm) and 5 μL of 10% SDS. This mixture was agitated by a bead beater for 30 seconds. After lysis, DNA purification was performed by extractions in phenol and chloroform. The DNA obtained was precipitated with isopropanol, dried and re-suspended in deionized water. After extraction, the integrity and quality of the DNA obtained was verified by agarose gel electrophoresis (1% w/v) followed by ethidium bromide staining and visualization using UV light.
Prior to sequencing, the isolates were clustered into groups using the whole genome profiling technique, BOX-PCR (Soto-Ramírez et al., 2010). BOX-PCR was performed using approximately 50 ng of genomic DNA added to 25 μL PCR reactions containing 1 mM dNTPs, 3 mM MgCl2, 1× Taq buffer, 0.4 mM primer BOX-1AR (5′-CTA CGG CAA GGC GAC GCT GAC G -3′), 10% DMSO (2.5 μL), and 2 U of Taq DNA polymerase. The amplification began with one cycle of denaturation at 95 ºC for 2 min that was followed by 35 cycles of 94 ºC for 2 s, 92 ºC for 30 s, 50 ºC for 1 min and 65 ºC for 8 min. After all of the cycles were complete, a final extension step was performed at 65 ºC for 10 min.
The resulting amplicons were separated on a 2% agarose gel for 2 hours and 40 min, stained with ethidium bromide and visualized under UV light. Clustering of the isolates was performed based on the BOX-PCR patterns observed after scanning the agarose gel images at Bionumerics (Applied Maths, Belgium), where the band profiles were compared and clustered by UPGMA based on a Pearson correlation analysis (Tacão et al., 2005).
Bacterial isolates representing the clusters defined by BOX-PCR analysis were used for identification based on the sequencing of approximately 500 bp covering the variable regions V6-V8 of the 16S rDNA gene. PCR products generated with primers P027 (5′-GAG AGT TTG ATC CTG GCT CAG -3′) and R1387 (5′-CGG TGT GTA CAA GGC CCG GGA ACG -3′) (Kathiresan and Bingham, 2001) were purified using the PowerCleanTM DNA Clean-Up Kit (MoBio Laboratories, USA). The resulting DNA fragments were sequenced using an automated sequencer with the reverse primer (R1387) as the basis for the sequencing reaction (ABI Prism377, PE Applied Bio-systems, Foster City, CA, USA). The obtained sequences were assessed for quality and subjected to similarity analysis using RDPQuery and BlastN analysis by GenBank (nr/nt). The best matches from the comparisons were retrieved from the databases to compose the dataset used in the phylogenetic approach. The sequences of the isolates were deposited in GenBank under the accession numbers EF488520 to EF488531.
For the phylogenetic analysis, the sequences were aligned and further clustered by a neighbor-joining analysis based on the Kimura-2 parameters from a study conducted using MEGA version 4.0 (Tamura et al., 2003), which aided in the determination of the preferred phylogenetic tree. The analysis was supported by bootstrap values and was based on an analysis of 1,000 subsamples.
Results and Discussion
In 1981, Bisaria and Ghose (Bisaria and Ghose, 1981) proposed that cellulose was the only renewable energy source capable of meeting the long-term demands of modern society and in a sustainable manner. However, cellulose is a complex source of organic carbon and often has a high molecular weight. For this reason, the use of cellulose requires the action of enzymes that assist in its degradation and that can function under varying conditions of temperature, pressure and salinity (Angelo, 2004; Taketani et al., 2010).
The combination of new methodologies and the exploitation of unexplored environments are the most important factors that may lead to an increase in the efficiency in the degradation of cellulose by enzymatic activities. Mesbah and Wiegel (2008) performed studies in saline lakes throughout the world using culture-independent technologies to identify cellulolytic genes in environmental DNA samples, which improved the description of cellulase-codifier genes. In this study, we analyzed the cellulose degradation capability of a collection of isolates retrieved from mangroves and obtained by enrichments with cellulose powder in an attempt to discover their biotechnological potential.
Enrichment and isolation of bacteria
The enrichment was successfully carried out in accordance with the methodology described in the materials and methods section. It was possible to observe the development of microorganisms in all culture media under the distinct levels of salinity imposed. Some important visual observations could be made in bottles where the enrichment was carried out in which flasks with 5% NaCl revealed the formation of biofilms on the glass walls. This finding corroborates previous observations made by Sá and Melo (2008), who described biofilm formation by cellulolytic bacteria in roots of Rhizophora mangle (a typical plant found in mangroves), and identified biofilm formation as an adaptation strategy to the changing salinity conditions in this niche. Concerning the isolation of bacteria, after 12 and 24 days of enrichment, the density of the bacterial communities was between 18 and 30 colonies per plate in the 10−4 dilution. Colonies were selected from these plates and composed a collection of 132 isolates (Table 1).
Table 1.
Composition of the collection of bacterial isolates used in genotypic characterization and the production of cellulases. MM indicates minimum medium and CP indicates the addition of cellulose powder.
| Enrichment | Codes | Nº of isolates | Activities | Endo/exoglucanase activities | |
|---|---|---|---|---|---|
|
| |||||
| endoglucanase | exoglucanase | ||||
| TSB5%+NaCl+CP | TNC | 33 | 12 | 4 | 1 |
| TSB5%+NaCl | TN | 33 | 7 | 4 | 1 |
| MM+NaCl+CP | MNC | 33 | 7 | 4 | 1 |
| MM+NaCl | MN | 33 | 6 | 6 | 0 |
Detection of the cellulolytic activities of the isolates
Characterization of the 132 isolates was performed using a test for the production of endoglucanase and exoglucanase evidenced by the degradation of CMC and microcrystalline cellulose (Avicel), respectively. The first important observation is that there was a higher occurrence of isolates with endoglycolytic activity (32 isolates) than exoglycolytic activity (18 isolates) (Table 1). This trend has been recurrently observed in many studies focusing on cellulolytic enzymes (Bisaria and Ghose, 1981; Leschine, 1995; Sinegani and Mahohi, 2010). Although it is believed that in the environment it would be advantageous for an organism to degrade cellulose in its entirety (endo- and exoglycolytic activity), this finding is not commonly observed. Morais et al. (Morais et al., 2010) observed that the combinations of endocellulase and exocellulase can lead to a rise in the cellulose degradation activity by synergy, working on both internal and external cleavage of the cellulose molecules. One possible explanation for the higher occurrence of endoglucanases is that bacteria produce only endoglucanases and do not use this carbon source as glucose for primary metabolism but instead make use of the oligosaccharides by alternative metabolism.
Comparing the cultivation media, the TSB was as efficient as MM for the isolation of cellulolytic bacteria, possibly indicating that the isolates present in these environments have endoglycolytic activity as a complementary form of their carbon nutrition. As described by Said and Pietro (2004), enzymes are considered biological catalysts composed of protein molecules and are produced by living cells. Therefore, the small amount of carbon compounds in the environment serve to supply the basal cell metabolism, which uses this feature for the production of enzymes that degrade cellulose and show high catalytic activity and selectivity to the specific substrate used (Bhat, 2000).
Characterization of the endoglycolytic activity of isolates submitted to the salinity variation
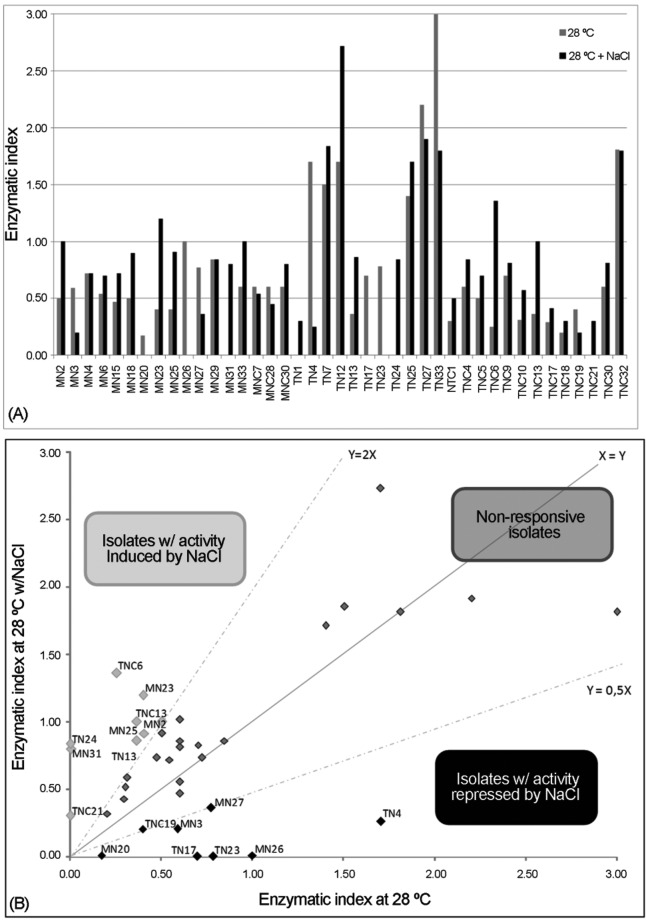
The 33 mangrove isolates with endoglycolytic activity (CMC degradation) were assessed for their sensitivity to changes in salinity. This test has revealed that of the 33 isolates evaluated, eight (TN4, MN26, MN27, TN23, TN17, MN3, TNC19 and MN20) have a reduced ability to degrade CMC when NaCl was added, while nine (TNC6, MN23, TNC13, MN25, TN13, TN24, MN31, TNC21 and MN2) isolates have an increased ability (Figure 1). This result suggests that, depending on the salinity present in mangroves (mainly imposed by the tidal regime), distinct bacteria can act on cellulose degradation. The mangrove is considered a saline environment (Hoguin, 2001), with values varying between 4 and 30% NaCl equivalent (Wilson, 2009). Thus, one may suggest that the variation in salinity can cause the selection of groups more adapted to high salinity, thus leading to a constant degradation level of carbon compounds throughout the salt gradient in this environment. Rejmánková and Houdková (Rejmánková and Houdková, 2006) have demonstrated through testing the degradation of cellulose that this activity was significantly slower in areas with high salinity. Sá and Melo (2008) found that biofilm formation and reduced endoglycolytic activity are induced in bacterial isolates from the rhizosphere of Rhizophora mangle, which is a tree species exclusively found in mangroves.
Figure 1.
Cellulase production by the isolates in mangrove sediment under two salinity conditions. (a) The results are presented as an index of enzyme activity, calculated by dividing the diameter of the halo by the diameter of the colony. (b) Relationship between the activities in the two conditions. Values indicating an increase or decrease in the two halos are represented by the lines y = 2× and y = 0.5×, respectively. Additionally, the names of the isolates with activity modulated by salinity are presented.
Genomic fingerprinting of isolates by BOX-PCR
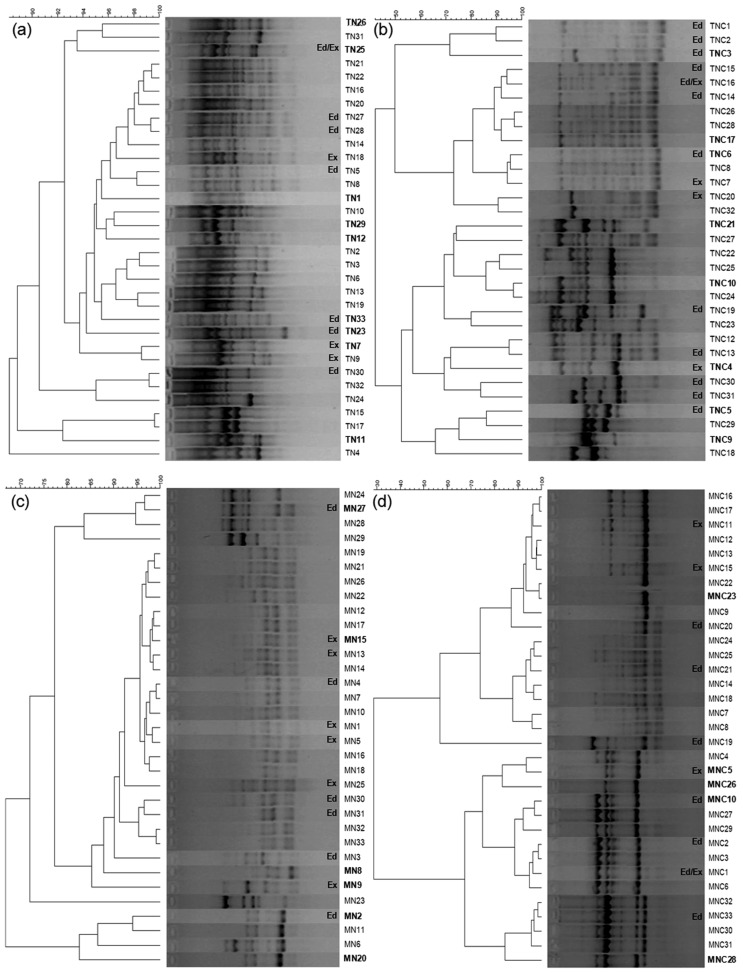
BOX-PCR fingerprinting revealed the selection pressure exerted by the use of distinct media during enrichments as well as the pressure exerted by the insertion of cellulose as a carbon source for bacterial multiplication (Figure 2). The diversity of fingerprinting patterns was higher in the TSB medium when compared with the isolates obtained in the MM. However, the selection due to cellulose was evidenced in both types of media. Moreover, the clustering analysis indicates the formation of more distinguishable and concise groups for isolates obtained from the enrichments in MM (Figures 2b and 2d) and using cellulose (FIGURES 2a and 2b). These results support our phylogenetic inference that is based on a limited number of isolates that represent the major groups determined by BOX-PCR.
Figure 2.
Fingerprint analyses of the isolates by BOX-PCR from isolates obtained from enrichments in TSB with glucose (a), TSB with cellulose (b), MM with glucose (c), and MM with cellulose (d). Similarity clusters were determined by UPGMA based on a Pearson correlation analysis of densitometric curves. The names of the isolates used in the phylogenetic reconstruction are in bold, and prefixes Ed and Ex indicate those with endoglucanase and exoglucanase activities, respectively.
A similar approach was previously used by Mendes et al. (2007) to cluster endophytic isolates from sugarcane, indicating that isolates with similar BOX-PCR patterns share a strict phylogenetic affiliation. For most of the clusters formed, we observed a similar taxonomic affiliation when the BOX-PCR patterns were clustered.
Identification of isolates by partial sequencing of the 16S rRNA
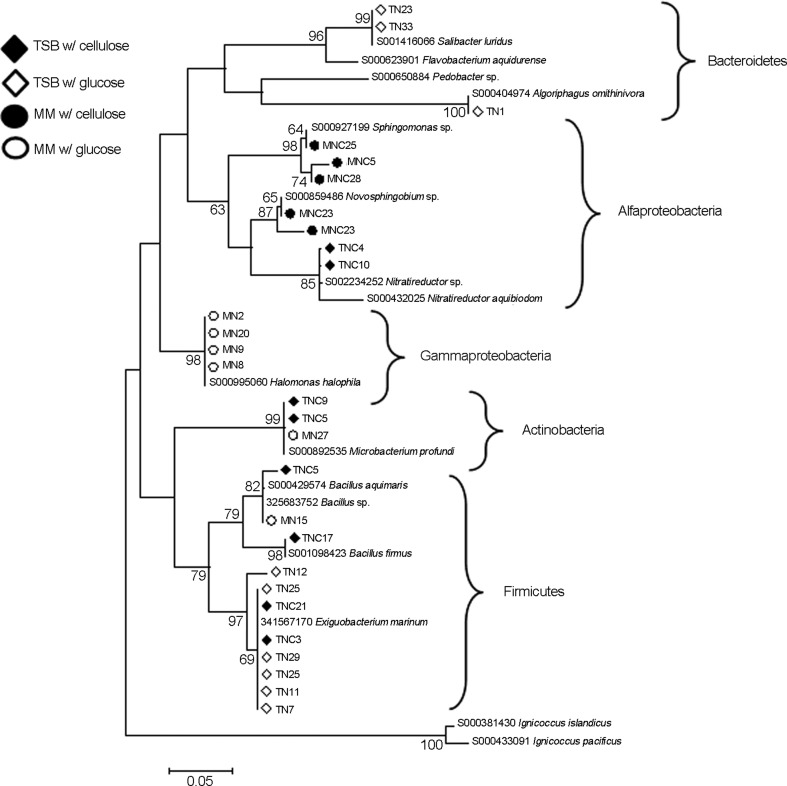
The identification of 28 selected isolates (12 positive for enzymatic activites - seven for endoglucanase, four for exoglucanase, and one positive for both activites) from the groups obtained using the technique of BOX-PCR resulted in the determination of the major phylogenetic groups of cellulolytic bacteria present in mangrove sediment. The comparison of the obtained sequences with those found in the databases (GenBank and RDP) demonstrated that the two most abundant groups were Alphaproteobacteria and Firmicutes, followed by Gammaproteobacteria, Actinobacteria and Bacteroidetes (Figure 3). Within the Alphaproteobacteria, the sequences of the isolates were affiliated with the genera Sphingomonas, Novosphingobium and Nitratireductor. The literature describes the growth of bacteria belonging to the class Alphaproteobacteria in the extremes of pH (5–12) and also shows the abundance of such groups in marine environments. However, the activity of these organisms in cellulose degradation in mangroves is described here for the first time. Concerning the Firmicutes, sequences from the isolates were similar to those for bacteria belonging to the genera Bacillus and Exiguobacterium, which have both been described as cellulose-degrading organisms in other environments (Lima et al., 2005). Gammaproteobacteria is represented by the genus Halomonas, which encompasses organisms adapted to saline environments, such as mangroves. According to Soto-Ramírez et al. (2007), the occurrence of the genus Halomonas in mangroves indicates the salinity tolerance, suggesting that in this study, this group could have been selected during the enrichment process. Bacteria affiliated with this genus were also found in oceanic water, where the abundance of such groups seems to be modulated by the variation in the salinity of superficial or deep-sea water (Radjasa et al., 2001). The isolates of Actinobacteria found in mangroves are affiliated with the genus Microbacterium, whereas the bacteria affiliated with Bacteroidetes seem to belong to the genera Salinibacter and Algoriphagus. These groups are not reported to degrade cellulose in the literature, and their role on cellulose consumption in mangroves remains to be elucidated.
Figure 3.
Phylogenetic relationships based on partial 16S gene sequences of bacterial isolates from mangrove sediment with the best matches from the databases (12, 27). The alignment was constructed by Mega 4.1 software (35) followed by clustering using neighbor joining and the Kimura-2 parameter. A bootstrap analysis was performed with 1,000 repetitions, and values indicate the percentage of clustering matching. The bar in the bottom of the figure scales the number of differences in base composition among the sequences.
Moreover, the taxonomical affiliation has shown the presence of known groups previously described to be involved in cellulose degradation as well as those not related to this feature. This finding indicates that studies focusing on endemic species of mangroves serve as a basis for obtaining new biotechnology tools with greater efficiency and applicability in the degradation of cellulose for various purposes.
Conclusions
The results of this study indicate that cellulolytic bacteria live in the mangrove sediment sampled at Ilha do Cardoso (Cananéia, SP). The applied methodology was efficient for obtaining a collection of bacteria involved in cellulose degradation in mangroves. Endoglucanase activity was more frequently observed among the isolates, and this activity was modulated according to the salinity. The main taxonomical groups of cellulolytic bacteria from mangrove sediments are Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, Firmicutes and Bacteroidetes. Overall, the results show that mangroves may harbor cellulolytic organisms with unique characteristics that may be further explored for cellulose degradation for distinct purposes.
Acknowledgments
This work was financially supported by the São Paulo Research Foundation (FAPESP, proc. number 2004/13910-6). Also, A.C.F. Dias received a graduate fellowship from FAPESP (Proc. 2008/54013-8). We also thank João L. Silva, for his support in mangrove expeditions and samplings.
References
- Akhtar N, Ghauri MA, Iqbal A, Anwar MA, Akhtar K. Biodiversity and phylogenetic analysis of culturable bacteria indigenous to Khewra salt mine of Pakistan and their industrial importance. Braz J Microbiol. 2008;39:143–150. doi: 10.1590/S1517-838220080001000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo RS. Fungi: an introduction to biology, biochemist and biotechnology. Brazil: 2004. Hydrolytic enzymes; pp. 263–265. [Google Scholar]
- Bhat MK. Cellulase and related enzymes in biotechnology. Biotech Advan. 2000;18:355–383. doi: 10.1016/s0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- Bisaria VS, Ghose TK. Biodegration of cellulosic materials: substrates, microrganisms, enzymes and products. Enzy and Microbial Tech. 1981;3:90–104. [Google Scholar]
- Blumer-Schuette SE, Irina Kateava I, Westpheling J, Adams MWW, Kelly RM. Extremely thermophilic microrganisms for biomass conversion: status and prospects. Curr Opini in Biotech. 2008;19:210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Dias ACF, Andreote FD, Dini-Andreote F, Lacava PT, Sá ALB, Melo IS, Azevedo JL, Araújo WL. Diversity and bio-technological potential of culturable bacteria from Brazilian mangrove sediment. World J Microbiol Biotech. 2009;25:1305–1311. [Google Scholar]
- Dias ACF, Andreote FD, Rigonato J, Fiore MF, Melo IS, Araújo WL. The bacterial diversity in Brazilian non-disturbed mangrove sediment. Anto van Leeuwe. 2010;98:541–551. doi: 10.1007/s10482-010-9471-z. [DOI] [PubMed] [Google Scholar]
- Dillon A. Enzymes as agents Biotechnology. 2004. Cellulases; pp. 243–270. [Google Scholar]
- Eveleigh DE. The microbial production of industrial chemicals. Sci American. 1981;245:155–178. [Google Scholar]
- Fengel D, Wegner G. Wood chemistry, ultrastructure, reactions. NCSU; 1989. Available at http://cnr.ncsu.edu/fb/extensionoutreach/techservices/woodchem.html. [Google Scholar]
- Flores-Mireles AL, Winans SC, Holguin G. Molecular characterization of diazotrophic and denitrifying bacteria associated with mangrove roots. Appl Environ Microbiol. 2007;11:7308–7321. doi: 10.1128/AEM.01892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Fuse N, Kodaira R, Kanda J. Cloning and characterization of a new exocellulase gene, cel3, in L. lactus. FEMS Microbiology Letters. 1999;172:231–237. doi: 10.1111/j.1574-6968.1999.tb13473.x. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrofic bacteria. Microbiol Reviews. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguin G, Vazquez P, Bashan Y. The role of sediment microrganisms in the productivity, conservation, and rehabilation of mangrove ecosystems: an overview. Biol and Fert of Soils. 2001;33:265–278. [Google Scholar]
- Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. Arapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr Microbiol. 2008;57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- Kathiresan K, Bingham BL. Biology of mangrove and mangrove ecosystems. Adv Mar Biol. 2001;40:81–251. [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Sthal DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Procee of the Natio Acade of Scien of the Unit Sta of America. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankau RA, Strauss SY. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science. 2007;317:1561–1563. doi: 10.1126/science.1147455. [DOI] [PubMed] [Google Scholar]
- Leschine S. Cellulose degradation in anaerobic environments. Annual Review of Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- Lima AOS, Quenice MC, Fungaro MHP, Andreote FD, Macheroni W, Jr, Araújo WL, Silva Filho MC, Pizzirani-Kleiner AA, Azevedo JL. Molecular characterization of a β-1,4-endoglucanase from a endophytic Bacillus pumilus strain. Applied Microbiology and Biotechnology. 2005;68:57–65. doi: 10.1007/s00253-004-1740-1. [DOI] [PubMed] [Google Scholar]
- Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM. Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol. 2007;73:7259–7267. doi: 10.1128/AEM.01222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah NM, Wiegel J. The anaerobic halophilic alkalithermophiles. New York Acad of Scienc. 2008;1125:44–57. doi: 10.1196/annals.1419.028. [DOI] [PubMed] [Google Scholar]
- Morais S, Heyman A, Barak Y, Caspi J, Wilson DB, Lamed R, Shoseyov O, Bayer EA. Enhanced cellulose degradation by nano-complexed enzymes: Synergism between a scaffold-linked exoglucanase and a free endoglucanase. J of Biotechnol. 2010;147:205–211. doi: 10.1016/j.jbiotec.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Radjasa OK, Urakawa H, Kita-Tsukamoto K, Ohwada K. Characterization of psychrotrophic bacteria in the surface and dee-sea waters from the Northwestern Pacific Ocean based on 16s ribossomal DNA analysis. Marine Biotech. 2001;3:454–462. doi: 10.1007/s10126-001-0050-1. [DOI] [PubMed] [Google Scholar]
- Rejmánková E, Houdková K. Wetland plant decomposition under different nutrient conditions: what is more important, litter quality or site quality? Biogeochemistry. 2006;80:245–262. [Google Scholar]
- Said S, Pietro R. Enzymes as Biotechnological Agents Brazil. 2004. Generalities on industrial enzyme application; pp. 1–7. [Google Scholar]
- Sá ALB, Melo IS. M.Sc. Dissertation. São Paulo, Brazil: Instituto de Biotecnologia. USP; 2008. Diversidade de rizobactérias endoglicoliticas isoladas de mangue vermelho (Rhizophora mangle) p. 61. [Google Scholar]
- Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Canad J Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- Sinegani AAS, Mahohi A. Soil water potential effects on the cellulase activities of soil treated with sewage sludge. Plant Soil Environme. 2010;56:333–339. [Google Scholar]
- Soto-Ramírez N, Sánchez-Porro C, Rosas S, González W, Quiñones M, Ventosa A, Montalvo-Rodríguez R. Halomonas Avicenniae sp. nov., isolated from the salty leaves of the black mangrove Avicennia germinans in Puerto Rico. Int J Syst Evol Microbiol. 2007;57:900–905. doi: 10.1099/ijs.0.64818-0. [DOI] [PubMed] [Google Scholar]
- Tacão M, Alves A, Saavedra MJ, Correia A. BOX-PCR is an adequate tool for typing Aeromonas spp. Antonie van Leeuwenhoek. 2005;88:173–179. doi: 10.1007/s10482-005-3450-9. [DOI] [PubMed] [Google Scholar]
- Taketani RG, Yoshiura CA, Dias ACF, Andreote FD, Tsai SM. Diversity and identification of methanogenic archaea and sulphate-reducing bacteria in sediments from a pristine tropical mangrove. Anto van Leeuwe. 2010;97:401–411. doi: 10.1007/s10482-010-9422-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software versión 4.0. Mol Biolo and Evolu. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tengerdy RP, Szakacs G. Bioconversion of lignocellulose in solid substrate fermentation. Bioche l Enginee J. 2003;13:169–179. [Google Scholar]
- Thauer R, Shima S. Biogeochemistry - methane and microbes. Nature. 2006;440:878–879. doi: 10.1038/440878a. [DOI] [PubMed] [Google Scholar]
- Wilson DB. Cellulases and biofuels. Curr Opi in Biotech. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Zahran HH. Diversity, adaptation and activity of the bacterial flora in saline environments. Biol and Fert of Soils. 1997;25:211–223. [Google Scholar]