Abstract
Purpose
MicroRNAs (miRNAs) can act as oncomirs or tumor-suppressor miRs in cancer. This study was undertaken to investigate the status and role of miR-34b in prostate cancer.
Experimental Design
Profiling of miR-34b was performed in human prostate cancer (PCa) cell lines and clinical samples by quantitative-real-time-PCR and in situ hybridization. Statistical analyses were done to assess diagnostic/prognostic potential. Biological significance was elucidated by performing a series of experiments in vitro and in vivo.
Results
We report that miR-34b is silenced in human PCa and the mechanism is through CpG hypermethylation. miR-34b directly targeted methyltransferases (DNMT) and deacetylases (HDACs) resulting in a positive feedback loop inducing partial demethylation and active chromatin modifications. miR-34b expression could predict overall and recurrence free survival such that patients with high miR-34b levels had longer survival. Functionally miR-34b inhibited cell proliferation, colony formation, migration/invasion and triggered G0/G1 cell cycle arrest and apoptosis by directly targeting the AKT and its downstream proliferative genes. miR-34b caused a decline in the mesenchymal markers Vimentin, ZO1, N-cadherin and Snail with an increase in E-cadherin expression, thus inhibiting epithelial to mesenchymal transition. Finally we demonstrated the antitumor effect of miR-34b in vivo. MiR-34b caused a dramatic decrease in tumor growth in nude mice compared to cont-miR.
Conclusion
These findings offer new insight into the role of miR-34b in the inhibition of PCa through demethylation, active chromatin modification and AKT pathways and may provide a rationale for the development of new strategies targeting epigenetic regulation of miRNAs for the treatment of PCa.
Keywords: MicroRNA-34b, Methylation, DNMT, HDAC, prostate cancer
Introduction
MicroRNAs (miRNAs) are non-protein-coding RNAs thought to regulate the expression of up to >90% of human genes (1). Growing evidence has strongly implicated the involvement of miRNAs in carcinogenesis (2, 3). Dysregulated miRNAs may function as oncogenes (4), such as the miR-17–92 cluster (5), or tumor suppressor genes such as miR-205 (6–8). A single miRNA can have hundreds of target mRNAs, highlighting the importance of this gene regulation system in cellular functions (9). The study of miRNAs has become the subject of intense interest, especially after the discovery of their fundamental role in a myriad of cellular and biological processes ranging from development to disease (10). From a clinical point of view, miRNAs have great potential as diagnostic and therapeutic agents. Since microarray analysis shows a general down regulation of miRNAs in tumors when compared with normal tissues (11). Owing to the remarkable tissue specificity of miRNAs, they have become a very useful tool for defining the origin of tumors in poorly differentiated cancers (12). The prognosis and survival of patients depend on the stage of the cancer at diagnosis. For this reason, one of the most important issues in clinical cancer research is to find early biomarkers of the tumorigenic process. MicroRNA signatures have been reported to be powerful tools for early diagnosis in renal cancer and have been found to distinguish between metastatic and non-metastatic tumors. The 5-year survival rate of patients with primary metastasis was reported to be 10% as compared with 70–90% in non-metastatic patients (13).
Among various epigenetic mechanisms of cancer related gene silencing, DNA hypermethylation of CpG sites within CpG islands is known to lead to the inactivation of many tumor suppressor miRNAs (14). Previous reports have shown that disruption of DNA methylation in cancer cell lines induces upregulation of substantial numbers of miRNAs (15, 16) and thus helps identify candidate tumor suppressor miRNAs that are silenced by methylation. For example, treatment of bladder cancer cells with histone deacetylase (HDAC) and DNA methyltransferase (DNMT) inhibitors induced demethylation and re-expression of miRNA-127 (17). Similarly miR-34b/c was found to be methylated and down regulated in colorectal and gastric cancers (16, 18). The DNA methylation profile of tumors can be used as a signature to define tumor type, clinical prognosis and treatment response (19, 20). MiRNAs transcribed from CpG islands undergo DNA methylation-associated repression with a similar chromatin context to protein coding genes (15, 16, 21). Epigenetic regulation of miRNA genes is tightly linked to chromatin signatures and transcriptionally active miRNA genes are characterized by active chromatin marks such as trimethylated histone H3 lysine 4 (3H3K4) (22). Epigenetic silencing of miRNAs is also involved in the acquisition of an invasive phenotype and the development of metastasis (23). Inactivation of tumor-promoting miRNAs (24, 25) or restoration of tumor-suppressor miRNAs (15, 17, 26) has great potential for use in cancer treatment.
Here we report that: (i) miR-34b is significantly down-regulated in prostate cancer (PCa) and the mechanism of silencing is through CpG hypermethylation, (ii) reconstitution of miR-34b induced partial demethylation and active chromatin modifications in upstream sequence of miR-34b gene, (iii) miR-34b acts as a tumor suppressor microRNA in PCa, (iv) miR-34b has antiproliferative effects and targets AKT pathway genes, (v) miR-34b has anti-migratory/-invasive effects and downregulates epithelial to mesenchymal transition (EMT) markers, (vi) miR-34b suppresses tumor growth in an in-vivo xenograft nude mouse model.
Materials and Methods
Cell culture, plasmids and probes
Human PCa cell lines PC3, LNCaP and a non-malignant renal cell line RWPE1 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and grown according to ATCC protocol and as described previously (7). These human-derived cell lines were authenticated by DNA short-tandem repeat analysis by ATCC. The experiments with cell lines were performed within 6 months of their procurement/resuscitation. Plasmids of 3´UTR target expression clones were purchased from GeneCopoeia, Rockville, MD. TaqMan probes and microRNA precursors were purchased from Applied Biosystems, Foster City, CA. The probes for methylation specific realtime PCR were synthesized by Applied Biosystems (Foster City, CA, USA), labeled with 6FAM reporter at the 5’ end and with MGB quencher at the 3’ end.
Quantitative real-time PCR and in situ hybridization
Tissue samples from radical prostectomy were obtained from the Veterans Affairs Medical Center, San Francisco, CA, USA. Written informed consent was obtained from all patients and the study was approved by the UCSF Committee on Human Research (Approval number: H9058-35751-01). Laser capture microdisection was employed to get total RNA. All reactions were run in a 7500 Fast Real Time PCR System and microRNA assays were performed in accordance with the manufacturer's instructions (Applied Biosystems, Foster City, CA). Relative expression was calculated using comparative Ct.
In situ hybridization (ISH) was performed as described previously (27). Detailed method is described in Supplemental Materials and Methods (M & M). ISH results for tissue array were graded according to quick score (percent cells stained × intensity of stain) and normalized to U6 levels.
Methylation analysis of miR-34b by sequencing and quantitative methylation-specific PCR (qMSP)
DNA was available for 32 pairs of laser capture microdisected samples. Out of 32, 19 pairs are from the same cohort for which microRNA expression was available. Methylation status of tissue samples was analyzed by quantitative methylation-specific PCR (qMSP) within the 1.0 kb region upstream of the miR-34b gene. For PCa cell lines methylation was determined by sequencing and percent methylation calculated. The sequences are given in supplemental Table 2.
Immunoblotting and Immunofluorescence
Immunoblotting was performed as described previously (7) also described in Supplemental M&M. For immunofluorescence, cells were transfected with precursors of miR-34b or cont-miR for 72 hours, washed and fixed with acetone-methanol (1:1) mixture. Cells were then blocked with 10% normal goat serum blocking solution (Zymed Laboratories, Carlsbad CA) and hybridized with the specific primary antibodies against EMT markers. Cells were washed and hybridized with fluorescein conjugated secondary antibody (1:1000) then washed and mounted with ProLong Gold antifade reagent with DAPI (Invitrogen-Life Technologies).
Luciferase Assays
For reporter assays, cells were transiently transfected with wild-type or mutant reporter plasmid and miR-34b or control-miR. Firefly luciferase activities were measured using the Dual Luciferase Assay (Promega, Madison, WI) 18 hr after transfection and the results were normalized with Renilla luciferase. Complementary sequences are given in supplemental Table 2.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the EZ-ChIP Kit (Upstate Biotechnology) as described previously (27). Immunoprecipitation was performed using antibodies purchased from Upstate Biotechnology. Power SYBR Green PCR Mastermix (Applied Biosystems) was used to perform real-time PCR with a 7500 Fast Real-Time PCR System (Applied Biosystems). Signals were also confirmed by conventional PCR and gel analyses. Primer sequences are given in supplemental Table 2.
Flow cytometry, cell viability, migration, clonability and invasion assays
FACS analysis for cell cycle and apoptosis was done 72 hours post-transfection using nuclear stain DAPI for cell cycle analysis or ANNEXIN V-FITC /7-AAD KIT (Beckman Coulter, Inc. Fullerton, CA) for apoptosis analysis according to the manufacturer’s protocol. Cell viability was determined at 24, 48 and 72 h by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. For colony formation assay, cells were seeded at low density (1000 cells/plate) and allowed to grow untill visible colonies appeared. Then, cells were stained with Giemsa and colonies were counted. Cytoselect 24-well cell migration and invasion assay kit (Cell Biolabs, Inc) was used for migration and invasion assays according to manufacturer’s protocol.
In vivo intratumoral delivery of miR-34b
The antitumor effect of miR-34b was determined by local administration of miR-34b precursor in established tumors. Each mouse was injected with 5.0×106 PC3 PCa cells subcutaneously and there were five mice in each group. Once palpable tumors developed (average volume 50mm3), 6.25 µg of synthetic miRNA-34b and/or control miRNA complexed with 1.6 µl siPORT Amine transfection reagent (Ambion, Austin, TX) in 50 µl PBS was delivered eight times intratumorally at 3-day intervals. Tumor growth was followed for 21 days from the first injection. All animal care was in accordance with institutional guidelines.
Statistical analysis
Statistical analyses were performed with StatView version 5 for Windows, GraphPad Prism 5 and MedCalc version 10.3.2. All quantified data represents an average of at least triplicate samples or as indicated. Error bars represent standard deviation of the mean. All tests were performed two tailed and p-values <0.05 were considered statistically significant. Receiver operating curves (ROC) were calculated to determine the potential of miR-34b or its methylation to discriminate between malignant and non-malignant samples. Chi-square tests were performed to determine the correlation between miR-34b expression, clinicopathological characteristics and disease progression analysis. Kaplan-Meier (log-rank test) analysis was also performed for survival analysis.
Results
Downregulation of miR-34b expression and its correlation with clinicopathological characteristics of patients
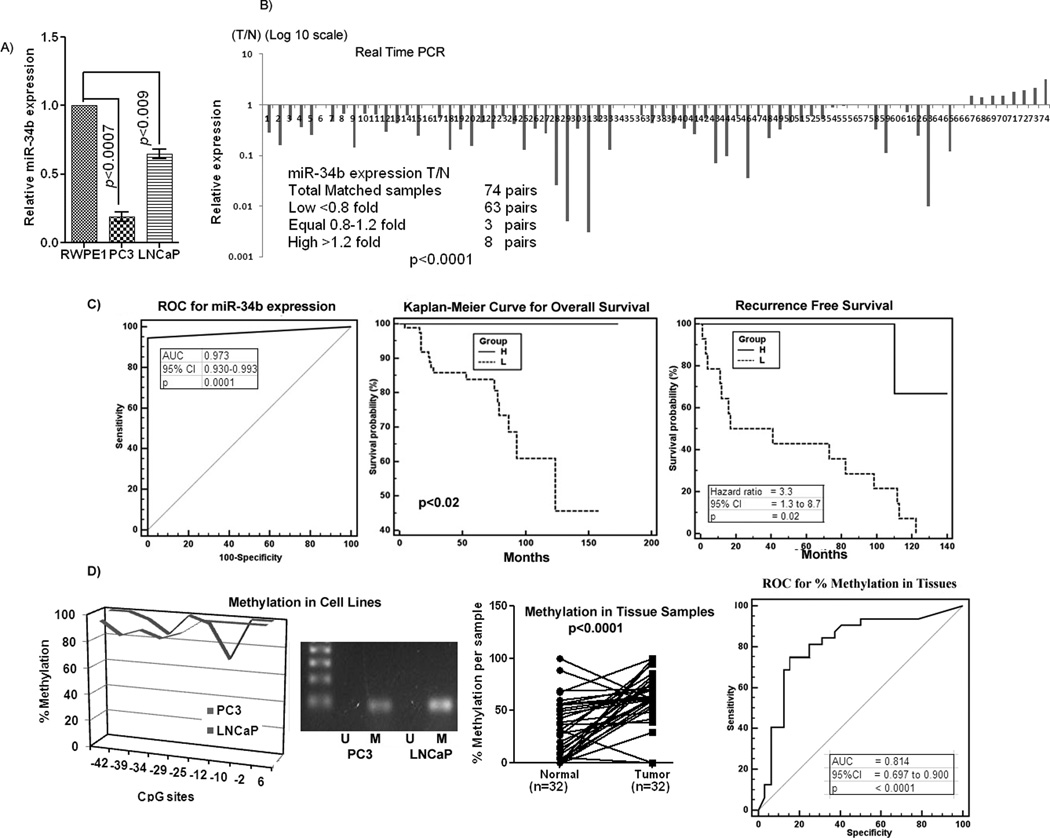
Expression of miR-34b was examined in prostate cell lines and tissue samples by miRNA-quantitative real time PCR (qRT-PCR). The expression of miR-34b was downregulated in cancer cell lines PC3 and LNCaP compared to the non-malignant cell line RWPE1 (Figure 1A). To examine the biological significance of miR-34b, its expression was analyzed in 148 laser-captured microdisected (LCM) matched human tissue samples (Figure 1B) along with an unmatched group of 27 benign prostatic hyperplasia (BPH) and 20 tumor samples (Supplemental Figure 1A). To validate the reproducibility of the data, we also performed in-situ hybridization on commercially available tissue arrays (US Biomax, Inc, Cat. No. PR956) to determine miR-34b expression in an independent cohort of tissues samples (Supplemental Figure 1B). Almost all carcinoma samples showed significant down regulation of miR-34b expression with respect to either normal or BPH samples (Figure 1B, Supplemental Figure 1A, B). Among the 74 pairs of matched samples, 63 pairs had either no or significantly lower expression of miR-34b (<0.8 fold) in tumors vs normal samples (p<0.0001), 3 pairs had equal expression (0.8–1.2 fold) whereas 8 pairs had higher miR-23b expression (>1.2 fold) in tumors compared to their matched normal samples. These results suggest that miR-34b is significantly downregulated and might be a putative tumor suppressor in PCa.
Figure 1. Downregulation of miR-34b expression, its potential diagnostic and prognostic significance and methylation status in PCa.
A) Quantitative RT-PCR analysis of miR-34b expression levels in PCa and a nonmalignant cell line. B) Quantitative real time PCR analysis of mir-34b expression in 74 pairs of matched Laser-Captured Microdissected (LCM) tissue samples. C) Receiver Operating curve (ROC) curve analysis and Kaplan-Meier analysis for overall and recurrence free survival based on miR-34b expression. (H and L-miR-34b High or Low). D) Mir-34b methylation status in cell lines and matched tissue samples and ROC for percent methylation in tissues. M-Methylated band, U-Unmethylated.
Clinical demographics of the study cohorts are summarized in Supplemental Table 1. Based on the relative expression of miR-34b, we grouped the samples into low and high miR-34b expression groups and assessed the correlation with clinicopathological variables like Gleason grade, pathological stage (pT) and biochemical recurrence (Supplemental Figure 1C). The time of recurrence was defined as the first postoperative PSA value >0.1 ng/ml PSA confirmed by at least one undetectable PSA level (detection limit <0.04 ng/ml) after surgery. Seventeen patients experienced a biochemical relapse according to this criterion. A significant correlation was observed between miR-34b expression and clinical variables. Decreased miR-34b expression was observed in 96% of cases with higher pathological stage (pT3–pT4) compared to 80% cases of low pathological stage (pT2) (p<0.001). To analyze the correlation with Gleason grade, we divided the samples into low (4–6), medium (7) and high (8–10) Gleason grade groups. Low expression of miR-34b was found in 73% cases of low, 90% cases of medium and 100% cases of high Gleason grade groups (p<0.0001). In PSA recurrence patients, 82% of cases had decreased miR-34b expression compared to 18% cases with high miR-34b expression (p<0.0001). These results reveal that the number of cases with low miR-34b expression increases from low grade, low pathological stage to high grade and high pathological stage. Furthermore, PSA recurrence patients also had significantly low miR-34b expression suggesting that miR-34b might have prognostic value in PCa (Supplemental Figure 1C).
Receiver operating curve (ROC) analyses were performed to evaluate the capability of miR-34b to discriminate between normal and tumor tissues using 148 laser captured microdisected tissue samples. An area under the ROC curve (AUC) of 0.973 (P<0.0001; 95% CI=0.930 to 0.993) (Figure 1C) was obtained suggesting that miR-34b expression can discriminate between malignant and non-malignant samples and might be useful as a diagnostic marker for PCa. To determine whether miR-34b expression has prognostic significance, we divided the matched samples into low (T/N<0.8 fold) and high (T/N>0.8 fold) miR-34b expression groups and performed Kaplan-Meier survival analysis. In Kaplan-Meier analysis, the miR-34b high expression group displayed significantly higher overall survival probability compared to the low expression group (Logrank Test p<0.02) (Figure 1C). We also analyzed recurrence free survival and risk of biochemical recurrence. Results indicated that miR-34b low expression cases had poor recurrence free survival (Logrank Test p<0.02) and a higher risk of biochemical recurrence (Hazard ratio=3.3, 95% CI = 1.3–8.7) (Figure 1C). These results suggest that miR-34b can predict the biochemical recurrence of PCa and has potential to be a diagnostic/prognostic biomarker, though a larger sample size is needed to confirm these results.
Mechanism of miR-34b silencing in prostate cancer is through CpG hypermethylation
To investigate the mechanism involved in the depleted levels of miR-34b in PCa we performed methylation analysis on the 1.0 kb upstream sequence of miR-34b. Primers are shown in supplemental Table 2. CpG rich regions were found (Supplemental Figure 1D) and further analyzed for methylation status in PCa cell lines and tissues. Our results show that this sequence is hypermethylated in PCa cell lines with an average methylation percentage of 94% (Figure 1D). We also investigated the methylation status of miR-34b in 32 pairs of matched laser captured microdissected tissue samples by methylation specific quantitative real time PCR. Our results indicated that the tumor samples had a higher methylation percentage compared to their normal counterparts (Figure 1D). For the 19 pairs of samples where DNA and microRNA was available, we found a significant inverse correlation between miR-34b expression and methylation. Samples with low miR-34b expression had higher average percent methylation and vice versa (p<0.0001) (Supplemental Figure 1E). We also performed ROC analysis to evaluate the ability of miR-34b methylation percentage to distinguish malignant from normal tissues. The AUC observed was 0.814 (p<0.0001, 95% CI=0.697 to 0.990) (Figure 1D) showing that the percentage of miR-34b methylation is able to distinguish malignant from normal tissues.
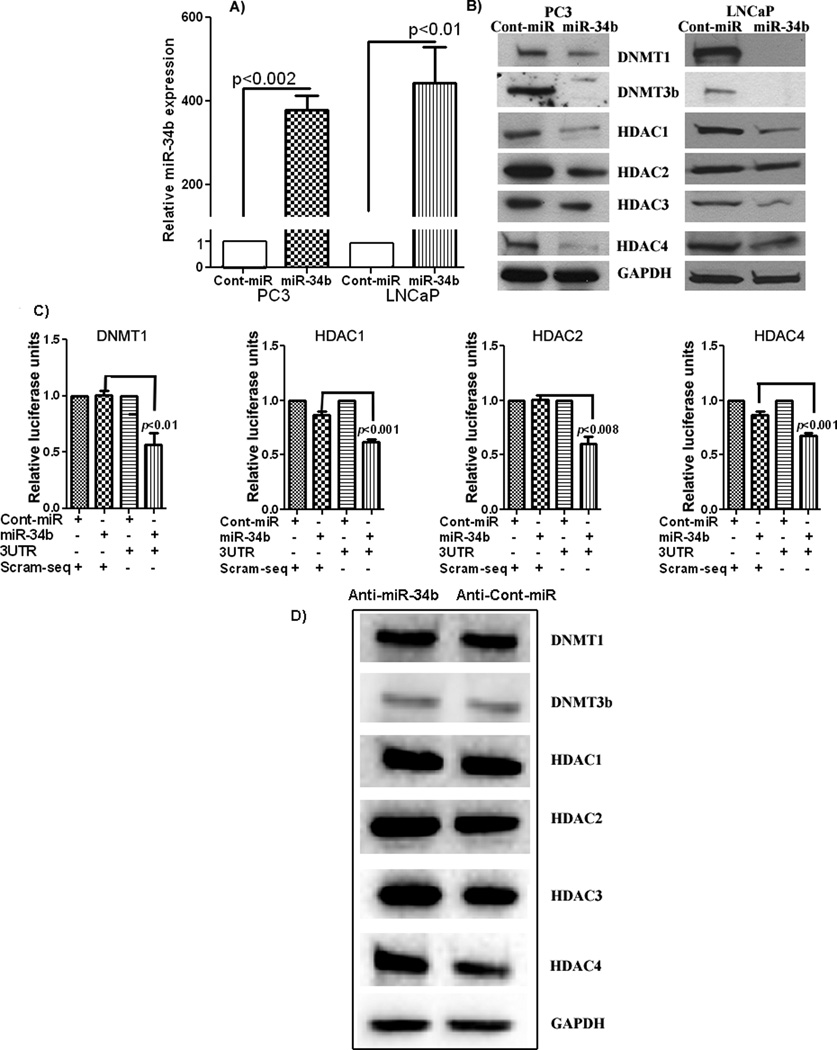
Reconstitution of miR-34b suppresses DNA-methyltransferases and histone deacetylases
Since miR-34b is epigenetically silenced in PCa and DNA methylation is linked to histone modifications, we asked whether reconstitution of miR-34b in cancer cells could downregulate DNA-methyltransferases or histone deacetylases. We overexpressed miR-34b in PC3 and LNCaP cells (Figure 2A). Our results show downregulation of DNMT1, DNMT3b, HDAC1, HDAC2, HDAC3 and HDAC4 protein expression in miR-34b overexpressing cells compared to negative control (Figure 2B). Complimentary sequences for miR-34b in the 3’UTR of these genes is given in Supplemental Table 2. To determine whether miR-34b targets these genes directly, we performed luciferase reporter activity assays. The results show that DNMT1, HDAC1, HDAC2 and HDAC4 are direct targets as the relative luciferase activity was significantly decreased with co-transfection of the wild type 3’UTRs of these genes along with miR-34b (Figure 2C). A decrease in luciferase activity was also observed in the case of DNMT3b and HDAC3 though it was not significant. Further we attenuated miR-34b expression by using anti-miR-34b in LNCaP cells since these cells had higher expression than PC3. Depletion of miR-34b in LNCaP cells rescued the protein expression of these genes (Figure 2D) which again confirms that miR-34b targets these genes in PCa.
Figure 2. Ectopic expression of miR-34b reduced expression of methyltransferases (DNMTs) and deacetylases (HDACs) in PCa cells.
A) Overexpression of miR-34b in PC3 and LNCaP cells. B) Ectopic expression of miR-34b inhibits DNMTs and HDACs at the protein level. C) Luciferase assays showing decreased reporter activity after co-transfection of wild type 3’UTR with miR-34b in PC3 PCa cells. The scrambled 3’ UTR sequence had no effect on reporter activity. D) Inhibition of miR-34b by antagoMirs rescued the expression of DNMTs and HDACs.
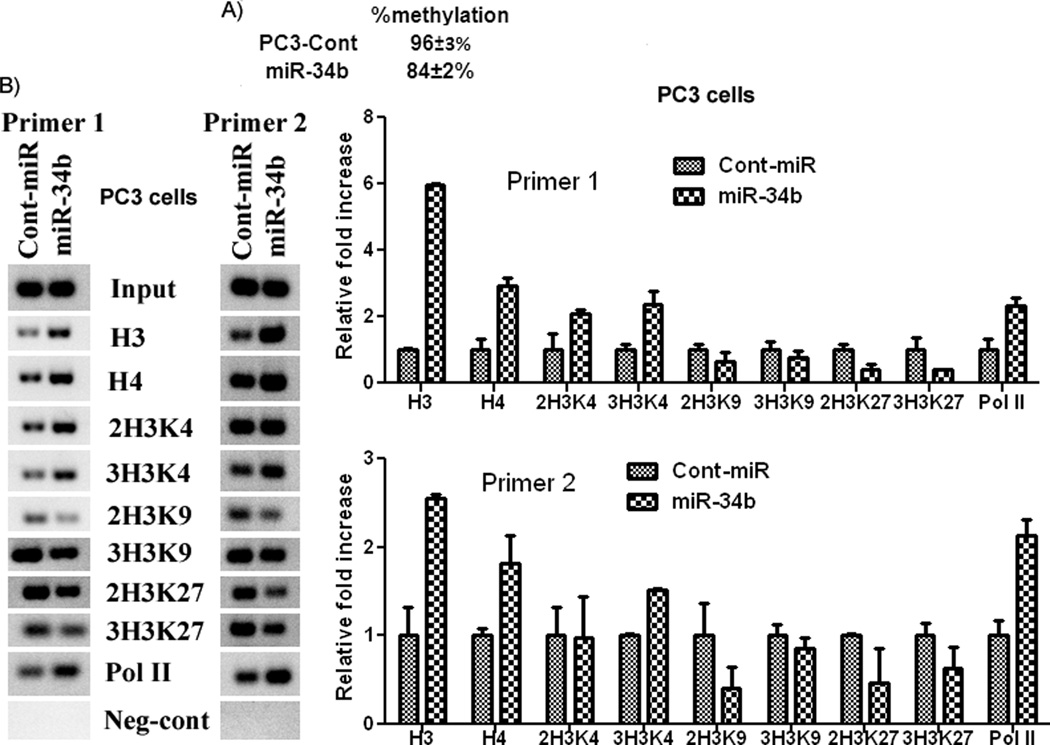
Ectopic expression of miR-34b induces partial demethylation and active chromatin modifications in 5’ upstream sequence of the miR-34b gene
Since miR-34b suppressed DNMTs, we looked to see if its reconstitution could also induce demethylation in PCa cells. Our results show that overexpression of miR-34b caused partial demethylation of CpG sites in the 5’ upstream sequence of the miR-34b gene as methylation decreased from 96±3% in controls to 84±2% in miR-34b transfected cells (Figure 3A). Epigenetic regulation of miRNA genes is tightly linked to chromatin signatures and transcriptionally active miRNA genes are characterized by active chromatin marks such as trimethylated histone H3 lysine 4 (3H3K4) (22). To investigate whether these active chromatin modifications are induced in epigenetically regulated miR-34b gene, we performed chromatin immunoprecepitation (ChIP) assay by PCR and quantitative real time PCR in the same sequence where we determined methylation. We found enrichment of active chromatin modifications and a modest decrease in repressive ones (Figure 3B). These changes are indicative of active genes and suggest that overexpression of miR-34b gene downregulates DNMTs and HDACs which in turn causes a positive feedback loop to induce demethylation and active modifications.
Figure 3. Induction of partial demethylation and active chromatin modifications.
A) Ectopic expression of miR-34b induces partial demethylation of CpG islands. B) Enrichment of active chromatin modifications by overexpression of miR-34b. ChIP assay in PC3 cells transfected with miR-34b or cont-miR was performed. Quantification of the immunoprecipitated upstream region of miR-34b was determined by quantitiative real-time PCR and normalized to input DNA. Signals were also confirmed by conventional PCR and PCR product for each sample was resolved on 2% agarose gel and visualized by staining with ethidium bromide.
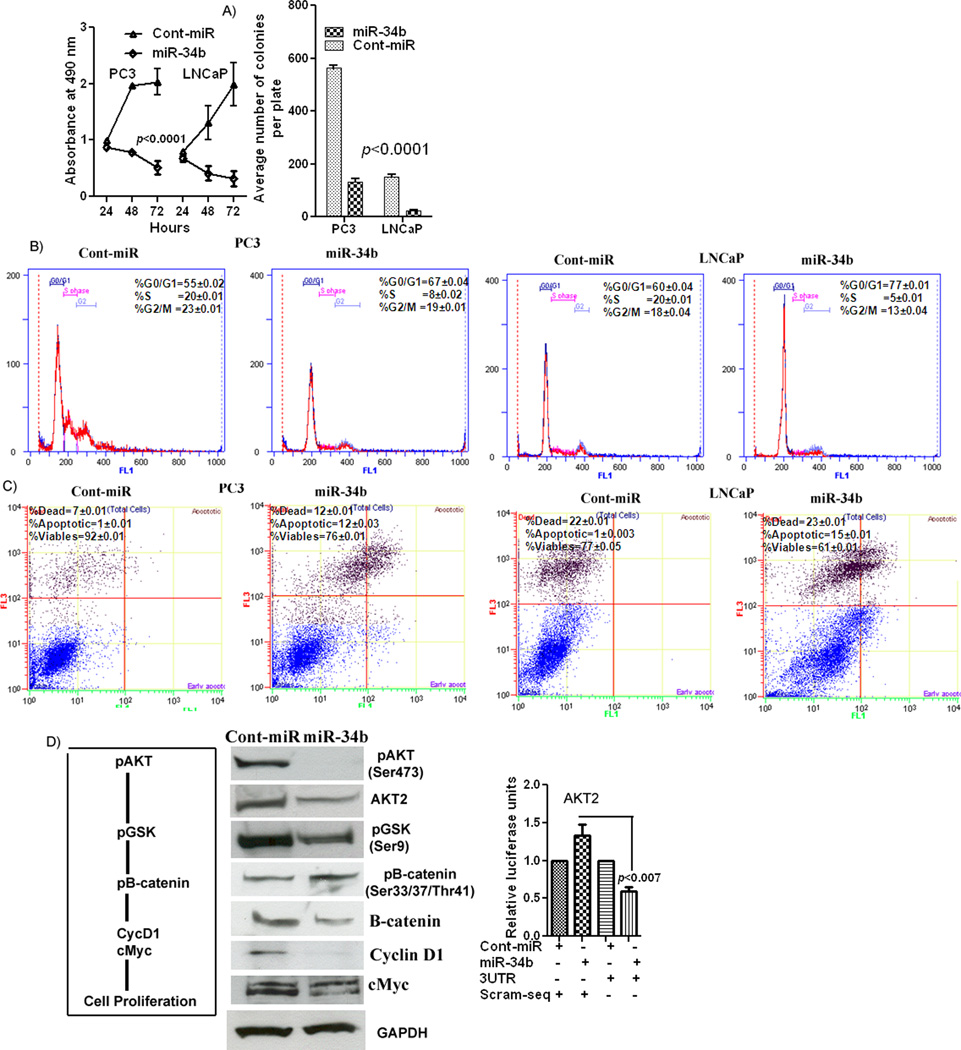
Functional significance of miR-34b in prostate cancer
Next we determined the functional significance of miR-34b in PCa by performing cell cycle, proliferation, clonability, migration and invasion assays. A significant decrease in cell proliferation was observed over time in PC3 and LNCaP cells overexpressing miR-34b (Figure 4A) as compared to conrols. The miR-34b-transfected cells also had low colony forming ability, as both size and number of foci in miR-34b overexpressing cells was reduced when compared to controls (Figure 4A). FACS (fluorescence activated cell sorting) analysis revealed that re-expression of miR-34b leads to a significant increase (12–17%) in the number of cells in the G0–G1 phase of the cell cycle while the S-phase population decreased from 20% to 5–8% suggesting that miR-34b causes a G0–G1 arrest in miR-34b transfected cells (Figure 4B). FACS analysis for apoptosis was performed using Annexin-V-FITC-7-AAD dye. The percentage of total apoptotic cells (early apoptotic + apoptotic) was significantly increased (12–15%) in response to miR-34b transfection compared to cont-miR (1%) with a corresponding 16% decrease in the viable cell population (Figure 4C). Collectively these results indicate a tumor suppressor role of miR-34b in PCa. To determine the pathway involved in the antiproliferative effect of miR-34b, we analyzed the protein expression of AKT proliferative pathway genes after miR-34b overexpression (Figure 4D). miR-34b significantly suppressed phosphorylation of AKT(Ser473) and its downstream target GSK3β(Ser9) which in turn gets activated. Activated GSK3β phosphorylates β-catenin and promotes its degradation. A decrease was also found in the downstream proliferative genes cMyc and CyclinD1 (Figure 4D). We also found that miR-34b directly targets AKT since there was a significant decrease in the relative luciferase units with cotransfection of the AKT 3’UTR along with miR-34b (Figure 4D). These results suggest that miR-34b exerts antiproliferative effects in PCa at least partly through the AKT pathway.
Figure 4. Functional significance and tumor suppressor role for miR-34b in PCa.
A) Proliferation and colony fromation of PC3 and LNCaP cells after miR-34b transfection was significantly reduced compared to cont-miR. B) Cell cycle analysis showing an increase in the G0/G1 phase and a decrease in S-phase cell population with miR-34b. C) Apoptosis assay showing induction of apoptosis by miR-34b. D) Luciferase reporter assay showing miR-34b directly targets proliferative marker AKT and Western blot showing downregulation of downstream Akt pathway genes at the protein level.
miR-34b downregulates EMT markers and suppresses cell migration and invasion independent of proliferation
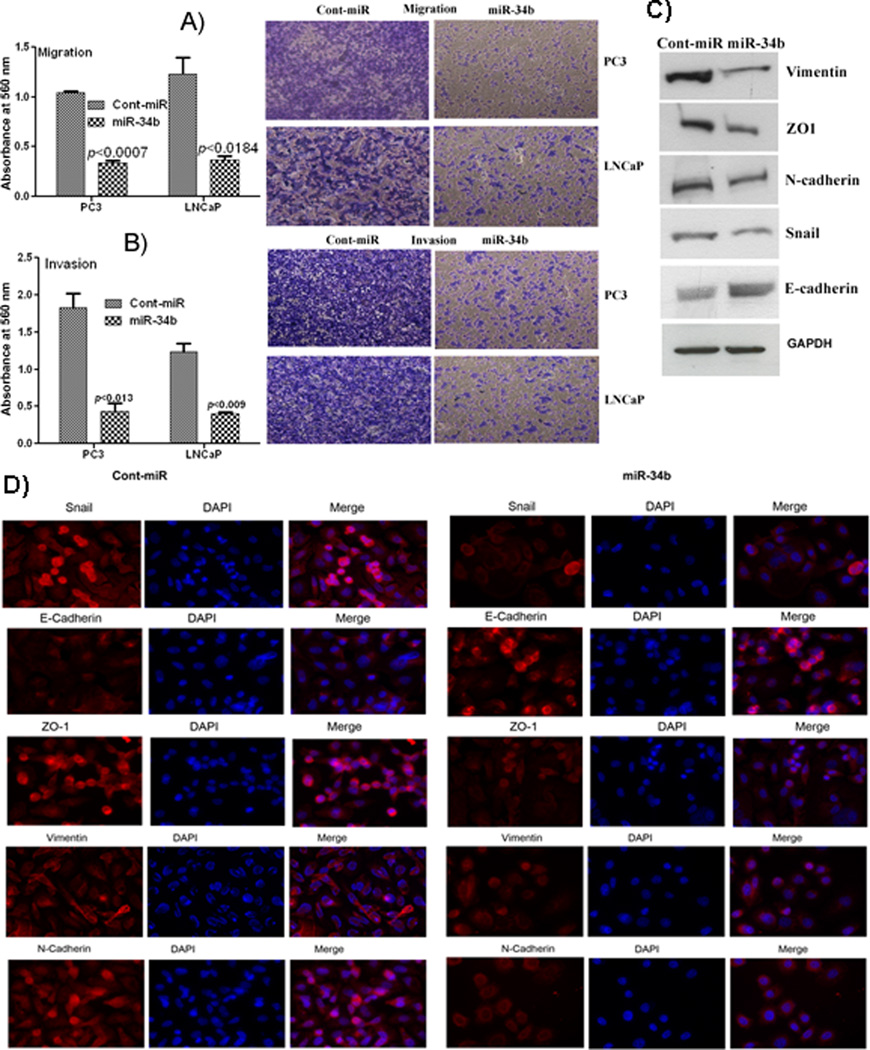
Cell migration and invasion assays revealed that miR-34b over-expressing cells were less proficient than controls in migrating or invading a barrier membrane with less absorbance observed at 560nm (Figure 6A–B). We examined the effect of overexpression of miR-34b on EMT markers and observed a decrease in Vimentin, ZO1, N-cadherin and Snail (mesenchymal markers) and an increase in E-cadherin (epithelial marker) (Figure 6C–D). The process of EMT is involved in epithelial-derived tumors causing them to become invasive and metastatic and these results show that miR-34b suppressed EMT markers.
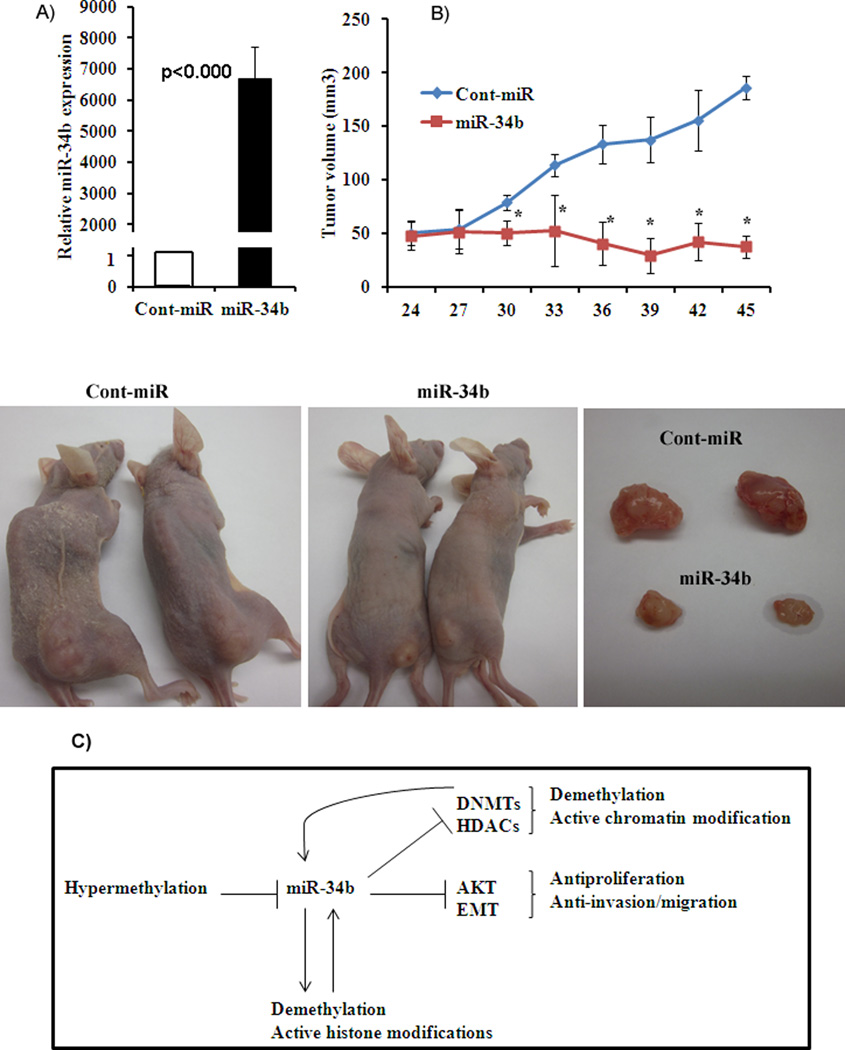
Figure 6. Antitumorigenic effect of miR-34b in vivo.
A) Average expression of miR-34b in excised tumors. B) Tumor volume following intratumoral injection of Cont-miR or miR-34b precursor into established tumors. *p<0.05. C) Schematic representation of pleiotropic tumor suppressor effect of miR-34b in PCa. miR-34b is silenced through hypermethlation in PCa. Reconstitution of miR-34b results in downregulation of DNA methyltransferases (DNMTs) and histone deacetylases (HDACs). This provides a positive feedback to further demethylate hypermethylated miR-34b and induce active chromatin modifications that are signatures of active genes. miR-34b overexpression also inhibits the AKT pathway and epithelial to mesenchymal transition (EMT) markers resulting in antiproliferative, antimigratory/anti-invasive effects in PCa.
Intra-tumoral delivery of miR-34b suppresses tumorigenecity in-vivo
Since miR-34b had an antitumor effect in-vitro, we performed in-vivo experiments to determine the effect of miR-34b by local administration into established tumors. We measured the expression of miR-34b in harvested tumors and found that miR-34b expression was significantly higher in tumors receiving miR-34b compared to controls (Figure 7A). The tumor volume of miR-34b injected mice was significantly lower (38 mm3) than those receiving control injections (186 mm3) (Figure 7B) at the termination of experiment. These results confirm the in vivo tumor suppressor effect of miR-34b in a PCa xenograft model.
Discussion
In this study we provide evidence that miR-34b is silenced in PCa and the mechanism is through CpG hypermethylation. MiR-34b downregulates DNA methyltransferases, histone deacetylases and induces partial demethylation and active chromatin modifications in PCa. Low expression of miR-34b can predict overall and recurrence free survival and can distinguish normal from malignant tissues. We also elucidated the tumor suppressor role of miR-34b in PCa and determined that the antiproliferative and antimigratory/invasive effects of miR-34b is partly through downregulation of the AKT pathway and EMT markers. We also show that miR-34b inhibits tumorigenecity both in vitro and in vivo, this being the first report documenting the detailed role of miR-34b in PCa.
One of the most common causes of the loss of tumor suppressor miRNAs in human cancer is the silencing of their primary transcripts by CpG island promoter hypermethylation (17, 28). In fact, DNA methylation–mediated downregulation of miRNAs by proximal CpG islands has been described by a number of groups (16, 17, 29). Further identification of additional methylation targets may clarify the specific molecular events involved in PCa progression, which hopefully will lead to prevention, diagnosis, and treatment of PCa at the molecular level. Here, we identified miR-34b is frequently silenced in tumors compared to normal samples and the mechanism of silencing is through DNA methylation in PCa tissues. Previous studies have reported CpG methylation of miR34b/c in colorectal cancer (16), oral squamous cell carcinoma (29), malignant melanoma where it correlated with metastatic potential (26) and various other cancers (30). Our study indicates that miR-34b methylation status can discriminate between normal prostate tissue and cancer samples making it a potential diagnostic marker for PCa. MicroRNAs possess several features that make them attractive candidates as new prognostic biomarkers and tools for the early diagnosis of cancer (13, 31, 32). In this study, we show that miR-34b expression could predict overall survival and recurrence free survival such that patients with high miR-34b levels had longer survival. Similarly promoter hypermethylation of miR-34b/c has been reported a common event in non-small cell lung cancer (NSCLC) and a potential prognostic factor for stage I NSCLC. Thus aberrant DNA methylation of miR-34b/c was correlated with a high probability of recurrence and associated with poor overall survival and disease-free survival of stage I NSCLC patients (33).
To link altered miR-34b expression with aberrant CpG methylation, we looked at the ectopic effect of miR-34b on DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) since their levels are reported to be elevated in PCa (34, 35). DNMTs catalyze the methylation of genomic DNA resulting in transcriptional repression. DNMTs are also known to recruit HDACs, leading to histone deacetylation and transcriptional repression (36, 37). DNMTs and HDACs are expressed at higher levels in PCa than the non-malignant prostate cells though the degree of expression varies among the cell lines based on the cell context (38–40). Androgen independent cell lines such as Du145 and PC3 express HDACs at higher levels than androgen dependent cell line such as LNCaP (38). Various reports have shown that microRNAs downregulate DNMTs (41) and HDACs (42). The microRNA-29 family directly targets DNMTs and restored normal patterns of DNA methylation in lung cancer cell lines (41). Similarly restoration of miR-9* levels in waldenstrom macroglobulinemia (WM) cells downregulated HDACs with an upregulation of histone-H3 and –H4 (42). Further epigenetic regulation of miRNA genes has been found to be tightly linked to chromatin signatures and transcriptionally active miRNA genes are characterized by active chromatin marks such as trimethylated histone H3 lysine 4 (3H3K4) (22). Our results showed that miR-34b suppressed DNMTs and HDACs in PCa cells resulting in partial demethylation and induction of active chromatin modifications that are the hallmarks of active genes. Therefore, downregulation of DNMTs and HDACs provides positive feedback that result in an active miR-34b gene (Figure 7C).
Our study also elucidated the tumor suppressor role and functional significance of miR-34b in PCa. Results showed that miR-34b has an antiproliferative effect and induced G0/G1 cell cycle arrest and apoptosis. To determine the antiproliferative and apoptotic effects of miR-34b, we focused on Akt-GSK-betacatenin-cMyc-Cyclin D1 proliferative pathway genes. Signaling through PI3K/Akt pathway controls cell proliferation and apoptosis (43, 44). Akt mediates these effects by modulating cell cycle and apoptosis regulatory proteins (45), and changes in expression of these genes have been linked to PCa (46). Other studies have reported that advanced human PCa was accompanied by the expression of phosphorylated Akt (47) and alterations in the serine/threonine kinase Akt/PKB pathway have been detected in a number of human malignancies (48). Akt has a wide range of downstream targets that regulate tumor-associated cell processes such as cell growth, cell cycle progression, survival, migration, epithelial–mesenchymal transition and angiogenesis (48). Our results confirmed that miR-34b directly targeted Akt and caused downregulation of downstream proteins pGSK, total beta-catenin, cMyc and cyclin D1 that are involved in cell proliferation/survival. These results suggest that antiproliferative effect of miR-34b is partly mediated through Akt pathway. We also observed a decrease in the migratory and invasive capabilities of PCa cells overexpressing miR-34b and found that it effected epithelial to mesenchymal transition (EMT) markers. EMT is a multi-faceted transdifferentiation program that enables tumor cells to acquire malignancy-associated phenotypes (49). This initial step of local invasion may be triggered by signals that carcinoma cells receive from the nearby stroma, causing them to undergo an epithelial-mesenchymal transition and subsequently metastasize to distant tissue sites (50). Considerable research has been focused on identifying the critical regulators of this metastatic process including both proteins and microRNAs (miRNAs) (50). Our results show that overexpression of miR-34b caused a decline in the mesenchymal markers Vimentin, ZO1, N-cadherin and Snail whereas there was an increase in the expression of E-cadherin, an epithelial marker. These results suggest that miR-34b suppresses migration and invasion independent of cell proliferation and that it has an anti-metastatic effect on PCa cells.
The anti-tumorigenic effects of miR-34b observed in this study were also confirmed in a PCa xenograft nude mouse model. In vivo experiments demonstrated a striking suppression in subcutaneous tumor growth in nude mice where miR-34b was directly administered to tumors compared to controls. Therefore, our study demonstrates that miR-34b has an important tumor suppressor role both in vitro and in vivo.
In summary, our study shows that miR-34b is an important tumor suppressor that is epigenetically silenced in PCa. Over expression of miR-34b reduced DNMTs and HDACs inducing partial demethylation and active chromatin modifications. MiR-34b expression is also able to predict overall and recurrence free survival of PCa patients. MiR-34b induces antiproliferative and anti-invasive effects partly by inhibiting the AKT pathway and EMT markers. This study documents the pleiotropic role of miR-34b in PCa. These findings offer novel insight into the association between dysregulation of miRNA and methylation/chromatin modification and provide a strong rationale for the development of new strategies targeting the epigenetic regulation of miRNAs for the treatment of PCa.
Supplementary Material
Figure 5. miR-34b impairs PCa cell migration and invasion independent of antiproliferative effect.
A) Migration and invasion assays of PC3 and LNCaP cells transfected with miR-34b or a cont-miR. B) Downregulation of epithelial to mesenchymal transition (EMT) markers at the protein level determined by Western blot and immunofluorescence in PC3 PCa cells transfected with miR-34b or cont-miR.
Translational Relevance.
This study documents the pleiotropic role of miR-34b in prostate cancer. Our results provide evidence that miR-34b acts as a tumor suppressor with diagnostic and prognostic significance in prostate cancer. Reconstitution of miR-34b downregulated DNMTs and HDACs resulting in partial demethylation and induction of active chromatin modifications. miR-34b had anti-proliferative and anti-invasive effect by impairing the AKT proliferative pathway genes and the EMT markers. We also confirmed the growth suppressive effect of miR-34b in in vivo xenografts. These findings offer novel insight into the association between dysregulation of miRNA and methylation/chromatin modification and provide a strong rationale for the development of new strategies targeting the epigenetic regulation of miRNAs for the treatment of prostate cancer.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
Financial Support: This study was supported by the Department of Veterans Affairs VA Merit Review, and Program Project and NIH / NCI RO1CA138642, RO1CA160079, T32-DK007790.
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 2.Shi XB, Tepper CG, White RW. MicroRNAs and prostate cancer. J Cell Mol Med. 2008;12:1456–1465. doi: 10.1111/j.1582-4934.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 13.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 14.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 15.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 16.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31:2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- 19.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 21.Urdinguio RG, Fernandez AF, Lopez-Nieva P, Rossi S, Huertas D, Kulis M, et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics. 2010;5:656–663. doi: 10.4161/epi.5.7.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 24.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 25.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010 doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 30.Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 31.Glud M, Klausen M, Gniadecki R, Rossing M, Hastrup N, Nielsen FC, et al. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129:1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 32.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2009;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011;11:490–496. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 34.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 116:66–76. doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 36.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Zou X, Berger AD, Twiss C, Peng Y, Li Y, et al. Increased expression of histone deacetylaces (HDACs) and inhibition of prostate cancer growth and invasion by HDAC inhibitor SAHA. Am J Transl Res. 2009;1:62–71. [PMC free article] [PubMed] [Google Scholar]
- 39.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2009;116:66–76. doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waltregny D, North B, Van Mellaert F, de Leval J, Verdin E, Castronovo V. Screening of histone deacetylases (HDAC) expression in human prostate cancer reveals distinct class I HDAC profiles between epithelial and stromal cells. Eur J Histochem. 2004;48:273–290. [PubMed] [Google Scholar]
- 41.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 44.Paez J, Sellers WR. PI3K/PTEN/AKT pathway. A critical mediator of oncogenic signaling. Cancer Treat Res. 2003;115:145–167. [PubMed] [Google Scholar]
- 45.Cully M, Shiu J, Piekorz RP, Muller WJ, Done SJ, Mak TW. Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res. 2005;65:10363–10370. doi: 10.1158/0008-5472.CAN-05-1633. [DOI] [PubMed] [Google Scholar]
- 46.Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–7474. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- 47.Malik SN, Brattain M, Ghosh PM, Troyer DA, Prihoda T, Bedolla R, et al. Immunohistochemical demonstration of phospho-Akt in high Gleason grade prostate cancer. Clin Cancer Res. 2002;8:1168–1171. [PubMed] [Google Scholar]
- 48.Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV. The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene. 2005;24:7482–7492. doi: 10.1038/sj.onc.1209088. [DOI] [PubMed] [Google Scholar]
- 49.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 50.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.