Abstract
Transcription factors are key regulators in both normal and pathological cell processes. Affecting the activity of these proteins is a promising strategy for understanding gene regulation and developing effective therapeutics. CoIII Schiff base complexes ([Co(acacen)(L)2]+ where L = labile axial ligands) have been shown to be potent inhibitors of a number of zinc metalloproteins including Cys2His2 zinc finger transcription factors. Inhibition by [Co(acacen)(L)2]+ of the target protein is believed to occur through a dissociative exchange of the labile axial ligands for histidine (His) residues essential for function. Here, we report a series of spectroscopic investigations with model peptides of zinc fingers that elucidate the interaction between [Co(acacen)(L)2]+ complexes and zinc finger transcription factors. Observed changes in NMR chemical shifts and 2D 1H-1H NOESY NMR spectra demonstrate the preference of [Co(acacen)(L)2]+ complexes to coordinate His residues over other amino acids. The conformation of [Co(acacen)(L)2]+ upon His-coordination was characterized by 1H NMR, near-UV circular dichroism, and electronic absorption. These studies reveal that the resulting His-coordinated [Co(acacen)(L)2]+ complex possesses an octahedral structure. The effects of [Co(acacen)(L)2]+ complexes on the zinc finger structure were assessed by the degree of hydrogen bonding (probed by 2D NMR) and secondary structure profiles measured by far-UV circular dichroism. These structural studies demonstrate the ability of [Co(acacen)(L)2]+ complexes to disrupt the ββα structure of zinc fingers, resulting in primarily random coil conformations. A mechanism is described wherein [Co(acacen)(L)2]+ complexes inhibit zinc finger transcription factor activity through selectively coordinating His residues in the zinc finger via dissociative ligand exchange and disrupting the ββα structural motif required for gene regulation.
Keywords: Cobalt, Schiff bases, Zinc Fingers, Spectroscopy, Transcription Factors
Introduction
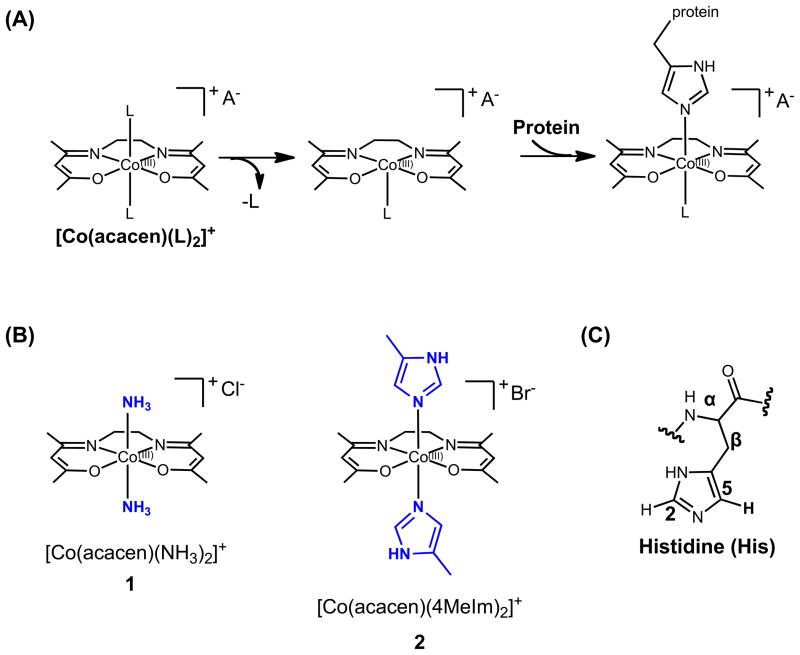
Advances in the understanding of metal-protein binding interactions have stimulated the development of metal-based protein inhibitors to modulate biological processes and alter disease progression.[1] CoIII Schiff base complexes bearing labile axial ligands ([Co(acacen)(L)2]+) inhibit histidine-containing proteins and enzymes including zinc finger (ZF) transcription factors (TFs) and metalloendopeptidases (Figure 1a).[1c,2] The mechanism of inhibition is believed to depend on the dissociative exchange of the labile axial ligands of the complex, enabling the [Co(acacen)(L)2]+ complex to coordinate the imidazole ring of histidine (His) residues and disrupt protein function.[2d, 3]
Figure 1.
Structures and proposed mechanism of inhibition of histidine-containing proteins by CoIII Schiff base complexes. (A) CoIII Schiff base complexes contain a CoIII metal center stabilized by a tetradentate acetylacetonatoethylenediimine ligand (acacen). The axial positions bear labile ligands that are believed to undergo dissociative ligand exchange allowing coordination of imidazole side chains of His residues. (B) CoIII Schiff base complexes used in the present studies, [Co(acacen)(NH3)2]+ (1) and [Co(acacen)(4MeIm)2]+ (2). (C) Naming scheme for histidine protons. Focus is placed on the methine protons of the imidazole referred to as H2 and H5.
His-containing ZF TFs regulate changes in gene expression through sequence-specific interactions with DNA. These proteins are known to mediate vital cell processes such as embyonic development and apoptosis.[4] Small molecule inhibitors of transcription are powerful chemical probes to study gene regulation, bypassing the difficulties associated with the current genetic methods of gene knockdown (RNAi) or knockout.[4] In addition to their role in normal physiology, aberrant activation of ZF TFs has been strongly implicated in the pathological processes including cancer metastasis and tumorigenesis.[4c, 5] Oncogenic TFs have been shown to regulate the production of multiple oncogenic proteins, and inhibition of one of these TFs may potentially disrupt several downstream protein targets of current chemotherapy.[4a,4c,5–6] Consequently, small molecule TF inhibitors are promising agents for potent therapeutic intervention.[5a, 6–7]
Recently, highly specific and potent inhibition of ZF TFs by [Co(acacen)(L)2]+ complexes with labile ammine ligands ([Co(acacen)(NH3)2]+ = 1, Figure 1b) was achieved by attachment of targeting DNA oligonucleotides to the acacen equatorial ligand.[1c, 2a–d] The 1/DNA conjugates selectively disrupt DNA-binding activity of the target TFs, namely the Snail[2a, 2c] and Gli[2b] families, that are both implicated in development and cancer progression. Through inhibition of the DNA-binding activity, the conjugates were shown to alter biological processes associated with the TFs in cancer cell lines in vitro (such as transcriptional repression) and embryo models in vivo (for example, neural crest formation in Xenopus laevis and denticle belt patterning in Drosophila).[2a, 2b] The ability of these 1/DNA conjugates to affect signaling pathways through TF inhibition strongly suggests that CoIII Schiff base complexes have potential as research tools and ultimately new therapeutics.
[Co(acacen)(L)2]+ complexes with labile ligands like 1 are thought to inhibit TFs by disrupting the structure of Cys2His2 ZFs in the DNA-binding domains.[2c, 2d] The proper folding of a Cys2His2 ZF into a functional structure is dependent on the presence of ZnII. Tetrahedral coordination of ZnII stabilizes the ZF into a ββα motif comprising a Cys2-containing antiparallel β-sheet in the C-terminus and His2-containing α-helix in the N-terminus.[8] The α-helices of tandem ZFs in TFs cooperatively bind the major groove of DNA in a sequence-specific manner to confer gene regulatory function.[8b, 9] Displacement of ZnII and insertion of the octahedral [Co(acacen)(L)2]+ complex in the ZnII binding site is proposed to disrupt the ββα structure, impairing DNA recognition and transcriptional activity.[2d] To date, a detailed characterization of the interaction between [Co(acacen)(L)2]+ complexes and Cys2His2 ZFs has not been performed. Elucidation of this interaction is necessary for the further development of this class of CoIII complexes as TF inhibitors.
This study investigates the interaction of [Co(acacen)(L)2]+complexes with ZF motifs at the molecular level using model peptides. Protein-ligand interactions are often evaluated with 3D structures determined by solid-state X-Ray crystallography or distance restraints from solution-state NMR. However, the fluxional conformation dynamics of the ZF peptides treated with [Co(acacen)(L)2]+ complexes (such as 1) made high-resolution 3D structures of the model peptides prohibitively difficult to obtain. In order to overcome this challenge, the model peptides were investigated by a series of spectroscopic methods to elucidate a comprehensive mechanism of inhibition. 2D NMR, circular dichroism (CD), and electronic absorption spectroscopy experiments were performed, and the results demonstrate that 1 selectively coordinates His residues at the axial positions. Consequently, 1 disrupts the structure of the ββα ZF motif necessary for function. In addition to providing valuable insight into the TF inhibitory activity of CoIII Schiff base complexes, the present work demonstrates the unique employment of spectroscopic methods to characterize interactions between metal coordination complexes and target proteins.
Results and Discussion
Model Peptides
The model peptides used in this study are summarized in Table 1. Two 26-mer model peptides were used to elucidate the interaction between 1 and the zinc finger motif: ZF4 derived from the fourth ZF of the five ZF-containing Human Snail 1 (exon residue 207–233),[10] and CP1, a consensus zinc finger peptide optimized for zinc binding.[11] The interaction between 1 and His in the zinc-binding region of zinc fingers was investigated using a short His-rich (HR) peptide fragments: a 10-mer model peptide derived from the His-rich region of ZF4 (residues 16–25) named HR1.
Table 1.
Sequences of the model peptides used to understand the interaction between [Co(acacen)(L)2)]+ and ZFs.A
| a) Peptides Containing ββα zinc finger motif
| |
|---|---|
| Name | Sequence |
| ZF4 | KSCPH CSRAF ADRSN LRAHL QTHSD V |
| CP1 | PYKCP ECGKS FSQKS DLVKH QRTHT G |
| b) Peptide Containing His-rich region of Zn-binding site | |
|---|---|
| Name | Sequence |
| HR1 | RAHLQ THSDV |
Two 26-mer peptides, ZF4 and CP1, were used to understand effects on the whole zinc finger motif and a shorter His-rich (HR) peptide, HR1 focused investigations on the His coordination. Residues involved in ZnII-binding are underlined and all histidines are italicized.
Selectivity of CoIII Schiff Base Coordination to Histidines
Previous studies of the protein inhibition of 1 have suggested that protein inhibition occurs by interactions between the metal center and His residues that are crucial for function.[2a–d, 3, 12] 2D 1H NMR spectroscopy experiments were conducted to determine whether 1 is selective for binding His over other amino acids.
NMR Chemical Shifts of His Protons
Although structural changes may affect chemical shifts of peptide protons due to altered hydrogen bonding, coordination of amino acids to transition metal complexes is expected to influence chemical shifts to a greater extent.[2d, 13] The chemical shifts in the aromatic regions of the 1H NMR and the 1H-1H-TOCSY spectra of the model peptides listed in Table 1 were analyzed in the presence (1-Peptide) or absence (apo-Peptide) of 1 (Figure 2 and SI Figure 1). Correlations from 2D-TOCSY experiments arise from mutually spin-spin coupled nuclei and can resolve overlapping peaks with different coupling environments such as the backbone NH and His imidazole protons.
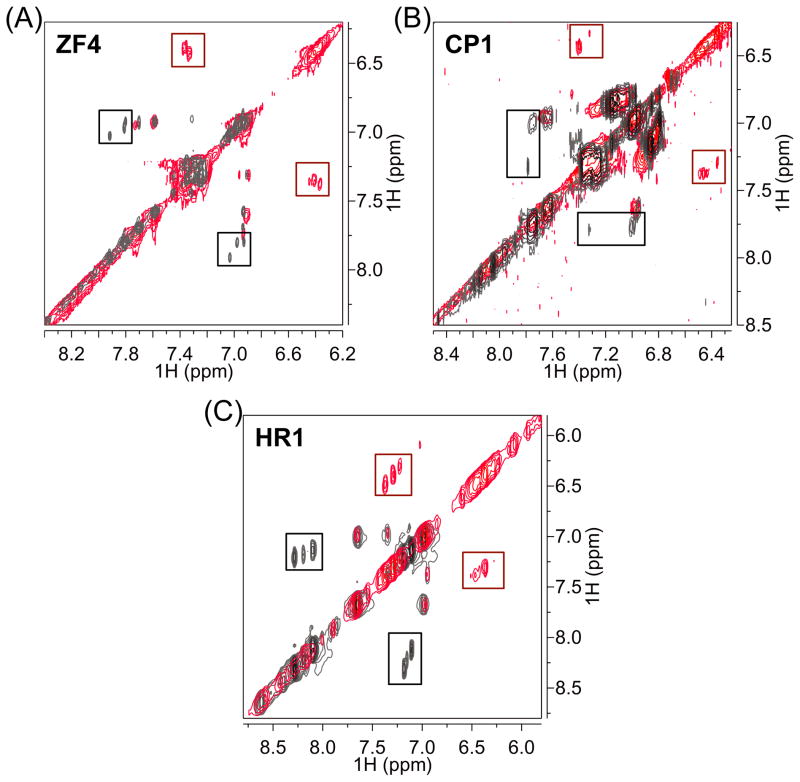
Figure 2.
Overlays of the downfield region of the 2D 1H-1H TOCSY spectra of ZF4 and CP1 at 15 ºC and HR1 at 10 ºC incubated with (red) without and 1 (gray). Boxes indicate crosspeaks between histidine side chain methine protons H2 and H5 for all His residues in each peptide. Notable upfield shifts of His proton resonances are observed in all peptides in the presence of 1 (red boxes) as compared to resonances of the free peptides (gray boxes).
Free ZF4 (apo-ZF4) is expected to adopt a random coil structure due to the absence of a ZnII ion to stabilize the structure.[8] In the spectra of free ZF4 (apo-ZF4) (Figure 2a, gray, and SI Figure 1a), peaks corresponding to the side chain methine protons H2 and H5 (See naming scheme in Figure 1c) of free His residues occur at 7.84 – 7.96 ppm and 6.96 – 7.02 ppm, respectively. In the spectra of ZF4 treated with 1 (1-ZF4) (Figure 2a, red), the H2 and H5 protons are shifted upfield to 7.25 – 7.28 ppm and 6.37–6.42 ppm respectively. Therefore, adding 1 to ZF4 induces changes in chemical shifts (Δδ) of approximately between 0.48 – 0.60 ppm for H2 and H5. Similar phenomena were observed for the CP1 and HR1 peptides (Figure 2b, 2c and SI Figure 1b, 1c), indicating that 1 significantly shifts His proton resonances upfield relative to the free peptides. The upfield-shift of the His imidazole resonances in the presence of 1 agrees with previous 1D 1H NMR studies investigating the interaction of 1 with a Cys3His peptide derived from the HIV-1 nucleocapsid protein, NCp7.[2d] Further, the shifts in 1H resonances of the ZF peptides can be correlated to the formation of a 1/Peptide adduct observed by ESI-MS (SI Figure 2).
In order to assess the coordination preference of 1 for His residues over other amino acids (especially the Cys residues involved in Zn II-coordination), the effects of the addition of 1 on the resonances of the amino acid residues within the model peptides were compared. 1H NMR resonances of the ZF4 and HR1 peptides in the apo-Peptides and 1-Peptides were assigned using 1H-1H 2D-TOCSY and 2D-NOESY experiments (SI Table 1–3). Addition of 1 to the CP1 peptide (1-CP1) led to a loss of several backbone NH 1H resonances relative to the apo-CP1 complex, precluding complete assignment of the 1-CP1 spectra. This loss of signal is likely due to conformational heterogeneity resulting from multiple coordination possibilities between 1 and the His residues of CP1. The difference in proton chemical shifts between the 1-Peptides and the apo-Peptides were determined for each assignable resonance (Δδ = δ(apo-Peptide) – δ(1-Peptide)) and plotted against the peptide sequence (Figure 3, SI Table 3, SI Figure 3, 4).
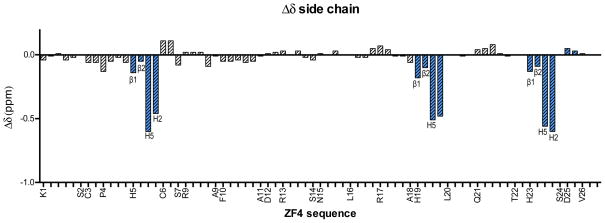
Figure 3.
The Δδ (δ(1-Peptide) – δ(apo-Peptide)) of amino acid side chain 1H chemical shifts (in ppm) plotted against peptide sequence of ZF4 at 15 ºC. For clarity, only the individual protons of the His residues are labeled while the remaining residues are labeled by the residue name at the corresponding Hβ/Hβ1. The remaining protons are plotted in order of closeness to the Hβ (e.g. K1 is plotted along the x-axis as Hβ1, β2, Hγ, Hδ, and Hε but labeled as K1 at the Hβ1 position). Fully labeled Δδ plots are available in SI Figure 5.The Δδ of the His protons are highlighted in blue for emphasis and labeled according to the Figure 1b. Coordination selectivity was observed by the significant effects of 1 on the His imidazole 1H resonances as compared to the other amino acid protons.
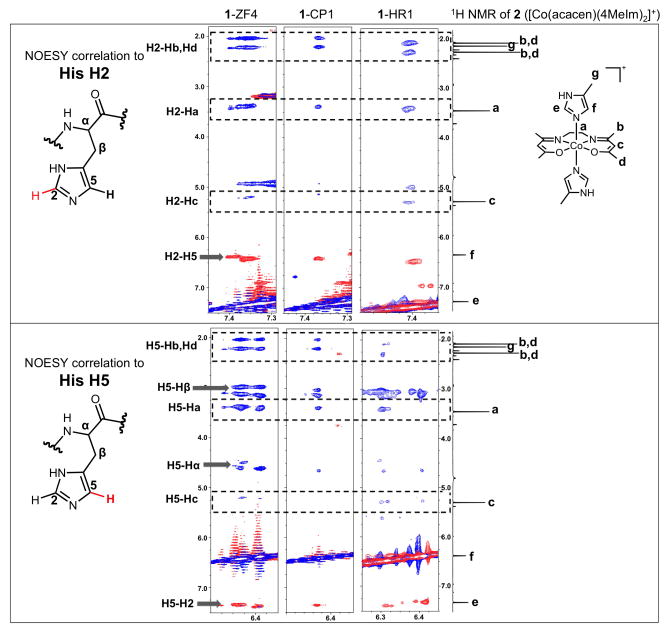
Figure 4.
Overlays of the 1H-1H NOESY spectra with 150 ms mixing time (blue) and 1H-1H TOCSY spectra (red) with 60 ms mixing time of His H2 (top panel) and H5 (bottom panel) protons of 1-ZF4 and 1-CP1 at 15 ºC and 1-HR1 at 10 ºC. For comparison, the 1D 1H NMR spectrum of 2 ([Co(acacen)(4MeIm)2]+), a small molecule model of [Co(acacen)(L)2]+ coordinated to two His, at 10 ºC is included along the right-hand edges of both sections of the figure. The intra-residue TOCSY and NOESY correlations of the His residues are labeled and indicated by arrows. In addition to the intra-residue correlations, NOE correlations are present with resonances that correspond to the protons in the acacen ligand of [Co(acacen)(L)2]+. These NOEs are indicated by dashed boxes that are inclusive of the homologous NMR peaks of 2. The presence of such NOEs demonstrates the close proximity of the protons of the acacen chelate to the His imidazole protons. The homology of 1H resonances to 2 validates the formation of a [Co(acacen)(His)x]+ species upon adding 1 to His-containing peptides.
The Δδ values of the model peptide protons can be used to assess the coordination preference of 1 for His over other residues in the peptide sequence. Relative to the Δδ values of non-His side chain protons, significant negative Δδ values of the three His side chain protons, particularly at the imidazole protons (|Δδ| > 0.45 ppm, upfield shift), are observed for ZF4 (Figure 3, SI Figures 3, 4). In addition, the Δδ magnitudes of the His side chain proteins are significantly greater than the Δδ magnitudes of the backbone NH and the CHα of the peptide. This indicates that the chemical shift effects at the His side chains are not solely due to a change in peptide structure. No notable differences are observed between the Δδ values of the two His in the Cys2His2 binding site (His19 and His23) and the third His of ZF4 (His5) at the concentration of 1 tested (2.2 molar equivalents of 1 to ZF4).
The pronounced shifts of the His side chain resonances in comparison to the other amino acid residues demonstrate the preferential coordination of His to 1 (Figure 3). These shifts are specifically observed at the His imidazole protons, and are markedly larger than the His CHα and CHβ protons (|Δδ| = 0.05 – 0.08 ppm and 0.05 – 0.18 ppm upfield shift, respectively), indicating that coordination of 1 occurs at the electron-donating imidazole nitrogens of the His and not the peptide backbone (Figure 3, SI Figure 3, 4).
Two effects may contribute to the observed chemical shift changes in ZF4 upon replacing ZnII with 1. One could be local electronic effects associated with coordination to the CoIII metal center. The other could be global structural changes that alter the electronic environments of the peptide protons. The latter may be observed if 1 stabilizes a structure that deviates from the expected random coil of the apo-ZF4. To determine the magnitude of the local electronic effects induced by 1 on the His2 region of the Cys2His2 ZnII-binding site, the truncated 10-mer derivative of ZF4, HR1, was evaluated in the absence (apo-HR1) or presence (1-HR1) of 1. Chemical shift changes observed in the HR1 model peptide must be associated primarily with local electronic effects rather than structural changes, since the HR1 peptide is too short to adopt significant secondary structure.
The Δδ values for HR1 show significant upfield shifts of the His protons from coordination to 1, as observed with ZF4 (SI Figure 5). The Δδ magnitudes are especially strong for the imidazole methine H2 and H5 protons. Significant changes in the chemical shifts of the arginine residue were not observed after binding to 1, further confirming that 1 is selectively coordinated by His residues. This pattern of chemical shift changes is also observed with ZF4 and HR1. The consistency and degree of the coordination demonstrated by the Δδ plots in the ZF4 and HR1 model peptides imply that His residues are the selective ligand target of 1, regardless of peptide structure and sequence. Previous work has shown that addition of a targeting domain to the acacen backbone of [Co(acacen)(L)2)]+ complexes confers specificity.[2c,2d,2f] This study gives the first direct evidence to demonstrate that [Co(acacen)(L)2)]+ complexes alone (without targeting domains) possess a degree of specificity through selective binding to His residues over other amino acids in a peptide sequence.
NOESY Correlations to His Protons
The interaction between His residues and 1 was further analyzed by the 1H-1H NOESY spectra of the His imidazole protons of the ZF4, CP1 and HR1 model peptides (Figure 4). Protons of molecules partaking in intra-residue and intermolecular interactions within NOE proximity (~ 4 Å) of His residues in the model peptides can be identified through NOE correlations to the His protons. The protons of the non-labile equatorial acacen chelate can be used to probe intermolecular interactions involving 1 with 1H NMR experiments. For help with assignment, the resonances of correlations arising from the 1H-1H NOESY spectra of ZF4, CP1, and HR1 in the presence of either ZnII (Zn-Peptide) or 1 were compared to the 1H spectrum of [Co(acacen)(4MeIm)2] (2 in Figure 1b, 4MeIm = 4-methylimidazole). Complex 2 was used as a small molecule model of [Co(acacen)(L)2]+ with two His residues at the axial positions.
As expected for both the Zn-Peptide and 1-Peptide NOESY spectra, intra-residue NOE correlations were observed between the His H5 and His Hβ (Figure 4, SI Figure 6a). Weak intra-residue NOE correlations were observed between the H2 and H5 of the imidazoles, but cannot be seen at the contour levels shown in this figure. In addition to the expected intra-residue His NOE peaks, the spectra of the 1-Peptides exhibit strong NOE correlations from the His H2 and H5 protons to resonances at 2.0, 2.2, and 3.5 ppm and weak NOE correlations at 5.2 ppm. The chemical shifts of these resonances correlate well to previous assignments of the protons of the acacen chelate[14] in the [Co(acacen)(L2)]+ complex.
The chemical shift correlations indicate that these NOE peaks represent intermolecular NOE from close proximity of the acacen ligand of 1 to the His imidazole protons. The NOE measurements support a model wherein His residues of the peptides coordinate the CoIII center of 1 while CoIII-coordination of the acacen equatorial chelate is retained (See scheme of NOE correlations in SI Figure 6b). Further, no detectable NOEs between other amino acid protons and the protons of the acacen chelate were observed in the full NOESY spectra of the 1-treated peptides validating the selectivity for interactions of 1 with the His imidazoles (SI Figure 7).
The chemical shifts of the observed NOE correlations to the His H2 and H5 protons were compared to 2. 4MeIm can be used as a simple model of the His side chain[15]; therefore, 2 was studied as a model of [Co(acacen)(L)2]+ axially coordinated to two His residues. The resonances of the His H2 and H5 NOE crosspeaks exhibit strong resemblance to the 1H spectrum of 2. The chemical shifts of the imidazole (protons e and f in Figure 4) and acacen protons (protons b, c, and d in Figure 4) of 2 correlate well to the chemical shifts of the His H2 (labeled as H2-H5, H2-a, H2-b, H2-c in Figure 4) and H5 (labeled as H5-H2, H5-a, H5-b, H5-c in Figure 4) crosspeaks. The only exception to the chemical shift correlation between 2 and the 1-Peptide NOESY crosspeaks is the methyl resonance of the 4MeIm ligand at 2.2 ppm. This discrepancy is expected since the 4-methyl group is replaced by a downfield-shifted Hβ in His residues. The agreement in chemical shifts between the His H2 and H5 NOEs and 2 confirms the presence of a CoIII-His coordination interaction (SI Scheme 1). Together with the chemical shift perturbations observed in the Δδ plots, the NOESY experiments demonstrate the selective intermolecular coordination of 1 with His residues within ZF model peptides. To our knowledge, these studies are the first application of 1H-1H NOESY experiments to detect selective binding of metal-based protein inhibitors to amino acids within peptide systems.
Conformation of His-Coordinated [Co(acacen)(L)2]+ Complexes
The changes in NMR chemical shifts and NOESY correlations demonstrate that 1 interacts with His residues. To gain further insight into the conformation of the His-coordinated [Co(acacen)(L)2]+ complex, the structure of the 1-HR1 adduct with respect to the CoIII metal center was investigated by CD and electronic absorption studies. HR1 was used as a peptide model to focus investigations on the interactions of 1 with the His in the Cys2His2 ZnII-binding site.
CD of the Acacen Ligand
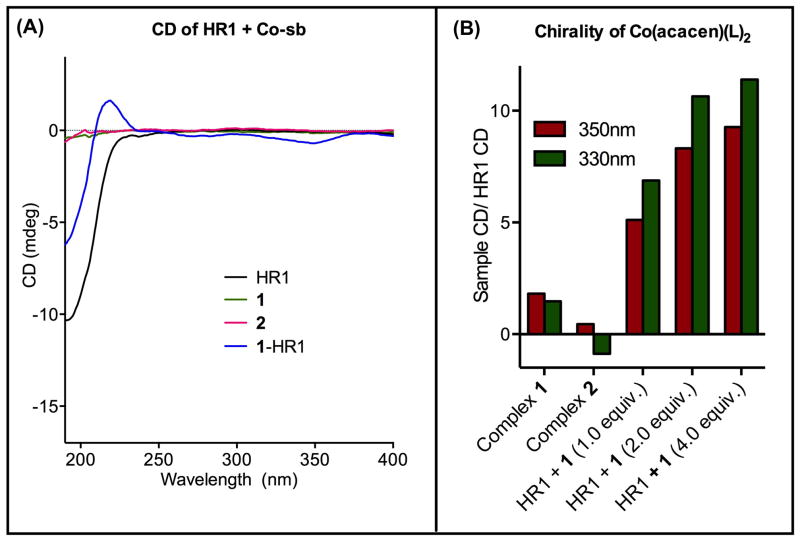
CD spectroscopy was used to evaluate the degree of chirality of [Co(acacen)(L)2]+ in the 1-HR1 adduct. The near-UV CD of the acacen electronic absorption band of 1-HR1 was measured and compared to the CD of the free HR1, 1, and the small molecule model, 2 (Figure 5).
Figure 5.
CD of 1-HR1 (1:HR1 = 1:1) and controls in the region of the electronic absorption bands of acacen at 25 ºC. A) The near-UV CD of 1-HR1 were compared to [Co(acacen)(4MeIm)2]Br, free HR1, and 1, negative controls for optical activity and chirality in the acacen absorption region. The presence of CD signal between 250 and 400 nm in the 1-HR1 validate His2 coordination that introduces chirality to 1. B) A plot of CD signal at 350 and 330 nm (the CD minimum and the λmax of the acacen absorption bands respectively) normalized to the signal of HR1 demonstrates the increase in optical activity of 1-HR1 with increasing concentrations of 1.
The CoIII-bound acacen ligand exhibits electronic absorptions in the near-UV region due to π-π* intraligand transitions.[16] Monodentate ligands with free rotation (such as NH3) at the axial positions result in achiral, and thus optically inactive [Co(acacen)(L)2]+ complexes. Coordination of a non-symmetric His2 ligand that precludes free rotation in the axial positions (such as a peptide with two coordinating His) is expected to introduce chirality to the resulting [Co(acacen)(L)2]+ species that can be detected by CD. In contrast to the NMR investigations, 2 was studied as a negative control in these CD studies since free rotation around the CoIII-4MeIm bond allowed by monodentate coordination would render the complex optically inactive. Although the HR1 peptide is optically active in the far-UV region (190–250 nm) due to the chiral peptide bonds, the peptide alone was found not to have any detectable CD in the near-UV region (>250 nm).
As expected, no detectable CD signal was observed for 1 or HR1. Upon treatment of HR1 with 1 to yield 1-HR1, negative CD signals were observed with minima at 280 nm and 350 nm. The appearance of these signals indicates that HR1 and 1 interact to introduce chirality to the [Co(acacen)(L)2]+ complex. No CD signal was observed in the 2 spectra in this region, attributing the observed optical activity to His-coordination specifically within a model peptide. Plots of the CD signals at 350 nm (CD minimum) and 330 nm (λmax of acacen absorption band) normalized to the signal of free HR1 (signal of the complex divided by signal of free HR1) (Figure 5b) show the dependence of signal intensity on the concentration of 1. At the lowest concentration of 1 analyzed (1:HR1 = 1:1), the observed signals at 350 and 330 nm are 3-fold greater than the CoIII complexes without peptide. The introduction of optical activity to 1 in the presence of HR1 suggests that the peptide coordinates to 1 through ligands with restricted rotation. Such rotational restriction likely results from simultaneous coordination of the two His residues of HR1 at the axial positions of 1.
Electronic Absorption of the Acacen Ligand
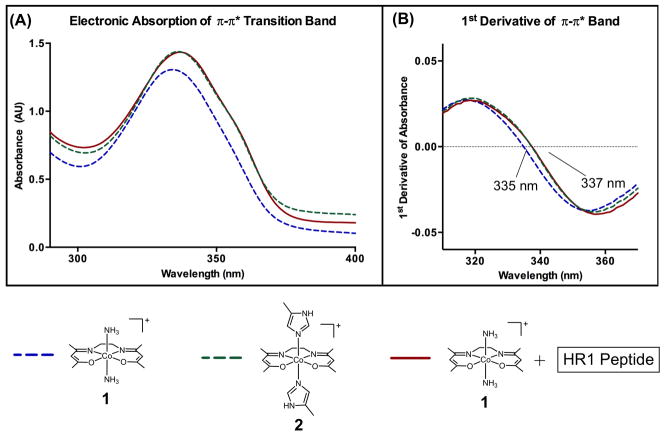
To characterize the conformation and geometry of the 1-HR1 adduct, its electronic absorption of was evaluated and compared to 1 and 2 (Figure 6). In contrast to the CD spectra, 1-HR1 exhibits an electronic absorption spectrum that closely resembles 2, demonstrating the presence of an octahedral [Co(acacen)(His)2]+ structure with the acacen in the equatorial plane and His2-coordination at the axial positions. The observed absorption bands agree with the previously published π-π* intraligand transition of CoIII-bound acacen, namely a maximum between 330–340 nm and a shoulder approximately 360 nm.[16]
Figure 6.
A) Electronic absorption spectrum of 1 + HR1 (1-HR1) near the acacen π-π* intraligand transition at 25 ºC in comparison to 1 and 2. The absorption band of 1-HR1 closely resembles 2, validating retention of the octahedral conformation upon His binding at the axial sites. B) First derivative of the π-π* transition band unambiguously assigns the λmax of 1-HR1 to 337 nm, equal to that of 2 and slightly red-shifted of the λmax of 1 at 335 nm.
In previous studies subtle differences in the π-π* transition λmax based on axial ligand identity were observed.[16] The λmax of the 1-HR1, 1 and 2 π-π* intraligand transition bands were determined with the first derivative plots of the electronic absorption spectra (Figure 6b). The λmax of the π-π* intraligand transition absorption band of 1-HR1 occurs at 337 nm, slightly red-shifted from the λmax of 1 at 335 nm and equal to the λmax of 2. These observations demonstrate the retention of the octahedral conformation of the CoIII center upon axial coordination of His within the HR1 peptide.
The electronic absorption data suggests that the discrepancy between the 2 and 1-HR1 CD spectra results from the difference in chiroptical properties not associated with change in the octahedral geometry or ligand conformation. Rather, the optical activity of 1-HR1 likely results from restriction in CoIII-imidazole bond rotation, that may result from His2 coordination as compared to the monodentate coordination of the 4MeIm.
The near-UV CD spectroscopy and electronic absorption studies presented here demonstrate the first use of these methods for both verification of His-coordination and [Co(acacen)(His)x]+ octahedral conformation in the studies of CoIII Schiff base complexes. Spectroscopic properties of [Co(acacen)(L)2]+ species were monitored in the near-UV wavelengths and were isolated from the far-UV signals of the peptide. These studies can be readily translated to the characterization of [Co(acacen)(L)2]+ species in the presence larger and more complex protein systems.
Structural Disruption of the Zinc Finger Motif by [Co(acacen)(L)2)]+ Complexes
The interaction of 1 with His residues is hypothesized to inhibit TFs via displacement of the tetrahedral ZnII ion with 1. This displacement is thought to disrupt the ββα structural motif required for sequence-specific DNA recognition.[2d] The structural effect of 1 coordinating to the full zinc finger motif ZF4 was investigated by chemical shift dispersion, exchange correlation NMR, and far-UV CD spectroscopy.
Structural Evaluation by NMR
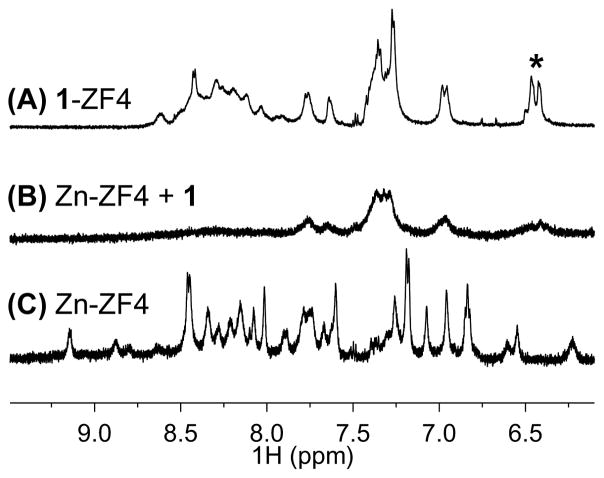
The effect of 1 on a ZnII-stabilized structure of ZF4 was evaluated by 1H NMR (Figure 7). Peptide backbone NH protons form hydrogen bonds within a structured peptide system that cause shifts in backbone NH proton resonances, dispersing the chemical shifts.[17] As expected, such dispersion is observed in the 1H NMR spectra of ZF4 in the presence of 2.2 equivalents of ZnII (Zn-ZF4), since ZnII stabilizes ZF motifs (Figure 8c) In contrast to Zn-ZF4, the spectra of 1-ZF4, exhibits a high degree of peak overlap suggests a low degree of structure commonly observed in random coil conformations (Figure 7a).[17–18] Upon addition of 1 to a solution of Zn-ZF4, loss in both signal and chemical shift dispersion is observed, suggesting that 1 is perturbing the ZnII-stabilized structure (Figure 7b). The resulting spectrum resembles 1-ZF4 (apo-ZF4 treated with 1). This may indicate that 1 is effectively displacing ZnII from the structure and forming a less structured adduct with 1. Perturbations in the well-defined spectra of Zn-CP1 are also observed upon addition of 1 (SI Figure 8). In contrast to ZF4, the spectrum of Zn-CP1 treated with the same equivalents of 1 bears less resemblance to 1-CP1 and more retention of the Zn-CP1 NMR peaks. This may result from tighter binding of ZnII to CP1 than to ZF4. CP1 contains an N-terminal aromatic residue (Phe11) that is believed to stabilize the ββα motif and ZnII—coordination.[11] The absence of such an aromatic residue in ZF4 could destabilize ZnII-coordination. The resulting higher ZnII binding affinity of CP1 could decrease the sensitivity of CP1 to perturbations by 1. Ongoing work is quantifying thermodynamics of metal-binding (of both Co(III) Schiff base complexes and ZnII) to various ZF model peptides to further elucidate the parameters (such as sequence) that influence binding affinities and stoichiometries. Nonetheless, these 1H NMR studies suggest that 1 can compete with ZnII to bind to ZF peptides and the interaction is influenced by the remaining peptide sequence and possibly the ZnII-binding propensities.
Figure 7.
1H NMR of the backbone NH of (A) 1-ZF4, (B) Zn-ZF4 challenged with 1 and (C) Zn-ZF4. Addition of 1 to Zn-ZF4 leads to a loss in signal and chemical shift dispersion and produces a spectrum that resembles 1-ZF4 (apo-ZF4 treated with 1).
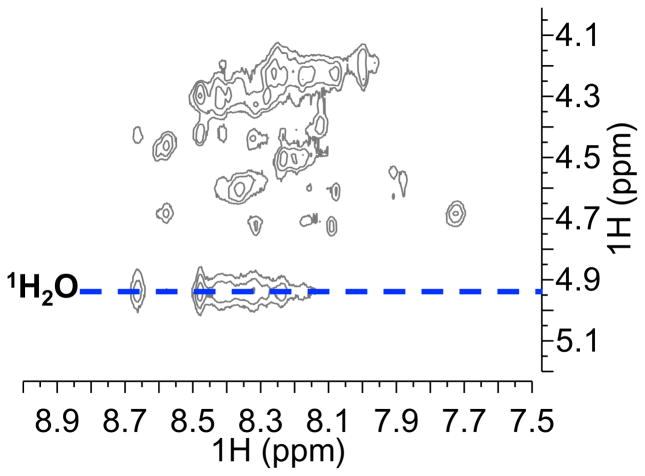
Figure 8.
1H-1H NOESY with 150 ms mixing time at 15 ºC in the backbone NH-CHα region of 1-ZF4. Rate of chemical exchange of backbone NH protons with water protons (δ 1H2O = 4.9 ppm, 15 ºC) was used to evaluate the degree of hydrogen bonding and consequently, structure. The position of the water peak is indicated by the blue dotted line. Peaks from the backbone NH of 1-ZF4 (as with apo-ZF4) exhibit detectable chemical exchange with water in comparison to those of Zn-ZF4 (SI Figure 9). This indicates a low degree of hydrogen bonding and from a disordered structure adopted by 1-ZF4.
NMR investigations of the ZF4 structure were conducted with 1H-1H NOESY experiments to assess the exchange rate of backbone NH protons with water (Figure 8, SI Figure 9). NOE correlations can be used to detect chemical exchange occurring at timescales shorter than the mixing time in the pulse sequence.[17–18] In an aqueous environment, backbone NH protons of a structured system involved in hydrogen bonding are expected to display low rates of chemical exchange with water. Loss of hydrogen bonding and increased disorder increases the rate of chemical exchange of the backbone NH protons with water protons. This high rate of exchange produces detectable water 1H-NH exchange correlations in the NOESY spectra. No detectable water 1H-NH correlations (δ 1H2O = 4.9 ppm, 15 ºC) were observed in the Zn-ZF4 NOESY spectra with 150 ms (SI Figure 9b) mixing time, confirming the presence of a well-ordered ZF4 structure stabilized by ZnII coordination. In contrast, strong water 1H-NH exchange correlations were observed in the 1-ZF4 NOESY spectra with the same mixing time, suggesting the absence of backbone NH hydrogen bonding due to a loss of structure (Figure 8). Similar water 1H-NH exchange correlations were observed in the apo-ZF4 NOESY spectra that is expected to adopt a random coil structure (SI Figure 9a). The NOESY data validate the hypothesis that replacement the 1-ZF4 adduct does not adopt significant structure.
Assessment of Secondary Structure by CD
CD spectroscopic analysis of peptides and proteins in far-UV wavelengths (190–250 nm) can reveal important secondary structure features. CD experiments were conducted with ZF4 to correlate the structural insight from the NMR experiments to secondary structure effects of 1 on the ββα motif of the peptide. The far-UV CD spectra of free ZF4 (apo-ZF4) or ZF4 in the presence of ZnII or 1 were monitored for signals characteristic of random coils (peak minimum at 198 nm) and ββα ZF motifs (minima at 208 and 220 nm and a maximum at 190 nm).[19]
ZnII was titrated into apo-ZF4 to evaluate the ability of ZF4 to adopt a ZnII-induced ZF structure (SI Figure 10). In the absence of ZnII, apo-ZF4 displays a minimum at 198 nm indicating that the free peptide is primarily random coil. Upon addition of ZnII, the 198 nm signal of apo-ZF4 is reduced in a concentration-dependent manner to reveal a spectrum characteristic of the ββα motif with minima at 208 and 220 nm and a maximum at 190 nm. The observed behavior of ZF4 is consistent with previously studied ZF peptides,[8a, 8b, 11b, 20] validating the peptide as an appropriate model of a folded ZF motif. A maximum structural effect was achieved at a 4:1 ZnII/ZF4 ratio. This ratio was employed in the subsequent titration experiment of 1 to ensure that the Zn-ZF4 model possessed a ZnII-stabilized ββα structure.
In previous work, addition of 1 was shown to induce the release of the ZnII ion in a ZnII-bound ZF peptide.[2d] CD signals of Zn-ZF4 (ZnII/ZF4 = 4:1) treated with 1 were monitored to determine the structural consequences of 1-induced displacement of the ZnII ion. Addition of 1 to Zn-ZF4 induced a concentration-dependent reduction of the 208 nm minimum and 190 nm maximum of the ββα motif and rise of the minimum at 198 nm of a random coil structure (Figure 9, SI Figure 11). The resulting CD spectrum closely resembles that of the apo-ZF4. It is worth noting that a small feature, namely a minimum at 227 nm is observed that deviates slightly from the shoulder observed in the apo-ZF4 spectra. This feature may result from the formation of disulfide bonds between the free cysteines of 1-ZF4 (See Supplementary Discussion 2). The CD data supports a mechanism of inhibition whereby His-coordination to 1 induces a loss of the functional ββα ZF structure by displacement of the tetrahedral ZnII and binding of the octahedral [Co(acacen)(L)2]+ complex.
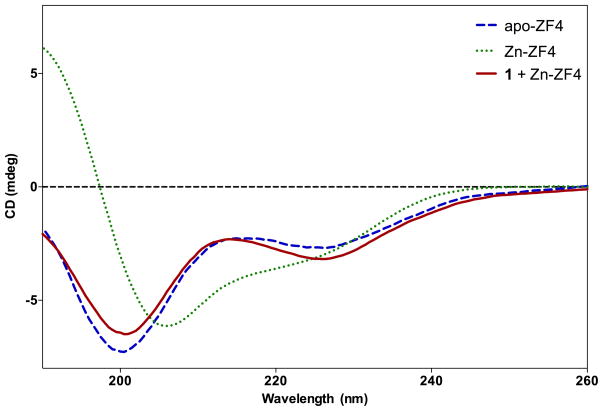
Figure 9.
CD of apo-ZF4 (free), Zn-ZF4 and Zn-ZF4 + 1 at 25 ºC. Apo-ZF4 is characterized by a minimum at 198 nm expected of a random coil. Zn-ZF4 exhibits a maximum at 190 nm, a minimum at 208 nm, and a shoulder at 224 nm, characteristic of a ββα motif. 1 addition to Zn-ZF4 disrupts the ββα motif as illustrated by a loss of the 208 minimum of the ββα motif and a shift towards the random coil 198 nm minimum.
Previous enzyme studies have shown that replacement of the labile axial ligands of [Co(acacen)(L)2]+ with N-heterocycles (such as the 4-methylimidazole ligands of 2) significantly reduces inhibitory activity.[3, 12] Recent studies have attributed the decreased activity to tighter axial coordination of such N-heterocycles in contrast to the labile ammines of 1, strongly suggesting that His-coordination occurs via dissociative ligand exchange at the axial positions.[14] In contrast to 1, [Co(acacen)(L)2]+ complexes like 2 with less labile ligands exhibited incomplete dissociative substitution at both axial positions in the presence of competing N-heterocycles, such as free imidazole (Im), resulting in mixed ligand complexes (e.g. [Co(acacen)(4MeIm)(Im)]+ rather than [Co(acacen)(Im)2]+).
CD analysis of Zn-ZF4 treated with 2 was conducted to determine the dependence of ZF4 structural perturbation by 1 on reactivity of the complex to axial ligand dissociation. While the lability of the ammine ligands of 1 allows for His-coordination at both axial sites, the previous studies suggest that the more substitutionally inert 2 would be likely to only coordinate one His residue. In contrast to effects observed with 1, addition of 2 to Zn-ZF4 yields a CD spectrum slightly shifted from the ββα signals but resembling Zn-ZF4 more than apo-ZF4 (SI Figure 12). No further structural changes detectable by CD were observed beyond the addition 0.25 equivalents of 2. The discrepancy between the CD spectra of ZF4 with 1 and 2 suggests that His-coordination at both axial positions is required to elicit complete disruption of the ββα motif and supports a dissociative ligand exchange mechanism for potent inhibitory activity.
The evaluations of backbone NH hydrogen bonding by NMR and secondary structure by far-UV CD experiments indicate that 1 disrupts the ZnII-dependent ββα structure of ZF4 in favor of a less ordered random coil conformation closely resembling apo-ZF4. The structural effects correlate to axial ligand lability by the reduced effect of 2 on ZF4 secondary structure. In conjunction with the observed His-coordination behavior of 1, these studies validate the proposed mechanism of inhibition. The CoIII Schiff base complex inhibits ZF proteins by coordinating His residues in the ZnII-binding site through dissociative axial ligand exchange thereby disrupting the ββα structural motif required for function.
Conclusion
The present work provides a comprehensive molecular understanding of the interactions between [Co(acacen)(L)2]+ complexes and ZF motifs that confer potent inhibition of transcription factors. Due to the disorder of the model peptides treated with 1, alternative techniques to traditional 3D structure elucidation were required to determine the binding site, selectivity, and mode of inhibition. NMR, CD and electronic absorption spectroscopy studies demonstrated that His residues coordinate to 1 at both axial ligand positions while retaining octahedral geometry. The coordination of His to 1 results in a loss of the structure stabilized by ZnII tetrahedral coordination. Consequently, protein function dependent on the zinc finger motif (such as sequence-specific DNA-binding of TFs) is inhibited. These investigations illustrate the remarkable specificity of [Co(acacen)(L)2]+ complexes for His over other amino acids revealing a degree of selectivity at the binding site even in the absence of a targeting molecule on the acacen backbone.
The knowledge gained in these studies has provided the analytical foundation to move from a qualitative understanding of the peptide-complex interaction to quantitative information by extension of the techniques employed. Although the model peptides in these investigations were derived from ZF TFs, the implications for the mechanism of action of [Co(acacen)(L)2)]+ complexes translate to other His-containing targets. Additionally, with the growing field of medicinal inorganic chemistry, new methods for understanding protein-metal interactions are required. The analytical approaches demonstrated in this work can be readily translated to other bioactive coordination complexes and promote the development and design of transition metal-based therapeutics.
Experimental Section
Materials
Two 26-mer ZF model peptides, ZF4 (KSCPHCSRAFADRSNLRAHLQTHSDV) and CP1 (PYKCPECG-KSFSQKSDLVKHQRTHTG), were purchased from GenScript USA Inc. (Piscataway, NJ, USA). Two His-rich (HR) peptide fragments, a 10-mer named HR1 (RAHLQT-HSDV) and a 7-mer (GHIRTHG) named HR2, were synthesized by standard solid-phase peptide synthesis techniques. Fmoc chemistry and HBTU/DIPEA activation on Wang resin were used. Following TFA cleavage, the peptides were purified by preparative HPLC using acetonitrile/water gradients in the presence of 0.05% TFA and 230 nm detection wavelength on an Atlantis T3 column (Waters). The identities and purities (>95%) of the products were confirmed by ESI-MS and analytical HPLC.
The two CoIII Schiff base complexes used in these studies, [Co(acacen)(NH3)2]+ (1) and [Co(acacen)(4MeIm)2]+ (2) (Figure 1) were synthesized as previously described.[14, 16] Materials for synthesis of the complexes, ZnSO4•7H2O, ZnCl2•6H2O and D2O were purchased from Sigma-Aldrich (St. Louis, MO, USA). Perdeuterated [D11]tris was purchased from Cambridge Isotope Laboratories (Andover, MA, USA). Purchased materials were used without further purification. Deionized water was obtained from a Millipore Q-Gard system equipped with a Quantum EX cartridge.
NMR Spectroscopy
NMR experiments for ZF4 and CP1 were performed on a Bruker Avance II 900 MHz spectrometer equipped with a cryogenic sensitivity-enhanced triple-resonance 5 mm inverse TCI cryoprobe. Samples of ZF4 and CP1 were prepared in peptide concentrations of 1.5 mM and dissolved in 90/10 (v/v) H2O–D2O solution buffered at pH 7.0 with 20 mM deuterated [D11]tris for solutions containing peptide alone or peptide and 1. ZnII treated peptides (Zn-Peptide) were prepared differently so as to minimize ZnII-coordination by tris buffer. Zn-Peptide samples were first prepared without buffer, then buffered to a pH 7.0 with a maximum capacity of 10mM deuterated [D11]tris The model peptides were coincubated with 2.2 equivalents of either ZnII (ZnSO4•7H2O) or 1 for at least 1 hour at 37 ºC and degassed for at least 2 hours prior to data acquisition. All NMR experiments for ZF4 and CP1 were performed at 15 ºC in 5 mm NMR tubes. The chemical shifts (in ppm) were measured downfield from an internal standard (4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS)). Solvent suppression for 1D, TOCSY and NOESY experiments was achieved using excitation sculpting using 180º water-selective pulses with gradients.[21] 1D experiments were accumulated using 32k data points with 256 scans. Gradient-based phase-sensitive 1H-1H 2D experiments were performed using mixing times of 20 or 60 ms for TOCSYs and 150 or 300 ms for NOESYs. 2D experiments were accumulated using 2048 data points in t2 for 64 t1 values with 16 scans. 1D NMR data were processed with TopSpin 2.1.6 software (Bruker Instruments) and analyzed by MestReNova 7.0.3 program (MestreLab Research S.L.). 2D NMR data were processed with NMRPipe[22] and analyzed by NMRViewJ (One Moon Scientific).
A combination of 1D, TOCSY and NOESY experiments was employed to assign the spectra of peptides treated with either ZnII or 1. To assign the chemical shifts of the amino acids, protons in the peptides were first classified into spin systems using 1H-1H TOCSY spectra acquired with 20 ms and 60 ms mixing times. Subsequently, sequence-specific assignments were determined using backbone NHι-CHαi and CHαi-NHi+1 correlations of the 1H-1H NOESY spectra with 150 ms and 300 ms mixing times.
NMR experiments for HR1 were performed on a Bruker Avance III 600 MHz spectrometer equipped with a broadband multinuclear 5 mm inverse BBI probe optimized for 1H applications. Samples were 5 mM in 2 or peptide concentration and dissolved in 90/10 (v/v) H2O–D2O solution buffered at pH 7.0 with 100 mM deuterated [D11]tris. For chemical shift determination experiments, peptides were coincubated with 2.2 equivalents of either ZnSO4•7H2O or 1 for at least 1 hour at 37 ºC and degassed for at least 2 hours prior to data acquisition. Peptide 1D, TOCSY and NOESY experiments for chemical shift evaluation were performed similarly to ZF4 and CP1 but at 10 ºC in 5 mm NMR tubes. Data was processed and analyzed similarly to ZF4 and CP1. Similar NMR experiments were performed on 2, a substitutionally inert [Co(acacen)(L)2]+ complex. Since the 4-methylimidazole ligand of 2 is intended to mimic the His imidazole side chain, 2 was implemented as a small molecule model of [Co(acacen)(His)2]+ and the chemical shifts were compared to those of the 1-Peptide adducts.
CD Spectroscopy
CD experiments were performed on a Jasco J-815 CD spectrometer at 25 ºC. For experiments evaluating the [Co(acacen)(L)2]+ intraligand transfer band, samples were either 125 μM in HR1 concentration (with, or without, varying concentrations of 1) or 62.5 μM [Co(acacen)(L)2]+ in the absence of peptide. The aqueous solutions were maintained at pH 7.0 with 10 mM tris buffer. For 1-HR1 samples, peptide and 1 were coincubated at varying molar equivalents (1:peptide = 1.0, 2.0, or 4.0) for at least 1 hour at 37 ºC. The spectra of 10 mM tris at pH 7.0 was subtracted from all the sample spectra. Experiments were performed at 25 ºC in 1 mm Hellma cuvettes. CD signal was measured from 190 to 400 nm with 5 accumulations at a scan rate of 1 nm/min.
For CD experiments evaluating zinc finger structure, samples were 33.6 μM in ZF4 concentration. The aqueous solutions were maintained at pH 7.0 with 5 mM phosphate buffer instead of tris buffer to minimize background signal in the far-UV region. For Zn-ZF4 samples, ZF4 was coincubated with varying molar equivalents of ZnII (ZnSO4•7H2O) and incubated at least 1 hour at 37 ºC. For samples treated with 1, Zn-ZF4 at 4.0 molar equivalents of ZnII-to-peptide was coincubated with varying molar equivalents of 1 for at least 1 hour at 37 ºC. For comparison, the same experiments were performed with 2 as with 1. Since 2 is less substitutionally labile than 1, its effects were tested to correlate structural perturbations to reactivity by dissociative ligand exchange. Experiments were performed at 25 ºC in 1 mm Hellma cuvettes. CD signal was measured from 190 to 260 nm with 5 accumulations at a scan rate of 1 nm/min. All CD data were processed and analyzed with CDPro software (Colorado State University).
Electronic Absorption Spectroscopy
1, 2 and 1-HR1 samples for electronic absorption spectroscopy were prepared in the same way as for CD spectroscopy experiments evaluating the [Co(acacen)(L)2]+ intraligand transfer band. Electronic absorption data were recorded on a Hewlett Packard HP 8453 diode array spectrophotometer at 25 ºC. First derivative plots were produced using GraphPad Prism 6 program (GraphPad Software, Inc.).
Additional experimental information of concentration-dependent 1H NMR, ESI-MS analysis, and free thiol analysis with Ellman’s reagent can be found in the Supplementary Information.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Natsuho Yamamoto, Allison S. Harney, Robert J. Holbrook, Daniel J. Feld, and Lauren M. Matosziuk for helpful discussions. The authors gratefully acknowledge Joseph Coomes, Viktorie Reichova, and Emily Testa for technical support. M. C. H. would like to acknowledge the National Science Foundation Graduate Research Fellowship. Portions of this work were completed at the Integrated Molecular Structure Education and Research Center, Keck Biophysics Facility, and Biomolecular NMR Center at Northwestern University; and the Center of Structural Biology at the University of Illinois, Chicago through the Chicago Biomedical Consortium. This work was supported by funding from the Center of Cancer Nanotechnology Excellence (CCNE) initiative of the National Institutes of Health’s National Cancer Institute under Award U54CA119341.
References
- 1.a) Haas KL, Franz KJ. Chem Rev. 2009;109:4921. doi: 10.1021/cr900134a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Meggers E. Chem Commun. 2009:1001. doi: 10.1039/b813568a. [DOI] [PubMed] [Google Scholar]; c) Heffern MC, Yamamoto N, Holbrook RJ, Eckermann AL, Meade TJ. Curr Opin Chem Biol. 2013;17:189–196. doi: 10.1016/j.cbpa.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Farrell N. Coord Chem Rev. 2002;232:1. [Google Scholar]; e) Thompson KH, Orvig C. Dalton Trans. 2006:761. doi: 10.1039/b513476e. [DOI] [PubMed] [Google Scholar]; f) Hambley TW. Dalton Trans. 2007:4929. doi: 10.1039/b706075k. [DOI] [PubMed] [Google Scholar]
- 2.a) Harney AS, Meade TJ, LaBonne C. PLoS One. 2012;7:e32318. doi: 10.1371/journal.pone.0032318. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hurtado RR, Harney AS, Heffern MC, Holbrook RJ, Holmgren RA, Meade TJ. Mol Pharm. 2012;9:325. doi: 10.1021/mp2005577. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Harney AS, Lee J, Manus LM, Wang PJ, Ballweg DM, LaBonne C, Meade TJ. Proc Natl Acad Sci U S A. 2009;106:13667. doi: 10.1073/pnas.0906423106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Louie AY, Meade TJ. Proc Natl Acad Sci U S A. 1998;95:6663. doi: 10.1073/pnas.95.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Takeuchi T, Bottcher A, Quezada CM, Simon MI, Meade TJ, Gray HB. J Am Chem Soc. 1998;120:8555. [Google Scholar]
- 3.Blum O, Haiek A, Cwikel D, Dori Z, Meade TJ, Gray HB. Proc Natl Acad Sci U S A. 1998;95:6659. doi: 10.1073/pnas.95.12.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Brayer KJ, Kulshreshtha S, Segal DJ. Cell Biochemistry and Biophysics. 2008;51:9. doi: 10.1007/s12013-008-9007-6. [DOI] [PubMed] [Google Scholar]; b) Ingham PW, McMahon AP. Genes & Development. 2001;15:3059. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]; c) Nieto MA. Nat Rev Mol Cell Biol. 2002;3:155. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 5.a) Darnell JE. Nat Rev Cancer. 2002;2:740. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]; b) Kasper M, Regi G, Frischauf AM, Aberger F. Eur J Cancer. 2006;42:437. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 6.Ng JMY, Curran T. Nat Rev Cancer. 2011;11:493. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg T. Curr Opin Chem Biol. 2008;12:464. doi: 10.1016/j.cbpa.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 8.a) Mortishire-Smith RJ, Lee MS, Bolinger L, Wright PE. FEBS Lett. 1992;296:11. doi: 10.1016/0014-5793(92)80392-t. [DOI] [PubMed] [Google Scholar]; b) Wolfe SA, Nekludova L, Pabo CO. Annu Rev Biophys Biomol Struct. 2000;29:183. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]; c) Lee MS, Cavanagh J, Wright PE. FEBS Lett. 1989;254:159. doi: 10.1016/0014-5793(89)81030-0. [DOI] [PubMed] [Google Scholar]
- 9.a) Colangelo CM, Lewis LM, Cosper NJ, Scott RA. J Biol Inorg Chem. 2000;5:276. doi: 10.1007/s007750050372. [DOI] [PubMed] [Google Scholar]; b) Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Structure. 1996;4:1171. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]; c) Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. Nature. 1993;366:483. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 10.Mingot JM, Vega S, Maestro B, Sanz JM, Nieto MA. J Cell Sci. 2009;122:1452. doi: 10.1242/jcs.041749. [DOI] [PubMed] [Google Scholar]
- 11.a) Blasie CA, Berg JM. Biochemistry. 2002;41:15068. doi: 10.1021/bi026621h. [DOI] [PubMed] [Google Scholar]; b) Krizek BA, Amann BT, Kilfoil VJ, Merkle DL, Berg JM. J Am Chem Soc. 1991;113:4518. [Google Scholar]
- 12.Takeuchi T, Bottcher A, Quezada CM, Meade TJ, Gray HB. Bioorg Med Chem. 1999;7:815. doi: 10.1016/s0968-0896(98)00272-7. [DOI] [PubMed] [Google Scholar]
- 13.a) Timari S, Kallay C, Osz K, Sovago I, Varnagy K. Dalton Trans. 2009:1962. doi: 10.1039/b816498c. [DOI] [PubMed] [Google Scholar]; b) Zamani K, Mobinikhaledi A, Foroughifar N, Faghihi K, Mahdavi V. Turk J Chem. 2003;27:71. [Google Scholar]
- 14.Manus LM, Holbrook RJ, Atesin TA, Heffern MC, Harney AS, Eckermann AL, Meade TJ. Inorg, Chem. 2013;52:1069–76. doi: 10.1021/ic302379j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Huvaere K, Skibsted LH. J Am Chem Soc. 2009;131:8049. doi: 10.1021/ja809039u. [DOI] [PubMed] [Google Scholar]; b) Yu XY, Cai SH, Xu X, Chen Z. Inorganic Chemistry. 2005;44:6755. doi: 10.1021/ic050739f. [DOI] [PubMed] [Google Scholar]
- 16.Bottcher A, Takeuchi T, Hardcastle KI, Meade TJ, Gray HB, Cwikel D, Kapon M, Dori Z. Inorg Chem. 1997;36:2498. [Google Scholar]
- 17.Raghothama S. Journal of the Indian Institute of Science. 2010;90:145. [Google Scholar]
- 18.Zhang YZ, Paterson Y, Roder H. Protein Sci. 1995;4:804. doi: 10.1002/pro.5560040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omichinski JG, Clore GM, Sakaguchi K, Appella E, Gronenborn AM. FEBS Lett. 1991;292:25. doi: 10.1016/0014-5793(91)80825-n. [DOI] [PubMed] [Google Scholar]
- 20.Frankel AD, Berg JM, Pabo CO. Proc Natl Acad Sci U S A. 1987;84:4841. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen BD, Meng X, Donovan KJ, Shaka AJ. J Magn Reson. 2007;184:263. doi: 10.1016/j.jmr.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.