Abstract
Diabetes Mellitus is affecting people of all age groups worldwide. Many synthetic medicines available for type 2 diabetes mellitus in the market. However, there is a strong requirement for the development of better anti-diabetes compounds sourced especially from natural sources like medicinal plants. The extracts from the leaves of neem (Azadirachta indica) is traditionally known to have anti-diabetes properties. Therefore, there is an increased interest to identify potential compounds identified from neem leaf extracts showing predicted binding property with the known diabetes mellitus type 2 protein enzyme target phosphoenol-pyruvate carboxykinase(PEPCK). The structure data for compounds found in the leaf extract of neem was screened against PEPCK using molecular docking simulation and screening techniques. Results show that the compound 3-Deacetyl-3-cinnamoyl-azadirachtin possesses best binding properties with PEPCK. This observation finds application for further consideration in in vitro and in vivo validation.
Background
Diabetes mellitus has become a universal metabolic prevalent that has effect the many biochemical events nearly in all age group population [1]. Good physical activity, a hygienic diet, avoiding from smoke and obsessive control from alcohol has an 82% esser rate of diabetes [2]. Diabetes is mainly characterized by the inadequate function of pancreas when enough insulin is not producing by the pancreas or the insulin which is produced by the pancreas is not well utilized in body. Diabetes is one of the leading causes of premature disorder and demise in worldwide [3]. Diabetes is reason for 60% of all deaths worldwide without including transmissible diseases [4]. At present diabetes mellitus has been affecting above 170 million individuals worldwide and hypothetically in the year 2030 it will be above 365 million [5]. Moreover the failure of beta cells, the other pathophysiological factor causative to the prevalence of type 2 diabetes mellitus is typically related to the inadequate secretion of insulin and target tissues show resistance to insulin [6]. From the past four decades the incidence of type 2 diabetes mellitus (T2DM) has been raised intensely. Type 2 diabetes mellitus has been described as: the resistance of insulin, hyperinsulinaemia and hyperglycaemia. Eminent hepatic gluconeogenesis producing surplus of glucose through aberrantly way is the vital indicator of hyperglycaemia in type 2 diabetes mellitus [7, 8]. There is clinical correlation to the insulin resistance causing the increase of glucose level this results due to the lack of inhibition of two enzymes involving in gluconeogenic process, PEPCK (phosphoenolpyruvatecarboxykinase) and G6Pase (glucose-6- phosphatase) subunit [9, 10]. Several enzymes catalyzed the gluconeogenesis pathway i.e. phosphoenolpyruvatecarboxykinase (PEPCK) and glucose-6- phosphatase (G6Pase) [11], PEPCK is present in two isoforms, a mitochondrial (PEPCK-M) and a cytosolic (PEPCK-C). Now PEPCK is deliberated a key enzyme in gluconeogenesis and glyceroneogenesis [12]. PEPCK-C is present on chromosome 3 in rat, on chromosomes 2 in mouse, and in humans on chromosome 20 [13–15]. PEPCK-C and PEPCK-M both are coded by separate genes [16]. PEPCK-C and G6Pase has vital role in gluconeogenesis in liver and kidney and also has some other metabolic roles such as glyceroneogenesis in liver and adipose tissues [17, 18]. Insulin is considered the most significant hormone that play role in the inhibition of gluconeogenesis. Primarily it acts by suppressing the expression PEPCK and G6Pase genes for the gluconeogenic enzymes. In the first rate limiting steps of gluconeogenesis, PEPCK catalyzes the reaction of oxaloacetic acid to phosphoenolpyruvate, and then in the final step of gluconeogenesis, G6Pase catalyzes the glucose 6-phosphate into glucose. Glucagon and glucocorticoids induced the expression of genes for PEPCK and G6Pase during fasting and periods of stress respectively [19–21]. A co-activator protein PGC-1 has been discovered that is also the important mediator for this process [22]. These signaling pathways that regulate the glucogenic enzymes are the potential targets for drug designing to restore the insulin sensitivity. An antidiabetic commercial drug, metformin has been used in clinical patients for more than 40 years to reduce the hepatic glucose production [23]. Medicinal plants have an important role in the treatment of diabetes particularly in the countries where resources are insufficient. Traditional medicines are mostly used in the majority of the world population. Rather than relying on the expensive imported drugs, the use of indigenous forms of medicines are encouraged [24]. As compared to the synthetic drugs these herbal drugs have minor side effects. Hence, it has become more important to isolate and identify the anti-hyperglycemic compounds from plants [25].
More than 150 plants are used for the management of diabetes mellitus by the WHO report [26]. Azadirachtaindica (neem) has engrossed universal importance in recent years due to its extensive medicinal properties. Neem has been widely used in Unani, Ayurveda and Homeopathic medicines. Neem has enormous range of biologically active chemical constituents that are different chemically and structurally. Over than 140 chemically active compounds have been identified from different parts of neem i.e. leaves, flowers, seeds, fruits, roots and bark are used traditionally for the cure of many diseases. Neem leaf and its active compounds have been identified to reveal the anti-inflammatory, immunomodulatory, antiulcer, anti-hyperglycaemic, antimutagenic, antioxidant, anticarcinogenic and antiviral properties [27]. Neem constituents are consisting of two groups: Isoprenoids and non-isoprenoids. The Isoprenoids comprise of diterpenoids and triterpenoids including protomeliacins, azadirone, limonoids and derivatives such as vilasinin, nimbin, salanin and azadirachtin. The non-isoprenoids consist of carbohydrates, proteins, sulphurous compounds, and polyphenolics including flavonoids, dihydrochalcone, coumarin and aliphatic compounds ete, [28]. A substantial and dose dependent decrease in blood pressure has been reported by an alcoholic extract of neem leaves [29]. Considering the above discussed medications by neem this study was designed to screen the active compounds of Azadirachtaindica (Neem) against the PEPCK protein by using molecular docking studies.
Methodology
This study was design for the in-silico investigation of Azadirachta indica (neem) leaves active chemicals against PEPCK protein. For the current study chemical structures of neem compounds were retrieved from PubChem. Few of them were drawn by using the Chemdraw software. 3D structure of PEPCK protein was retrieved from Protein Data Bank (PDB). This study was conducted using MOE (Molecular Operating Environment) software [30].
Preparation of Protein Structure:
The target protein was downloaded from Protein Data Bank (PDB) (PDB: http://www.rcsb.org/pdb/home). 1KHB is the PDB id of target protein. By using MOE (Molecular Operating Environment) software all water molecules were removed. Hydrogen atoms were added to the receptor molecule. Receptor molecule was optimized by doing 3D protonation and energy minimization. Energy minimization was performed using the AMBER99 force field and gradient was 0.05. Receptor was minimized till the root mean square gradient reaches below 0.05. Then minimized receptor molecule was used for docking study.
Preparation of Ligand Structure:
Neem leaves chemicals were retrieved from PubChem in 2D format. Few chemical compounds structures were not available on PubChem. Their 2D structures were found from literature study and their structures were drawn using ChemDraw software and saved in 3D format. Hydrogen atoms were added to the ligand structures and energy minimization was performed using the MMFF94X force field at the gradient 0.05.Then the ligands were saved in .mol2 format.
Construction of Database:
In MOE, a database of all the ligands was constructed. Database was saved in .mdb format. This database was docked against the target receptor protein.
Molecular Docking:
Molecular docking was performed with the database of ligands against PEPCK protein by setting the docking parameters. Ligands were docked by selecting the pocket from receptor protein in MOE. Output database file was saved in .mdb format. The minimum S scored complexes was taken to check the binding interactions with the active sites. The best hydrogen bonding and π-π interactions were analyzed by the ligX option in MOE.
Results & Discussion
Docking Analysis:
All ligands of Azadirachta indica (neem) were docked with the active sites of PEPCK enzyme and top ranked conformations of each ligand were selected from output database. These ligands were saved in a separate database. Compound 01 was ranked as top docked molecule on the basis of S-Score obtained from MOE docking algorithm. Thus, it can be inferred that this compound can serve as potential inhibitor against PEPCK enzyme. All other compounds were having S-Score close to each other. All compounds were studied in detail to obtain their interaction information that can be important for the inhibition of PEPCK enzyme. Interaction diagrams were obtained using MOE ligand interaction analysis feature.
Binding Interactions of Ligands and Protein:
The most active compound 01 was ranked as first on the basis of docking score. It is clear from the (Figure 1) that this compound was bound deep into the binding cavity of PEPCK making interactions with the residues Asn292, Met 295, Leu293, Asn297, Gly289, Thr343, Pro528, Phe530, and Gly531.On the basis of docking score compound 02, compound 03, compound 03, compound 04, compound 05, compound 06, compound 07, compound 08, compound 09 and compound 10 were ranked close to each other as shown in Table 1 (see supplementary material). Among all these compounds, Compound 03, compound 05, compound 06 and compound 10 showed the interactions with the Cys288 residue of active site of enzyme PEPCK. In all the listed compounds, compound 01, compound 02, compound 03, compound 05, compound 06, compound 09 and compound 10 were having interactions with the Asn292, Gly289 residues (compound 09 was not having interaction with Gly289). Compound 03, compound 04, compound 05, compound 07, compound 09 and compound 10 were having interactions with Arg405, Lys290.Compound 04 was not bound to the Lys290 residue. Compound 03, compound 05, compound 07, compound 09 and compound10 were have fine interactions with Asp310, Asp311 and Thr291 sites of enzyme. Compound 04 and compound 07 were also having interactions with Tyr235 residue. Compound 03, compound 07 and compound 10 showed interactions with Ser286 residue. Compound 03 and compound 09 were having interactions with Lys244 in binding site of protein molecule. Compound 03 is the only compound which was having interaction with Glu329 residue. Compound 03 and compound 10 were also bound to the Ala287 residue. Compound 04 was also having interaction with Asn403. Compound 05 was having interaction with Phe530 and compound 06, compound 09 and compound 10 were having interactions with Val335 in binding site of protein. Compound 07 was also bound to the Arg87 residue and compound 06, compound 09 and compound 10 were having interactions with Arg436 residues. Compound 08 was having binding site interactions with Pro337 and Pro402 residues. Compound 09 was also having interactions with the Gly338, His264 and Phe333 residues in binding site of PEPCK molecule.
Figure 1.
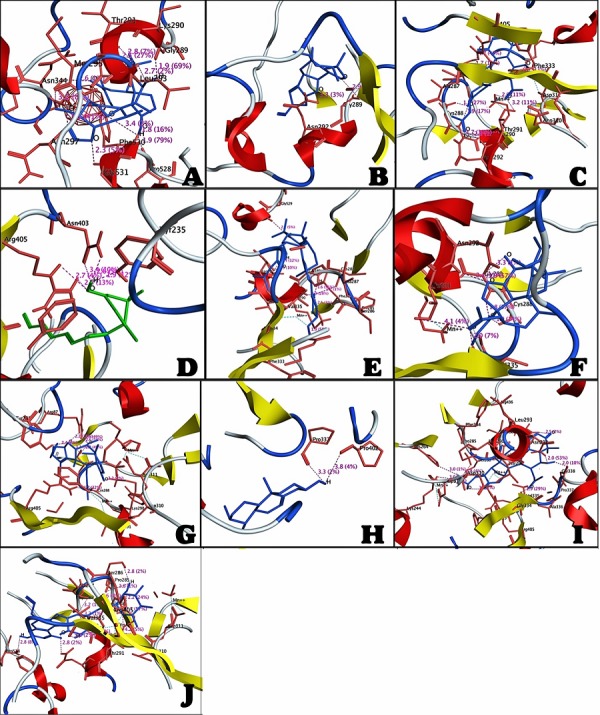
A) The interaction between the compound 01 and active sites of PEPCK; B) The interaction between the compound 02 and active sites of PEPCK; C) The interaction between the compound 03 and active sites of PEPCK; D) The interaction between the compound 04 and active sites of PEPCK; E) The interaction between the compound 05 and active sites of PEPCK; F) The interaction between the compound 06 and active sites of PEPCK; G) The interaction between the compound 07 and active sites of PEPCK; H) The interaction between the compound 08 and active sites of PEPCK; I) The interaction between the compound 09 and active sites of PEPCK; J) The interaction between the compound 10 and active sites of PEPCK.
Conclusion
There is an increasing need for the identification, screening and design of anti-diabetes compounds from natural sources like the leaf extracts of neem. Hence, PEPCK was screened using the structure data for compounds found in the leaf extract of neem using molecular docking simulation and screening techniques. The compound 3-Deacetyl-3-cinnamoyl-azadirachtin is found to have the best binding properties with PEPCK. This observation finds application for further consideration in in vitro and in vivo validation.
Supplementary material
Footnotes
Citation:Jalil et al, Bioinformation 9(20): 1031-1035 (2013)
References
- 1.Gupta R, et al. Afr J Tradit Complement Altern Med. 2007;5:1. [PMC free article] [PubMed] [Google Scholar]
- 2.Mozaffarian D, et al. Arch Intern Med. 2009;169:798. doi: 10.1001/archinternmed.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. http://www.who.int/ mediacentre/factsheets/fs312/en/
- 4. http://www.diabetesatlas.org /content/foreword.
- 5.Wild S, et al. Diabetes Care. 2004;27:1047. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 6.Ginsberg HN. J Clin Invest. 2000;106:453. doi: 10.1172/JCI10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitrakou A, et al. N Engl J Med. 1992;326:22. doi: 10.1056/NEJM199201023260104. [DOI] [PubMed] [Google Scholar]
- 8.Perriello G, et al. Diabetes. 1997;46:1010. doi: 10.2337/diab.46.6.1010. [DOI] [PubMed] [Google Scholar]
- 9.Nakae J, et al. Nat Genet. 2002;32:245. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, et al. Biochem J. 2004;378:839. doi: 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson RW, Reshef L. Annu Rev Biochem. 1997;66:581. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 12.Utter M, Kurahashi K. J Amer Chem. 1953;75:78. [Google Scholar]
- 13.Fulchignoni-Latuad MC, et al. Mamm Genome. 1992;3:42. doi: 10.1007/BF00355841. [DOI] [PubMed] [Google Scholar]
- 14.Lem J, Fournier RE. Somat Cell Mol Genet. 1985;11:633. doi: 10.1007/BF01534728. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, et al. Genomics. 1993;15:219. doi: 10.1006/geno.1993.1040. [DOI] [PubMed] [Google Scholar]
- 16.Chakravarty K, et al. Crit Rev Biochem Mol Biol. 2005;40:129. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 17.Iynedjian PB, et al. J Biol Chem. 1975;250:5596. [PubMed] [Google Scholar]
- 18.Parry DM, Brosnan JT. Can J Biochem. 1980;58:1298. doi: 10.1139/o80-174. [DOI] [PubMed] [Google Scholar]
- 19.Dickens M, et al. J Biol Chem. 1998;273:20144. doi: 10.1074/jbc.273.32.20144. [DOI] [PubMed] [Google Scholar]
- 20.Granner D, et al. Nature. 1983;305:549. doi: 10.1038/305549a0. [DOI] [PubMed] [Google Scholar]
- 21.Lamers WH, et al. Proc Natl Acad Sci USA. 1982;79:5137. doi: 10.1073/pnas.79.17.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barthel A, Schmoll D. Am J Physiol Endocrinol Metab. 2003;285:E685. doi: 10.1152/ajpendo.00253.2003. [DOI] [PubMed] [Google Scholar]
- 23.Hundal RS, et al. Diabetes. 2000;49:2063. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romila Y, et al. Biological and Environmental Sciences. 2010;6:167. [Google Scholar]
- 25.Akanksha, et al. Indian J Exp Biol. 2010;48:294. [PubMed] [Google Scholar]
- 26.Marles RJ, Farnsworth NR. Phytomedicine. 1995;2:137. doi: 10.1016/S0944-7113(11)80059-0. [DOI] [PubMed] [Google Scholar]
- 27.Subapriya R, Nagini S. Curr Med Chem Anticancer Agents. 2005;5:149. doi: 10.2174/1568011053174828. [DOI] [PubMed] [Google Scholar]
- 28.Kumar PS, et al. Annals of Biological Research. 2010;1:24. [Google Scholar]
- 29.Koley KM, Lal J. Indian J Physiol Pharmacol. 1994;38:223. [PubMed] [Google Scholar]
- 30.Khan M, et al. Bioinformation. 2013;9:710. doi: 10.6026/97320630009710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


