Summary
The universally conserved Signal Recognition Particle (SRP) system mediates the targeting of membrane proteins to the translocon in a multistep process controlled by GTP hydrolysis. Here we present the 2.6Å crystal structure of the GTPase domains of the E. coli SRP protein (Ffh) and its receptor (FtsY) in complex with the tetraloop and the distal region of SRP-RNA, trapped in the activated state. The structure reveals the atomic details of FtsY recruitment and, together with biochemical experiments, pinpoints G83 as the key RNA residue that stimulates GTP hydrolysis. Insertion of G83 into the FtsY active site orients a single glutamate residue provided by Ffh (E277) triggering GTP hydrolysis and complex disassembly at the end of the targeting cycle. The complete conservation of the key residues of the SRP-RNA and the SRP protein implies that the suggested chemical mechanism of GTPase activation is applicable across all kingdoms.
Introduction
The Signal Recognition Particle (SRP) is a universally conserved component of the cellular protein targeting machinery. It binds to ribosome nascent chain complexes (RNCs) that expose a SRP signal sequence and delivers them to the target membrane via interactions with the SRP receptor. The RNC is then transferred to the translocon, which mediates the co-translational transport of the emerging polypeptide across the lipid bilayer and also guides the insertion and folding of membrane proteins (Doudna and Batey, 2004; Egea et al., 2005; Saraogi et al., 2011, Akopian et al., 2013a).
In bacteria, SRP consists of the ∼50 kDa signal recognition particle protein Ffh and a 4.5S SRP RNA. The methionine-rich M-domain of Ffh interacts with SRP RNA and the signal sequence (Zopf et al., 1990; Keenan et al., 1998; Batey et al., 2000). The NG domain has a Ras-like guanosine triphosphatase (GTPase) fold with an SRP-specific insertion (insertion box domain, IBD) and an N-terminal helical bundle that mediates interactions with the ribosome (Freymann et al., 1997; Halic et al., 2004; Schaffitzel et al., 2006) (Figure S1). In E. coli, the 100 nucleotide SRP RNA forms an elongated, mostly helical hairpin structure with a GNRA type tetraloop. The Ffh M-domain binding element is located close to the tetraloop, while the distal binding site is located 100 angstrom (Å) away from the tetraloop, towards the 5′-3′ end of the hairpin (Ataide et al., 2011). The prokaryotic SRP receptor (FtsY) is a single protein consisting of an NG domain homologous to Ffh (Montoya et al., 1997) and an additional acidic domain (A domain) that interacts with the SecYEG translocon (Weiche et al., 2008). Ffh and FtsY belong to the signal recognition, MinD, and BioD (SIMBI) class of GTPases and their GTPase activity is regulated by dimerization (Wittinghofer and Vetter, 2011). In the heterodimer, both polypeptides contribute nearly identical residues to a composite active site that binds two guanosine triphosphate (GTP) nucleotides (Egea et al., 2004; Focia et al., 2004).
The SRP RNA serves as a platform for the association of Ffh and FtsY in the membrane targeting process and is also required for efficient GTPase activation (Peluso et al., 2000; 2001; Zhang et al., 2008). Ffh mediates initial interactions between SRP and the RNC: The M-domain recognizes the emerging signal sequence and the NG domain contacts ribosomal protein L23 on the opposite side of the exit tunnel (Gu et al., 2003; Halic et al., 2004; Schaffitzel et al., 2006). The targeting process begins with the formation of an early complex stabilized by an interaction between FtsY and the tetraloop (Zhang et al., 2007; Shen and Shan, 2010). Subsequent formation of the closed complex involves the GTP-mediated dimerization of FtsY and Ffh via their NG-domains (Egea et al., 2004; Focia et al., 2004; Zhang et al., 2008). The GTP-bound NG-domain heterodimer then undergoes a large-scale repositioning to the distal region of the SRP RNA, which is facilitated by a helical linker between the RNA-bound M-domain and the NG domain of Ffh (Ataide et al., 2011). This SRP remodeling is required for efficient GTPase activation, which leads to the dissociation of the complex and handover of the cargo (the ribosome nascent chain complex) to the translocon (Jagath et al., 2001; Siu et al., 2007; Estrozi et al., 2011).
While structural and biochemical evidence shows that both tetraloop (Jagath et al., 2001; Egea et al., 2004; Focia et al., 2004; Siu et al., 2007; Estrozi et al., 2011) and distal interactions (Ataide et al., 2011; Shen et al., 2012) with the SRP RNA are required for the efficient formation of an Ffh:FtsY complex, GTP hydrolysis and protein translocation, the lack of high resolution models has so far limited our understanding of the SRP RNA in the targeting process. We have determined the 2.6Å crystal structure of two Ffh:FtsY NG domain heterodimers, bound to the tetraloop and the distal region of SRP RNA in presence of guanosine diphosphate (GDP) and aluminum tetrafluoride (AlF4), a mimic of the GTP hydrolysis transition state (Wittinghofer, 1997). In combination with the biochemical evidence presented herein, the complex reveals the structural basis for the regulation of the SRP pathway at several stages, including the recruitment of FtsY and the RNA-mediated GTPase-activation upon SRP remodeling.
Results
GDP-aluminum fluoride stabilizes the interaction between the Ffh and FtsY NG heterodimer and SRP RNA
Binding of transition state analogs such as GDP-AlFx to GTPases increases the affinity for their cognate GTPase activating proteins (GAPs) and therefore promotes the formation of a hydrolysis competent complex (Gasper et al., 2009). Since it was established that the RNA is directly responsible for GTPase activation in the SRP system (Ataide et al., 2011) we decided to test whether the presence of AlFx enhances binding of the Ffh and FtsY NG domain heterodimer to SRP RNA. Dimerization of Ffh:FtsY NG-domains in the presence of either of GMPPCP or GDP AlFx was confirmed by analytical size exclusion chromatography. Then, a constant amount of a shortened SRP RNA construct (described in detail below) was incubated with increasing concentrations (1:1 to 1:10 fold molar ratio) of proteins in the presence of either nucleotide analog. In presence of GDP-AlFx, but not GMPPCP, the RNA elution volume shifts, indicating the formation of a larger complex (Figure 1, S2). Hence, the affinity of the NG heterodimer for the RNA is increased in presence of the transition state mimic, relative to the ground state analog. Based on the RNA concentration of 5 μM and the dilution effect on the SEC column, we estimate the dissociation constant of the NG-heterodimer:RNA complex in the presence of GDP:AlFx in the order of 0.5-1μM.
Figure 1. Analytical size exclusion chromatography of NG-heterodimer:RNA complexes.
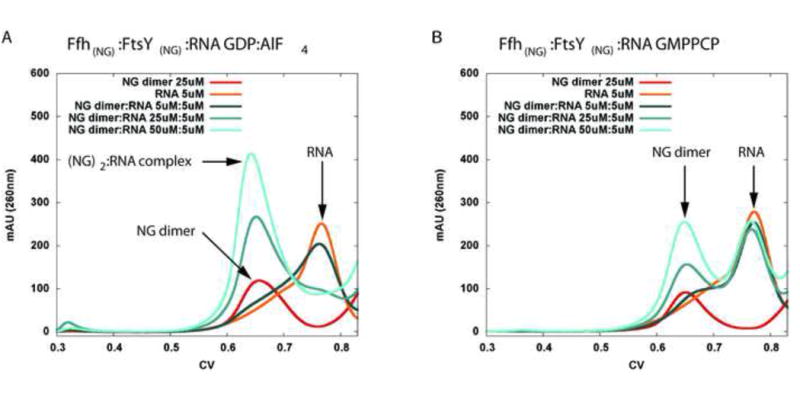
a) Activated complex with transition state analogue GDP:AlF4 bound. Tenfold molar excess of protein complex over RNA (50 uM : 5 uM) shows full complex formation as seen by a thourough shift of the RNA elution volume. b) In contrast, no shift in the RNA or NG-dimer elution volumes is observed if the protein complex is formed with the non-hydrolyzable preactivation state GTP analog GMPPCP. Traces for both proteins and the complex without RNA are shown in figure S2.
Overview of the crystal structure
In order to obtain further insights into the mechanism of receptor recruitment and GTPase activation by the SRP RNA we co-crystallized the NG domains of FtsY and Ffh with GDP:AlFX and shortened SRP RNA (also used in the gel filtration experiments above). To facilitate crystallization, the RNA construct was truncated to the tetraloop and the distal region identified previously as responsible for the binding of the NG heterodimer and GTPase activation by structural and biochemical experiments (Ataide et al., 2011; Shen et al., 2012) (Figure 2). Based on our current understanding of complexes involved in SRP targeting (Akopian et al., 2013a) omitting the M-domain and the RNA region it interacts with is unlikely to perturb physiological interactions between the SRP RNA and the NG domains of Ffh and FtsY. Crystals diffracted to 2.6Å and the structure was solved by molecular replacement (Table 1). Coordinates and structure factors have been deposited in the Protein Data Bank (accession code 4c7o). The asymmetric unit consists of a single SRP RNA molecule that interacts simultaneously with two Ffh:FtsY NG-domain heterodimers. One is bound to the tetraloop, and the second interacts with the distal region of the SRP RNA (Ataide et al., 2011). Two GDP-AlFx complexes are bound in a head-to tail orientation at each NG-domain interface, and a flipped-out RNA base (corresponding to base G83 of full length E. coli SRP RNA) projects into the NG-domain interface, pointing toward the FtsY-bound AlF4 moiety, while the G83 binding site of the tetraloop-bound heterodimer is occupied by a free nucleotide from the crystallization buffer.
Figure 2. Crystal structure of Ffh:FtsY NG heterodimers bound to truncated SRP RNA in distal- and tetraloop positions.
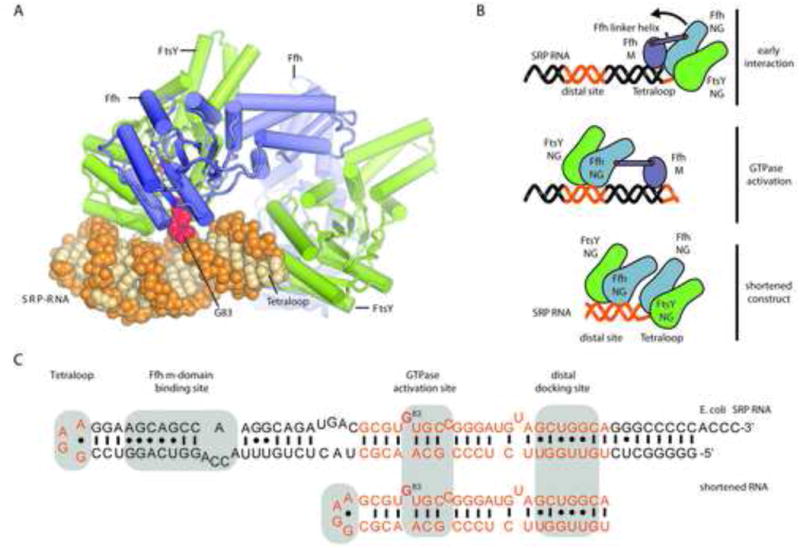
a) SRP RNA is shown as orange spheres and the GTPase-activating flipped out G83 is highlighted in red. The SRP receptor (SR or FtsY) is shown in green and the Ffh in blue. Two GDP-AlF4 complexes bound at each dimer interface are shown as sticks, colored by atom. Details of the structural elements of the GTPases are shown in Figure S1 b) Schematic representation of the structural rearrangements during substrate recognition and GTPase activation. (top) Initial interactions between SRP and the ribosome are mediated mainly via the N-domain of Ffh and protein L23. FtsY transiently associates with the tetraloop (orange) in a GTP-independent fashion, before a tight heterodimer between the homologous NG domains of FtsY and Ffh is formed upon binding of two GTP molecules (middle). The flexible linker tethering M-domain and NG domain of Ffh allows the relocation of the heterodimer to the distal binding site (orange) on the SRP RNA, where a flipped-out base stimulates GTPase activity (bottom). c) Secondary structure diagrams of E. coli SRP RNA and the shortened construct employed in this study. The regions present in the shortened construct are highlighted in orange. Tetraloop, distal docking site and GTPase activating site are shaded in grey. The GTPase-activating G83, which is rotated out of the stack of the helix is highlighted in red.
Table 1. Crystallographic data collection and refinement statistics.
| Wavelength (Å) | 1.00 |
| Resolution range (Å) | 49.22 - 2.6 (2.693 - 2.6) |
| Space group | P 21 21 21 |
| Unit cell | 56.72 166.33 154.59 90 90 90 |
| Total reflections | 195365 |
| Unique reflections | 45637 (4506) |
| Multiplicity | 4.28 |
| Completeness (%) | 99.32 (99.21) |
| Mean I/sigma(I) | 13.38 (2.77) |
| Wilson B-factor | 61.98 |
| R-factor | 0.1632 (0.2701) |
| R-free | 0.2255 (0.3576) |
| Number of atoms | 10375 |
| macromolecules | 9709 |
| ligands | 164 |
| water | 501 |
| Protein residues | 1214 |
| RMS(bonds) | 0.010 |
| RMS(angles) | 1.34 |
| Ramachandran favored (%) | 96 |
| Ramachandran outliers (%) | 0.088 |
| Clashscore | 17.94 |
| Average B-factor | 87.50 |
| macromolecules | 89.10 |
| solvent | 67.70 |
statistics for the highest-resolution shell are shown in parentheses.
The structure captures a snapshot of two distinct stages of the SRP cycle: (1) The closed GTPase heterodimer that is formed after cargo recognition, just before it detaches from the tetraloop, and (2) the activated state of GTP hydrolysis that occurs after the heterodimer has re-localized to the distal region of the SRP RNA (Ataide et al., 2011; Zheng et al., 2011; Shen et al., 2012) (Figure 2).
The tetraloop is recognized by FtsY helix 2 and Insertion Box Domain (IBD) helix B
The interaction between the tetraloop and FtsY –although transient in nature (Shen and Shan, 2010; Estrozi et al., 2011)- is crucial for successful complex formation and targeting in vivo and in vitro (Peluso et al., 2000; Siu et al., 2007). The presence of the SRP RNA accelerates the formation of the GTPase heterodimer (Peluso et al., 2000; 2001; Siu et al., 2007; Zhang et al., 2008; Gasper et al., 2009) via the tetraloop. As proposed earlier (Peluso et al., 2000; Zhang et al., 2008) the SRP RNA acts as an initial attachment point that increases the local concentration of FtsY and orients it in the correct orientation to form an early complex with Ffh (Zhang et al., 2008; Estrozi et al., 2011). Upon GTP binding, the NG heterodimer rearranges into the closed, tightly associated state observed in the crystal structure (Egea et al., 2004; Focia et al., 2004).
The presented crystal structure reveals the atomic details of the SRP RNA-FtsY interaction at the tetraloop, which are in agreement with the contacts observed for the early complex visualized by electron microscopy (Estrozi et al., 2011). The contacts involve three clusters of universally conserved residues (Figure 3, S3); First, a highly basic patch on FtsY helix 2 (Shen and Shan, 2010) forms salt bridges with the tetraloop backbone via residues K399, R402 and to a lesser extent K406. Biochemical experiments demonstrated that these residues are critical for complex formation (Egea et al., 2004; Shan et al., 2004; Shen and Shan, 2010) and also play a role in binding to the SRP RNA on the distal site (see below and Figure S6). The second recognition patch forms aromatic interactions with bases two to four of the tetraloop and consists of the hydrophobic residues L407 and phenylalanine F365 of helix 2 and IBD helix B, respectively. The hydrogen-bonding pattern between D366 (IBD helix B) and the exocyclic amine of the second base in the tetraloop motif is specific for a guanosine in this position. The exocyclic amine of the second tetraloop base also hydrogen bonds to the backbone carbonyl oxygen of the preceding S362. RNA sequence-independent hydrogen bond interactions occur between IBD helix B D359 and the ribose of the second tetraloop residue.
Figure 3. Detailed view of the tetraloop FtsY interaction.
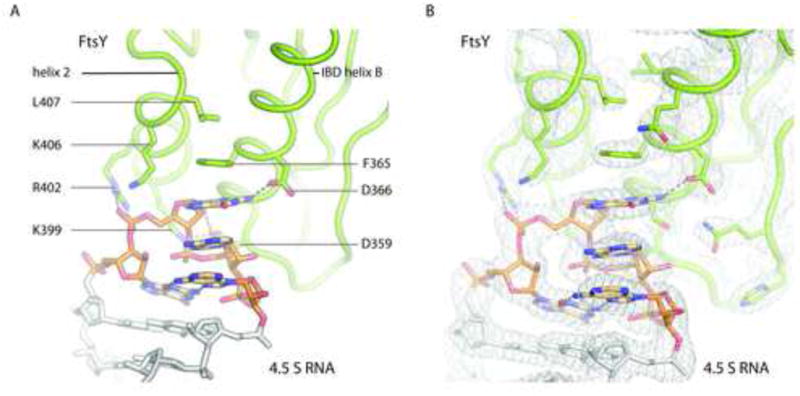
a) FtsY helix 2 contributes mainly electrostatic interaction with the backbone of the SRP RNA while residues in IBD helix B (Phenylalanine 365 as well as Aspartate 366 and 359) form specific interactions mainly with the base on top of the RNA stack in position to of the GNRA type tetraloop. b) Electron density at the contact area exemplifies quality of the electron density maps in well-ordered regions. The 2mFo-dFc electron density map shown as grey mesh is countered at 1.5 σ. See also S3, S4, S5 and S6
These interactions with the SRP tetraloop optimally position FtsY for formation of the closed complex with the NG domains of the Ffh. Due to steric effects the closed complex detaches from the SRP RNA tetraloop in the context of the ribosome (Halic et al., 2006b), as discussed below.
Two conserved sites provide an interface for FtsY at the distal region of the SRP RNA
The interaction between FtsY and the SRP RNA at the distal region was first described in the structure of E. coli FtsY and Ffh in complex with D. radiodurans SRP RNA, determined at 3.94Å resolution (PDB identifier 2XXA) in presence of the non-hydrolysable GTP analog GMPPCP(Ataide et al., 2011). The binding interface can be subdivided into two sites, the docking site and the GTPase activation site, which are separated by one helical turn of the SRP RNA (Figure 2c). Both sites of the RNA are highly conserved among bacteria and across all kingdoms of life (Figure 2, S4).
The interactions at the SRP RNA docking site are mediated by helix B and the preceding loop of the FtsY insertion box domain (IBD) and the minor groove of the RNA helix (Figure 2a, Figure S1). Within experimental accuracy, these interactions are equivalent to the previous structure (Figure 4).
Figure 4. Overview of the distal interaction site.
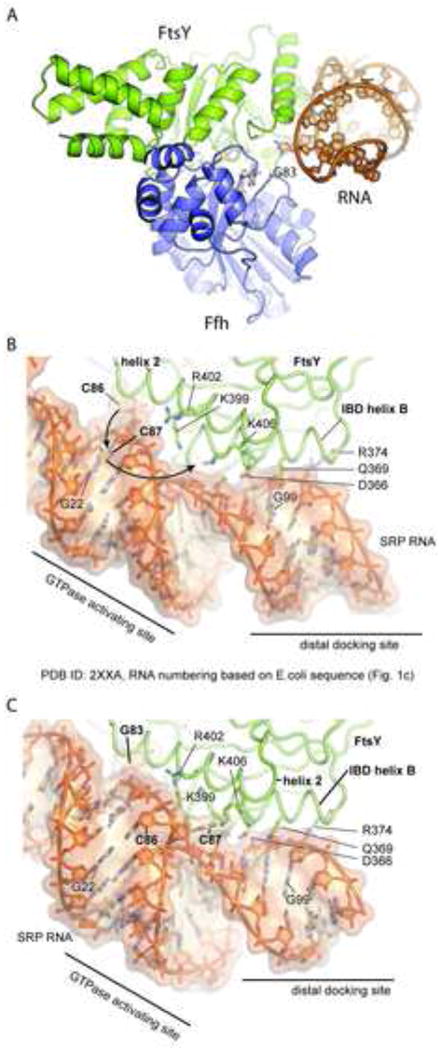
a) Base G83 is sandwiched between the GTPase domains. RNA is almost exclusively bound by FtsY (buried surface area FtsY:RNA ∼1000 Å2 Ffh:RNA ∼90 Å2) See also Figure S3 and 4 b-c) detailed view of the distal interaction site as found in 2xxa (b) and 4c7o (c). The SRP RNA is mainly interacting with FtsY IBD elements (helix B and to a lesser extent strand B). While the binding of the distal docking site nearly identical between the GMPPCP- bound complex shown in b) and the structure determined with bound transition state mimic shown in panel c), dramatic differences in the configuration of the GTPase activating site are found: An unwinding of approximately 100 degrees of the distal RNA brings the catalytic base (G83) close to the active site. This RNA conformation also orients the bulge region around bases C86 and C87 in the proximity of the secondary nucleotide-binding site of FtsY allowing C87 to bind into the pocket formed by residues K406, F365 and E396.
However, the RNA backbone in the current structure is partially untwisted and G83 is inserted into the cleft between the G-domains of FtsY and Ffh. Another flipped-out base, C87, occupies a secondary binding site on the surface of FtsY, where it interacts specifically with K406. The untwisted region of the RNA backbone is stabilized by interactions of K399 and R402 with the phosphate groups. In the previous structure, C87 is base-paired to G22 and C86 is flipped out (E. coli numbering), occupying the binding site between the two proteins (Ataide et al., 2011). Inspection of the secondary structure diagram of the SRP RNA in this region (Figure 2c) reveals that both conformations may exist at equilibrium.
Interestingly, the same residues of FtsY that mediate tetraloop binding also contribute to the distal interaction and engage in very similar interactions at the two sites. For example, the hydrogen-bonding pattern between D366 and the unpaired exocyclic amine of G12 echoes the specific recognition at the top of the tetraloop base stack. Moreover, salt bridges to the RNA backbone are mediated at both sites by helix 2 residues R402 and K406 (Figure 3a, Figure 4c). With the exception of the interaction between D366 and the highly conserved G12 (Figure S4), binding of FtsY to the SRP RNA relies on the general electrostatic features (Figure S5) of an RNA helix rather than sequence specific contacts. Mutations in the basic patch residues K399, K402 or K406 (Figure 3 and Figure 4) involved in RNA interactions at the tetraloop and at the distal docking site of the SRP RNA affect both: initial complex formation and the observed GTPase rate. (Figure S6).
The distal region is critical for the stimulation of GTPase activity
Since we now have structural evidence that the RNA may exist in two conformations with either C86 or C87 flipped out of the helix and both C86 (Ataide et al., 2011) and G83 (presented here) have been observed inserted into the GTPase active site in the GMPPCP and GDP-AlF4 complexes, respectively (Figure 4 and Figure 5a,b), either of these may be implicated in GTPase activation. To explore the functional roles of C86, C87 and G83, we assayed the ability of various SRP RNA mutants to stimulate GTP hydrolysis and translocation.
Figure 5. Schematic representation of GTPase:RNA binding modes.
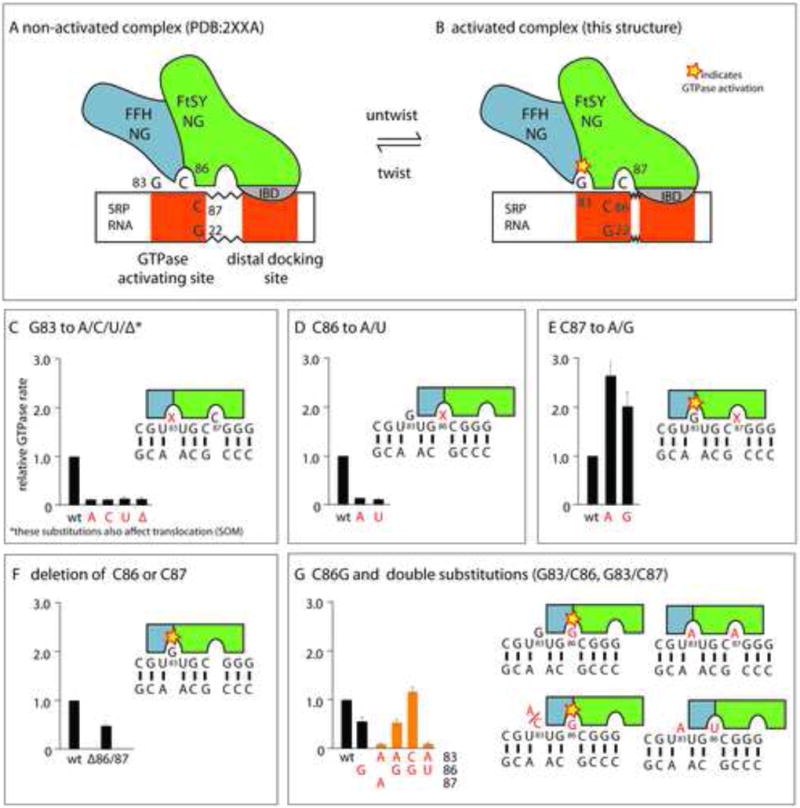
a) non-activated and b) the activated SRP-FtsY complex. In the activated complex presented here, the RNA is slightly untwisted, and as a result, G83 is inserted into the active site, rather than C86, as observed previously, while the interaction at the distal docking site is virtually identical. c) Biochemical characterization demonstrates that the presence and identitiy of G83 is crucial for GTPase activation and also for translocation (see Figure S6). d) C86 A or U substitutions strongly reduce GTPase activation, consistent with the observation that a flipped out base at this position impedes access to G83 (Figure 2,3). e) In contrast to C86 A or U substitutions, C87A and G substitutions increase GTPase activation. A flipped out base at position 87 leaves G22 available for basepairing to C86 and therefore improves accessibility of G83. f) Deletion of either C86 or C87 results in a modest reduction of GTPase rate, in agreement with the loss of interaction surface area (Figure 2c) that weakens the interaction between RNA and FtsY. g) Substitution of C86 with a guanosine only reduces GTPase activation moderately, compared to the C86 A or U substitution panel (d). The C86G substitution rescues GTPase activation by SRP RNA with a G83 substitution (panel c), thus indicting that a flipped-out G86 may insert into the active site to stimulate GTP hydrolysis. Data represent mean +/- SD (c-f: n=3, g: n=2)
Deletion as well as any base-substitution of G83 has a deleterious effect on the GTPase activity reducing it to levels observed in absence of RNA (Figure 5c). This demonstrates that not only the presence but also the identity of the base at position 83 is critical for the ability of the SRP RNA to stimulate GTP hydrolysis. The mutations also have a pronounced effect on translocation efficiency, which is strongly reduced compared to wild type RNA (Figure S6). Further reduction of translocation activity is observed in absence of RNA (Shen et al., 2012), consistent with additional roles of the RNA in in the targeting reaction such as accelerating SRP-FtsY assembly via the tetraloop.
Base-substitutions of C86 and C87 have opposed effects on GTPase rate: While C87A increases GTPase stimulation, C86A reduces observed rates to those observed in absence of RNA (Figure 5d,e) (Ataide et al., 2011). Neither of the C86 or C87 substitutions have an appreciable effect on translocation efficiency (Figure S6). Deletions of C86, as demonstrated previously (Ataide et al., 2011), or C87 also reduce the ability to stimulate GTPase activity. However, the reduction is not as pronounced, resulting in a 50% lower GTPase rate compared to wild type (Figure 5f).
The existing structural data are consistent with biochemical observations and can be rationalized with the following model (Figure 5a,b): According to the secondary structure diagram (Figure 2c), either C86 or C87 may base pair to G22, as observed in this structure and the previous one(Ataide et al., 2011)(Figure 4b,c). If the flipped-out C is deleted, several interactions with FtsY are lost, which may affect binding affinity. Moreover, a flipped-out base at position 86 sterically impedes access to the GTPase-activating G83 (Figure 4b). Therefore, a substitution that shifts the equilibrium towards a flipped out base at position 87 improves access to G83, and a substitution that shifts the equilibrium towards a flipped out base at position 86 impedes access (Figure 5d,e). This model not only explains the inhibitory effect of the C86 substitutions, but also the stimulatory effect observed for C87 substitutions. Notably, mutation of C86 to G results in only a two-fold reduction in GTPase rate. In addition, double mutations show that the deleterious effects of G83 substitution or deletion on GTPase activation can be compensated by a C86G substitution (Figure 5g). Taken together, these results indicate that a structural state with base 86 inserted into the active site exists in solution, but activation only occurs if the inserted base is a guanosine.
Insertion of G83 rearranges conserved active site residues and water molecules
Compared to the apo-form of FtsY (PDB 1FTS) and the GDP-bound form of Ffh (PDB 1JPJ), both proteins undergo concerted rearrangements during hetero-dimerization in the presence of non-hydrolysable GTP analogs (PDB 1OKK, 1RJ9) (Egea et al., 2004; Focia et al., 2004). A rigid-body shift of the N domain relative to the conserved core of the G-domain is accompanied by rearrangements of the conserved GTPase sequence motifs G1-G5 (Figure S1) to form an almost symmetrical binding site for two nucleotides at the subunit interface. The 3′ OH groups of the FtsY and Ffh-bound nucleotides, which are essential for association and GTP hydrolysis in trans donate a hydrogen bond to the γ-phosphate of the opposite nucleotide (Egea et al., 2004; Focia et al., 2004). Large rearrangements occur in the G2/switch 1 (SW1) and G3/switch 2 (SW2) regions of both proteins as well as the G4 and G5 regions of FtsY, relative to the nucleotide-free state (Freymann et al., 1997; Montoya et al., 1997; Egea et al., 2004; Focia et al., 2004). These rearrangements bring several conserved sidechains into the active site: Arginines R141 and R333 of the Ffh- and FtsY G2 regions hydrogen-bond to the α- and γ-phosphate groups. The arrangement of the arginine residues is asymmetric, as they coordinate a single water molecule (C) between the two nucleotides. The G1 aspartate residues 138 and 330 are positioned by hydrogen bonds to a water molecule in the coordination shell of the magnesium ion and a second water molecule (N) close to the γ phosphate, that is thought to act as a nucleophile in GTP hydrolysis (Egea et al., 2004; Focia et al., 2004) (Figure 6a).
Figure 6. Active site comparision and conservation levels of SRP.
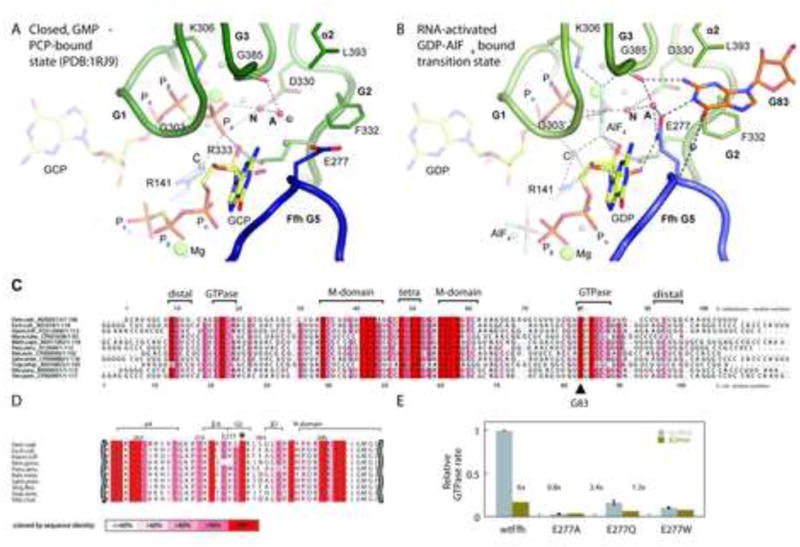
a) Closeup of the active site in the GMPPCP bound form (PDB 1RJ9). Glutamate 277 in the closing loop points away from the active chamber. The closing loop of Ffh is shown as a dark blue cartoon, and the P-loop (G1), SW1 (G2) and SW2 (G3) regions of FtsY are show as a green cartoon (see also S1 and S5). The nucleophilic (N) and auxiliary (A) water molecules implicated in GTP hydrolysis (Focia et al., 2006) are highlighted in red. Dashed lines represent hydrogen bonds. b) RNA bound complex with GDP:AlF4. The flipped out base G83 of the SRP RNA makes specific contacts with FtsY and Ffh residues, organizing a network of hydrogen bonds that finally activates the nucleophilic water (N) for hydrolysis of the FtsY bound GTP. c) Sequence alignment of bacterial SRP RNAs. Conservation is indicated by color intensity, based on an alignment of more bacterial sequences (Figure S4). G83 (E. coli numbering) and several other residues on both strands of the GTPase-activating region are highly conserved. Significant conservation is also observed for residues of the distal binding site. d) Alignment of bacterial Ffh sequences. The universally conserved E277 (E. coli gene numbering) is indicated by (*) and has 100% sequence identity, also across sequences from different kingdoms (full alignment in figure S3b) e) Effects of Ffh E277 mutations on stimulated GTP hydrolysis by SRP. E277A and E277W substitutions reduce GTPase rate below levels observed in absence of the activating G83, and even the conservative E277Q mutation reduces GTPase rate approximately fivefold. The difference in activity observed in presence of full length (grey bars) or truncated SRP RNA (Ataide et al., 2011) lacking the distal binding site (green bars) shows that the Ffh mutants cannot be effectively stimulated by SRP RNA. Shortened RNA is assayed once; data for wt RNA represent the mean of two experiments +/- SD.
In contrast to the large conformational changes during dimerization, the structure of the RNA-bound GTPase heterodimer in complex with RNA and the transition-state analog GDP-AlF4 does not reveal additional large scale rearrangements (Figure 6b). Nevertheless, the small differences between the structure determined here and the structures of the NG heterodimer in presence of the ground state analogs GMPPCP (PDB 1RJ9, 1OKK) provide important clues to the mechanism of RNA-mediated GTPase activation (Egea et al., 2004; Focia et al., 2004). The active site region of the structure presented here - including the flipped-out base, ligands and water molecules - is well defined, with temperature factors significantly below the average of the structure, also for the observed solvent molecules (Table 1). The flipped-out RNA base G83 reaches into a binding pocket formed predominantly by residues of FtsY in the vicinity of the γ-phosphate (mimic) of the FtsY-bound nucleotide. G83 is stacked between FtsY F332 (G2/SW1 motif) and L393 (helix 1). The 2′ hydroxyl group of G83 is within hydrogen bonding distance of FtsY E396, which is in turn positioned by R386 of the FtsY G3 region, and the 5′ phosphate of G83 accepts a hydrogen bond from K278 of Ffh. In addition to these stacking- and backbone interactions, G83 forms a Watson-Crick-like hydrogen bond pattern with residues from FtsY and Ffh that is specific for the guanine base: The exocyclic N2 amine donates a hydrogen bond to the backbone carbonyl of FtsY G385 (G3/SW2 motif), the cyclic N1 amine donates a hydrogen bond to the carboxyl group of E277 in the Ffh G5 motif (closing loop) and the O6 carbonyl group of G83 interacts with the main chain amine of Ffh E277 via a bridging water molecule. Thus, the insertion of the flipped-out base may stabilize a catalytically active conformation of the FtsY active site motifs G2 and G3, as well as the Ffh G5 motif. In the light of the specificity of the interaction, it is not surprising that G83 is completely conserved in bacterial SRP RNA sequences (Figure 6c). Moreover, a conserved, unpaired guanine can be identified at the corresponding position of archaeal or eukaryotic SRP RNA sequences (Andersen et al., 2006; Hainzl et al., 2007) (Figure S4).
Compared to the GMPPCP bound state (Figure 6a), the sidechain of Ffh E277 is positioned by hydrogen bonds to G83 and the 3′ hydroxyl group of the Ffh-bound nucleotide to interact with the auxiliary (A) water molecule in the activated state (Figure 6b). The auxiliary water in turn hydrogen bonds to the nucleophilic water (N) and the water molecules concertedly move closer to the γ phosphate position occupied by the AlF4 (Figure 6a,b). As a result, the auxiliary water molecule is exposed to the carboxyl groups of the E277 side chain and the backbone carbonyl of FtsY G385 (G3/SW2 motif). To investigate the role of this residue in propagating the signal from the bound RNA to trigger GTP hydrolysis we have introduced mutations at this position and measured the rates of RNA-stimulated GTP hydrolysis (Figure 6d,e). The E277A and E277W mutations reduce relative GTPase rate below levels observed in absence of G83, and even the conservative E277Q mutation leads to a fivefold reduction. An essential role of E277 is consistent with the observation that E277 is highly conserved in bacteria (Figure 6d) and across kingdoms (Figure S3b). These results suggest that the insertion of G83 positions E277 of Ffh to polarize the auxiliary water molecule (A) allowing it to partially accept a proton from the nucleophilic water, therefore activating it for attack on the γ-phosphate of the FtsY-bound nucleotide (Figure 6b).
Since the interaction with the flipped-out base of the SRP RNA occurs specifically on one side of the GTPase heterodimer (Figure S5), GTP hydrolysis is likely to begin with the FtsY bound nucleotide. This is supported by the observation that the chemical environment of the auxiliary water molecule near the Ffh bound nucleotide is significantly less polarizing (Figure S5e,f). Moreover, the asymmetrically positioned central water molecule (Egea et al., 2004; Focia et al., 2004) links the α - and γ -phosphates of the FtsY-bound nucleotide to FtsY R333 and Ffh R141. This may stabilize the build-up of negative charges during GTP hydrolysis and also provides a possible route for coupling of GTP hydrolysis at the two sites (Powers and Walter, 1995, Shan et al. 2004).
Discussion
Membrane protein targeting by SRP requires the spatial and temporal coordination of several events (Figure 7b): Initially, SRP binds to the ribosome near the exit tunnel via interactions of the Ffh NG domain with ribosomal protein L23 (Gu et al., 2003) and the Ffh M-domain recognizes the emerging signal sequence (Zopf et al., 1990; Schaffitzel et al., 2006; Halic et al., 2006a). Upon signal sequence binding the SRP molecule adopts a discrete orientation (Estrozi et al., 2011), in which the tetraloop of the RNA binds the FtsY and orients it for GTP-dependent association with Ffh, which recruits the RNC to the membrane (Batey et al., 2000; Peluso et al., 2000; Bradshaw et al., 2009). Next, the Ffh and FtsY NG domain complex is detached from the tunnel exit and the tetraloop (Halic et al., 2006b) freeing up the L23 region of the ribosome for binding of the translocon (Schaffitzel et al., 2006; Halic et al., 2006a; 2006b). Translocon binding enables repositioning of the NG heterodimer to the distal region of the SRP RNA where GTP hydrolysis is triggered, leading to the disassembly of the Ffh:FtsY complex (Ataide et al., 2011; Shen et al., 2012).
Figure 7. a) Schematic representation of GTPase activation and b) steric regulation in the SRP cycle.
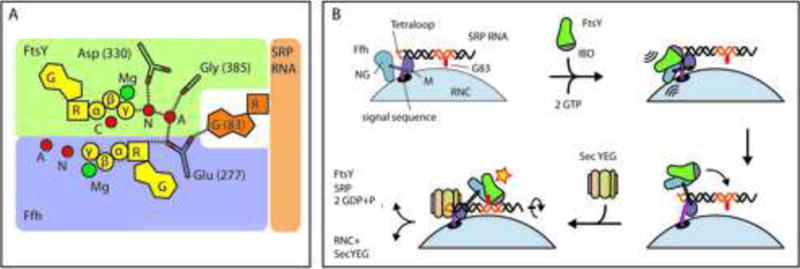
(a) Insertion of G83 reorients Glu 277 of Ffh and repositions active site water molecules. The auxiliary water molecule (A) is polarized by interactions with several carboxyl and carbonyl groups in its environment and may therefore partially accept a proton from the nucleophilic water molecule (N) activating it for attack on the γ -phosphate. The central water molecule (C) bridges the α- and γ -phosphates of the FtsY-bound nucleotide and stabilizes the negative charges. (b)(1) SRP recognizes the signal sequence on the nascent peptide chain emerging from the RNC via the M-domain of Ffh. (2) FtsY binds to the SRP RNA tetraloop and is oriented for interaction with Ffh. The formation of the closed, GTP-bound complex supports the detachment of the NG heterodimer from the tetraloop, see also S7 (3) However, access to the distal site is blocked in absence of SecYEG (Ataide et al., 2011, Shen et al., 2012)., (4) Binding of SecYEG may induce a rotation of the SRP RNA that facilitates SRP remodeling and insertion of the catalytic G83 into the active site of FtsY
Although we have an increasingly accurate mechanistic understanding of this process, several important questions remain; i) How does the SRP RNA position FtsY to accelerate the association with Ffh? ii) Why does the closed complex dissociate from the tetraloop? iii) How does the RNA stimulate GTPase activity? iv) How is RNA-mediated GTP hydrolysis delayed until the RNC has been transferred to the translocon?.
Interactions with the SRP RNA tetraloop position FtsY for binding to Ffh
Biochemical data and cryo-EM show that the Ffh:FtsY complex initially assembles on the tetraloop of the SRP RNA and departs from it upon formation of a tight GTP activated heterodimer. However, in the absence of the ribosome the activated complex visualized in this study has maintained interactions with the tetraloop. Several lines of evidence suggest that the observed contacts are representative for the initial interactions between FtsY and the tetraloop, which occur during RNC recruitment to the membrane; i) very similar interactions have been observed for the early complex between FtsY and Ffh visualized on the translating ribosome by cryo-electron microscopy (Estrozi et al., 2011), ii) these interactions are consistent with mutational and biochemical studies (Egea et al., 2004; Spanggord et al., 2005; Zhang et al., 2008) which identified regions of FtsY responsible for the tetraloop binding, iii) the tetraloop interaction site (helix 2, IBD helix B) is spatially separated from the active site and does not change in response to nucleotide binding and dimerization (Montoya et al., 1997; Egea et al., 2004) (PDBs 1FTS, 1RJ9). Therefore, we reason that the interaction of FtsY with the tetraloop is independent of the functional state of the GTPase heterodimer.
To obtain further insights into the mechanism of NG heterodimer detachment from the tetraloop, our high-resolution structure can be combined with information from previous cryo-EM studies. Superimposing the crystal structure to the ribosome bound SRP RNA (Schaffitzel et al., 2006; Halic et al., 2006a) reveals that formation of the tetraloop bound closed complex would lead to clashes of the Ffh N-domain with the ribosomal surface (Figure S7). Therefore, the GTP-dependent dimerization supports the dissociation from the tetraloop in the ribosomal context. This agrees with the reduced affinity of the closed complex for the ribosome (Shan et al., 2007; Zhang et al., 2009) and the detachment of the Ffh NG domain upon receptor binding (Halic et al., 2006b). In agreement with the transient character of the tetraloop:FtsY interaction (Shen and Shan, 2010) in the SRP targeting cycle the interaction surface between FtsY and the tetraloop is small (∼270Å2) compared to the distal site (∼1000 Å2) or the M-domain RNA binding site (∼680 Å2).
Insertion of SRP RNA G83 into the active site of FtsY triggers GTP hydrolysis
In this work we visualized the activated complex of the GTPase heterodimer bound to the distal region of the SRP RNA (Figure 2). The complex is stabilized by the presence of GDP-AlF4 in the active site, which mimics the transition state of GTP hydrolysis and significantly increases the affinity of the NG heterodimer for the RNA. At 2.6Å resolution, the structure allows us to characterize the distal docking site of FtsY on the SRP RNA (Figure 4) and to deduce how the flipped out nucleotide rearranges the active site to stimulate GTP hydrolysis (Figure 5,6). The flipped out base of G83 inserts on one side of the NG heterodimer into a binding pocket on FtsY and contacts the G2 and G3 motifs of the FtsY active site, close to the γ-phosphate mimic of FtsY bound GTP. Although most interactions involve FtsY residues, G83 also reorients the completely conserved E277 (Figure 6, S3b) provided in trans by Ffh to interact with the auxiliary water molecule. Consequently, the auxiliary water molecule is exposed to several carbonyl- and carboxyl groups. This suggests a chemical mechanism in which the auxiliary water molecule partially accepts a proton from the nucleophilic water and consequently activates it for nucleophilic attack on the γ-phosphate of the FtsY-bound nucleotide (Figure 6, Figure 7a). A catalytic mechanism involving a second water molecule has recently been proposed for the related Ras GTPase: Ras residue Q61 occupies a position close to the reoriented Ffh E277 sidechain and has been implicated in the stabilization OH-and H3O+ ions generated in the reaction (Martın-Garcıa and Mendieta-Moreno, 2012). In agreement with such a mechanism, two water molecules are also observed in GTP bound Ras (PDB 1CTQ), and the Ras Q61A mutation decreases GTPase activity, while the Q61E mutation increases it (Martın-Garcıa and Mendieta-Moreno, 2012).
The asymmetric interaction between the NG heterodimer and the SRP RNA is reflected by the differences in the charge distribution on the surface of the proteins and by sequence differences near the active site (Figure S5). Moreover, the relative positions of the water molecules differ between the two-nucleotide binding sites of the heterodimer. The chemical environment of the auxiliary water molecule at the Ffh-bound nucleotide is significantly less polarizing than at the FtsY-bound nucleotide, indicating that although GTP hydrolysis is coupled (Powers and Walter, 1995), it may be triggered in an asymmetric manner, beginning with the FtsY-bound nucleotide (Figure S5e,f, Figure 7a). As observed previously (Focia et al., 2006), the central water molecule (C) bound at the interface of the NG heterodimers, is positioned asymmetrically relative to the two nucleotides. It bridges the α- and γ-phosphates of the FtsY-bound nucleotide but does not contact the Ffh-bound nucleotide. The position of the central water relative to the FtsY-bound nucleotide mirrors the position of the Arginine Finger R789 in the complex of Ras with its GTPase activating protein, GAP334 (PDB 1WQ1) (Scheffzek et al., 1997). The central water is within hydrogen bonding distance of the two asymmetrically positioned arginine sidechains of FtsY (R333) and Ffh (R141), suggesting that it plays a similar role in the stabilization of negative charges during GTP hydrolysis (Figure 6, Figure S5, Figure 7a).
The final stage of the SRP targeting process involves GTP hydrolysis followed by the dissociation of the SRP:FtsY complex. To prevent premature release of the cargo, GTP hydrolysis must be delayed until the signal sequence has been handed over to the translocon. Superposition of the Ffh:FtsY heterodimer to the distal region of a ribosome-bound SRP RNA (Halic et al., 2006a; Estrozi et al., 2011) reveals that the access to the GTPase site with the flipped out G83 is sterically precluded. This is consistent with the observation that RNCs displaying signal sequence inhibit the binding of the GTPase dimer to the distal site as well as GTPase activation. The inhibition can be overcome by the addition of the translocon (Shen et al., 2012, Akopian et al., 2013b). Therefore, binding of the translocon to the SRP during signal sequence handover may induce a rotation the SRP RNA that would allow the insertion of the flipped out G83 base into the GTPase active site of the FtsY:Ffh complex, triggering GTP hydrolysis and complex dissociation (Figure 7b).
Conclusions
The structural investigation combined with biochemical experiments reported in this study reveal the molecular basis of the SRP receptor recruitment to the tetraloop of the SRP RNA and the GTPase activation at the distal site of the SRP RNA. Since the protein and RNA residues involved in the chemistry of GTPase activation are completely conserved, the identified chemical mechanism is likely to apply to SRP targeting across all kingdoms. Future work on this fascinating and complex system is likely to focus on the remaining mechanistic questions, in especially the role of the translocon during cargo handover.
Experimental Procedures
Protein and RNA expression and purification
The shortened RNA construct was generated by PCR from wild type SRP RNA from E. coli and refolded as described previously (Ataide et al., 2011). His-tagged Ffh (residues 1-298) and FtsY (residues 196-498) for crystallization and SEC binding assays were expressed in E. coli and purified essentially as described (Ataide et al., 2011). The Ffh:FtsY complex was formed by incubation of equimolar amounts of the two proteins for 3 days at 4oC in complex formation buffer (50mM HEPES-KOH pH7.5, 50mM NaCl, 2mM MgCl2, 2 mM TCEP) supplemented with 1mM GDP, 20mM NaF and 2mM AlCl3. The complex was purified on a Mono-Q column as described previously for the SRP:FtsY complex.. Full length Ffh for biochemical assays was produced as above and E277A/W/Q mutations were introduced by PCR. The mutations were confirmed by plasmid sequencing and protein mass spectrometry and do not alter the elution properties in analytical SEC. Wild type SRP RNA, and proteins for biochemical assays were expressed and purified as described (Peluso et al., 2001). Mutant SRP RNA was prepared by in vitro transcription as previously described (Ataide et al., 2011).
Crystallization conditions and structure determination
Purified Ffh:FtsY NG heterodimers were combined with an equimolar amount of refolded RNA. For crystallization, 1μL of well solution containing 18% PEG 3350, 100mM Bis-Tris pH 7.2, 140mM NH4OAc and 10mM MgCl2, was added to 1μL of sample and crystals were grown by vapor diffusion at 19°C in siting drops. Crystals were stabilized with 25% glycerol and flash-frozen in liquid nitrogen. Data were collected at the Swiss Light Source, PSI, Villigen on the Beamline X06SA. Data reduction and scaling were performed using the XDS package (Kabsch, 2010). The structure was solved by molecular replacement with a models of the Ffh-FtsY NG heterodimer (PDB 2CNW) using PHASER (McCoy et al., 2007). Iterative rebuilding of the model was performed in COOT (Emsley et al., 2010) and ONO (Jones et al., 1991) with several rounds of XYZ, individual ADP and TLS refinement in PHENIX (Adams et al., 2002). TLS groups were defined automatically and updated manually. NCS restraints were applied between chains A/C and B/D during refinement along with secondary structure restraints to limit distortions at the more flexible periphery of the complex. Small thermal motions are observed for the GTPase centers and protein-RNA interfaces, while the N-domains and the termini of the RNA display increased temperature factors. All crystallographic figures were generated with Pymol (Schrödinger).
Analytical size exclusion chromatography
Purified Ffh and FtsY NG-domains were combined in equimolar amounts (approx. 300 μM each) and incubated overnight with 1 mM nucleotide analogue (1mM GMPPCP or 1 mM GDP, 2mM AlCl3 and 4.5 mM NaF) in complex formation buffer (50 mM Hepes pH7.5, 50 mM NaCl, 1 mM TCEP). GDP:AlF4 bound NG-dimer:RNA complexes formed rapidly at room temperature. All complexes were set up by incubating 5 μM refolded RNA (Ataide et al., 2011) with 2.5-50 μM NG-dimer 10-30 minutes prior to size exclusion chromatography. 50 μL of each complex were injected onto a Superdex 200 3/150 (GE Healtcare) equilibrated in NG-SEC buffer (50 mM Hepes KOH pH 7.5, 150 mM Potassium Acetate, 1mM TCEP and 25 μM of the corresponding nucleotide analogue).
GTPase activity
Measurements of the stimulated GTPase reaction between SRP and FtsY were carried out as described before (Ataide et al., 2011). In general, reactions containing 100 nM Ffh, 200 nM SRP RNA, and varying concentrations of FtsY were initiated by adding 100 μM GTP (doped with γ-32P-GTP). Reactions were then quenched by 0.75 M KH2PO4 (pH 3.3) at different time points. The product (Pi) was separated from unreacted GTP by thin layer chromatography (TLC) and quantified by autoradiography.
Translocation Assays
Efficiency of the co-translational protein targeting and translocation were determined as described (Powers and Walter, 1997; Shan et al., 2007). Reactions contained 10 μL in vitro translation mixtures synthesizing 35S-methionine labeled preprolactin (pPL), to which 67 nM Ffh, 333 nM wildtype or mutant SRP RNA, 333 nM FtsY, and 0.5 eq/μL of salt washed, trypsin digested microsomal membrane was added to a total volume of 15 μL. Reactions were analyzed by SDS-PAGE followed by autoradiography.
Alignments
Alignments were prepared using CLUSTAL W (Larkin et al., 2007) and visualized using ALINE (Bond and Schüttelkopf, 2009).
Accesion numbers
The coordinates of the structure reported in this paper have been deposited in the Protein Data Bank under ID code 4c7o.
Supplementary Material
Highlights.
Tetraloop interactions suggest a mechanism of detachment from the ribosome
The activated complex structure reveals residues that stimulate GTP hydrolysis
The identified key residues in the SRP RNA and FtsY are universally conserved
Structural and biochemical data suggest a mechanism of GTPase activation
Acknowledgments
We acknowledge support by the Swiss National Science Foundation (SNSF), the National Center of Excellence in Research (NCCR) Structural Biology program of the SNSF, and European Research Council grant 250071 under the European Community's Seventh Framework Programme. N. S. was supported by the Boehringer-Ingelheim Fonds. We thank Daniel Boehringer and Marc Leibundgut for critical discussions of the manuscript and Stephan Imseng for contributions to the initial stages of the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan S. Signal Recognition Particle: An Essential Protein-Targeting Machine. Annual Review of Biochemistry. 2013a;82(1):693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Dalal K, Shen K, Duong F, Shan S. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. The Journal of Cell Biology. 2013b;200(4):397–405. doi: 10.1083/jcb.201208045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C. The tmRDB and SRPDB resources. Nucleic Acids Research. 2006;34:D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Bond CS, Schüttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu Rev Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Current Opinion in Structural Biology. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrozi LF, Boehringer D, Shan SO, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia PJ, Gawronski-Salerno J, Coon JS, Freymann DM. Structure of a GDP:AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of a peripheral nucleotide interaction site. J Mol Biol. 2006;360:631–643. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. Rna. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc Natl Acad Sci U S A. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Jagath JR, Matassova NB, de Leeuw E, Warnecke JM, Lentzen G, Rodnina MV, Luirink J, Wintermeyer W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. Rna. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Martın-Garcıa F, Mendieta-Moreno JI. The Role of Gln 61 in HRas GTP Hydrolysis: A Quantum Mechanics/Molecular Mechanics Study. Biophysical Journal. 2012;102(1):152–7. doi: 10.1016/j.bpj.2011.11.4005. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science-AAAS-Weekly Paper Edition. 1995 doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. Embo J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraogi I, Akopian D, Shan SO. A tale of two GTPases in cotranslational protein targeting. Protein Sci. 2011;20:1790–1795. doi: 10.1002/pro.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Schrödinger The PyMOL Molecular Graphics System [Google Scholar]
- Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. The Journal of Cell Biology. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Stroud RM, Walter P. Mechanism of Association and Reciprocal Activation of Two GTPases. PLoS Biol. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Shan SO. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci U S A. 2010;107:7698–7703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Arslan S, Akopian D, Ha T, Shan SO. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature. 2012;492:271–275. doi: 10.1038/nature11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu FY, Spanggord RJ, Doudna JA. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. Rna. 2007;13:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Weiche B, Bürk J, Angelini S, Schiltz E, Thumfart JO, Koch HG. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol. 2008;377:761–773. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A. Signaling mechanistics: aluminum fluoride for molecule of the year. Current Biology. 1997 doi: 10.1016/s0960-9822(06)00355-1. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JLJ. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kung S, Shan SO. Demonstration of a multistep mechanism for assembly of the SRP-SRP receptor complex: implications for the catalytic role of SRP RNA. J Mol Biol. 2008;381:581–593. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Yamamoto R, Sokabe M, Tanaka I, Yao M. Crystallization and preliminary X-ray crystallographic analysis of eIF5BΔN and the eIF5BΔN-eIF1AΔN complex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:730–733. doi: 10.1107/S1744309111015910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. Embo J. 1990 doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


