Abstract
Dendritic spines are the elementary structural units of neuronal plasticity and the cascades that promote dendrite spine remodeling center on Rho GTPases and downstream effectors of actin dynamics. In a model of hormone replacement therapy (HT), we sought the effect of estradiol (E) and progesterone (P) on gene expression in these cascades in laser captured serotonin neurons from rhesus macaques with cDNA array analysis. Spayed rhesus macaques were treated with either placebo, E or E+P via Silastic implant for 1 month prior to euthanasia after which the midbrain was obtained, sectioned and immunostained for TPH. TPH-positive neurons were laser captured using an Arcturus Laser Dissection Microscope (PixCell II). RNA from laser captured serotonin neurons (n=2 animals/treatment) was hybridized to Rhesus Affymetrix GeneChips. There was a significant change in 744 probe sets (ANOVA, p < 0.05), but 10,493 probe sets exhibited a 2-fold or greater change. Pivotal changes in pathways leading to dendrite spine proliferation and transformation included 2-fold or greater increases in expression of the Rho GTPases called CDC42, Rac1 and RhoA. In addition, 2-fold or greater increases occurred in downstream effectors of actin dynamics including PAK1, ROCK, PIP5K, IRSp53, WASP, WAVE, MLC, cofilin, gelsolin, profilin and 3 subunits of ARP2/3. Finally, 2-fold or greater decreases occurred in CRIPAK, LIMK2 and MLCK. The regulation of RhoA, Rac1, CDC42, ROCK, PIP5k, IRSp53, WASP, WAVE, LIMK2, CRIPAK1, MLCK, ARP2/3 subunit 3, gelsolin, profilin and cofilin was confirmed with nested qRT-PCR on laser captured RNA (n=3 animals/treatment). The data indicate that ovarian steroids target gene expression of the Rho GTPases and pivotal downstream proteins that in turn, would promote dendritic spine proliferation and stabilization on serotonin neurons of the dorsal raphe nucleus.
Keywords: serotonin, macaques, estrogen, progesterone, Affymetrix array, apoptosis, cell cycle, dorsal raphe
Introduction
The serotonin system modulates a wide range of neural outcomes from emotion to intellect to metabolism and it is a target of pharmacotherapies, steroid hormones, cytokines, neuropeptides and trophic factors, all of which impact the generation and efficacy of serotonin neurotransmission. Recently, it has been suggested that antidepressants and other pharmacotherapies may act by promoting neuronal plasticity (1). The underlying structural element of neuronal plasticity in the adult nervous system is the dendritic spine (2). In addition, dendritic spines are the morphological basis for excitatory neurotransmission (3, 4). Hence, dendritic spine proliferation on serotonin neurons would have a profound effect on serotonergic function and neurotransmission.
A significant body of literature has demonstrated that estrogen (E) increases dendritic spines in hippocampal neurons (5, 6). We hypothesized that ovarian steroids support neuronal plasticity by enhancing dendritic spine proliferation on serotonin neurons. In the adult nervous system, this would facilitate excitatory input to the serotonin neurons and in turn, increase serotonin neural activity.
The extension of dendrites requires a multiplex intracellular cascade that culminates in dynamic actin reorganization (2). This reorganization requires extension and branching of actin at the cell membrane as well as degradation of actin further back in the spine. Upstream of actin dynamics are effector proteins that are activated by Rho GTPases. The most well accepted of these are CDC42, Rac 1 and Rho A. These small molecules activate cascades that lead to filopodia extension, spine head enlargement and spine shortening, all of which are necessary for the production of a mature dendritic spine.
This laboratory has devoted effort toward understanding the actions of ovarian hormones in serotonin neurons and their terminal fields with a macaque model of surgical menopause. Serotonin neurons express estrogen receptor beta (ERβ) and progestin receptors (PR) (7, 8). We found that E, plus or minus P supplementation, regulates the expression of pivotal serotonin-related genes and proteins in the monkey dorsal raphe in a pattern suggestive of increased serotonin production, increased serotonin turnover, increased neural firing and decreased degradation (9–11).
We recently reported that ovarian steroids regulate gene expression in laser captured serotonin neurons in a manner that would increase cellular resilience and decrease apoptosis (12). Further examination of the proteins implicated by gene expression showed that in normal, undamaged midbrain, ovarian steroids act through the caspase independent pathway and decrease translocation of apoptosis inducing factor (AIF) from the mitochondria to the nucleus (13).
The additional issue of steroid supported dendritic spine proliferation may be extremely important for menopausal women grappling with issues surrounding hormone therapy (HT). Women experience ovarian failure and loss of ovarian steroid production around 50 years of age. Thus, with extended life spans, a woman may live 35–40 years without ovarian steroids. If dendritic spines on serotonin neurons shrink or atrophy due to lack of steroid supported gene expression, then geriatric depression, anxiety, fretfulness, decreased coping skills and increased vulnerability to stress can be predicted outcomes.
Therefore, in this study, we sought novel genes related to dendritic spine protrusion that are regulated by estrogen (E) and progesterone (P) in laser captured serotonin neurons of rhesus monkeys using the Rhesus Affymetrix cDNA array and nested quantitative (q) RT-PCR. Fifteen pivotal gene changes predicted by the microarray were confirmed by qRT-PCR.
Materials and Methods
The Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee approved this study.
Animals and treatments
Nine adult female rhesus monkeys (Macaca mulatta) were spayed (ovariectomized only) by the surgical personnel of ONPRC between 3 and 8 months before assignment to this project according to accepted veterinary surgical protocol. All animals were born in China, were aged between 7–14 years by dental exam, weighed between 4 and 8 kg, and were in good health.
Animals were either treated with placebo (ovx-control group; n=3), or treated with estrogen (E) for 28 days (E group; n=3), or treated with E for 28 days and then supplemented with P for the final 14 of the 28 days (E+P group; n=3). The placebo treatment of the spay-control monkeys consisted of implantation with empty Silastic capsules (s.c.). The E-treated monkeys were implanted (s.c.) with two 4.5-cm E-filled Silastic capsules (i.d. 0.132 in.; o.d. 0.183 in.; Dow Corning, Midland, MI). The capsule was filled with crystalline estradiol (1,3,5(10)-estratrien-3,17-b-diol; Steraloids, Wilton, NH). The E+P- treated group received E-filled capsules, and 14 days later, received one 6-cm capsule filled with crystalline P (4-pregnen-3,20 dione; Steraloids). All capsules were placed in the periscapular area under ketamine anesthesia (ketamine HCl, 10mg/kg, s.c.; Fort Dodge Laboratories, Fort Dodge, IA).
The monkeys were euthanized at the end of the treatment periods according to procedures recommended by the Panel on Euthanasia of the American Veterinary Association. Each animal was sedated with ketamine, given an overdose of pentobarbital (25 mg/kg, i.v.), and exsanguinated by severance of the descending aorta.
Steroid Hormone Assays
Assays for estradiol and progesterone were performed utilizing a Roche Diagnostics 2010 Elecsys assay instrument and the serum levels achieved by the implants were previously reported (12)
Tissue preparation for Laser Capture Dissection (n=9; 3 animals/treatment group)
The left ventricle of the heart was cannulated and the head of each animal was perfused with 3 liters of 1× cold RNA-later buffer (Ambion Inc., Austin, TX) plus 20% sucrose. The brain was removed from the cranium, dissected into blocks and frozen at −80°C. The pontine midbrain block was placed in a cryostat (Microm HM500OM) and brought to −20°C. Thin sections (7 µm) through the dorsal raphe nucleus were thaw mounted onto plain glass slides and frozen at −80°C. The next morning, the sections were processed in a rapid, RNAse free immunohistochemical assay for tryptophan hydroxylase (TPH). The sections were immersed in cold acetone for 1 minute, cold ethanol for 30 seconds, cold PBS for 3 minutes, and then covered with normal rabbit serum (NRS, 1/500) containing 1% RNAse inhibitor for 10 minutes. The NRS was blotted and the sections were covered with sheep anti-TPH (1/300; Chemicon, Temecula, CA) containing 1% RNAse inhibitor for 30 minutes, then immersion washed in PBS for 3 minutes, and covered with biotinylated rabbit anti-sheep serum (1/120; Vector Laboratories, Burlingame, CA) containing 1% RNAse inhibitor for 20 minutes. The sections were then immersed in PBS for 3 minutes, covered with Vector ABC reagent for 30 minutes, immersed in cold 0.2 M Tris (pH 8.2), immersed in cold diaminobenzidine (DAB) containing H2O2 (30% solution diluted 1/5000) and dehydrated in 100% ethanol for 2 minutes. Finally, the slides were immersed in xylene for 2 minutes and then dried under vacuum for 1 hr prior to laser capture. Serotonin neurons appeared darkly stained and were captured with an Arcturus Laser Dissection Microscope (PixCell II). After capture to the microcap film (Capsure macro-211), the films were removed from the caps and immersed in lysis buffer. Up to 10 films with 1000–3000 pulses each were collected into one microtube containing 200 µl of lysis buffer. Approximately 150,000 laser pulses were conducted for a pool (12–15 microtubes were pooled). Individuals interested in obtaining other areas of the brain for similar studies are encouraged to contact the senior author.
RNA extraction from laser captured neurons
Two pools were prepared from two placebo, two E-treated and two E+P treated animals. One of the pools was used for hybridization to the microarray and one pool was set aside for qRT-PCR. An additional pool was prepared from another placebo, E-treated and E+P treated animal for qRT-PCR. Thus, 2 animals per group were used for hybridization and 3 animals per group were used to confirm gene changes with qRT-PCR.
The tubes containing the lysis buffer and films were vortexed to dislodge the captured material from the films. Each tube was adjusted to 350 µl of lysis buffer and then 350 µl of 70% ethanol was added and mixed well. The samples were extracted with the RNAeasy microRNA kit from Qiagen according to the directions. The final eluates were pooled and evaporated in a Speedvac. The RNA was suspended in 12 µl of TE (0.01MTris+0.005M EDTA). The quantity of RNA in the resuspended sample was determined with the Ribogreen Quantitation Kit (Molecular Probes, Eugene, OR) or with a Nanodrop Spectrophotometer (ND 1000 V3.3, Wilmington, DE). The integrity of the RNA was examined with the Agilent Bioanalyzer using the pico-chip according to the directions of the manufacturer.
Affymetrix hybridization
Labeled target cRNA was prepared from 6 pools of laser captured serotonin neurons (n=2 animals/treatment group). The cRNA was hybridized to Rhesus Affymetrix GeneChip arrays. The Affymetrix GeneChip® Rhesus Macaque Genome Array interrogates over 47,000 M. mulatta transcripts. The array contains 52,024 rhesus probe sets and was designed using public data sources including data from the University of Nebraska (R. Norgren), the Baylor School of Medicine’s rhesus macaque whole-genome shotgun assembly (October 1, 2004), and GenBank® STSs, ESTs, and mRNAs up to March 30, 2005. Additionally, probe sets were designed to interrogate rhesus transcripts orthologous to the 3’ end of human transcrips (GeneChip® Human Genome U133 Plus 2.0 Array and RefSeq sequences up to March 2005), sixteen viral sequences from human and other primate species, three different rhesus r(ribosomal)RNAs, and thirteen rhesus mitochondrial genes along with control and reporter sequences. Microarray assays were performed in the Affymetrix Microarray Core of the OHSU Gene Microarray Shared Resource.
Data Analysis with GeneSifter Software
The data was processed with Affymetrix GCOS interface software and compressed into CHP files and uploaded to GeneSifter (VisX Labs, Seattle, WA). The software calculated the mean signal intensity and the standard error of the mean for each treatment group. Probe sets that were undetectable in all treatment groups were eliminated. The remaining probe sets were filtered and those probes sets exhibiting a 2-fold or greater change between the treatment groups were subjected to KEGG analysis. The use of a 2-fold filter was chosen based upon a survey of other microarray studies. Probe sets with robust signal intensities that showed a 2-fold or greater change with E or E+P and that were consistent across multiple representations on the chip were reported. Individuals interested in obtaining the raw data are encouraged to contact the senior author.
Nested Quantitative (q) RT-PCR
A second laser capture pool was made from the 6 animals used for the microarrays, plus an additional 3 animals, yielding 9 laser capture pools (3 animals/treatment group) for qRT-PCR confirmation of 15 pivotal genes which were: RhoA, Rac1, CDC42, ROCK, PIP5k, IRSp53, WASP, WAVE, ARP3, gelsolin, profilin, cofilin, LIMK2, CRIPAK1 and MLCK. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), GUSB and PIAA were measured for reference. GAPDH was used for normalization of the data.
Complementary DNA (cDNA) synthesis was performed using Oligo-dT 15 and Random hexamer primer (Invitrogen Life Technologies, Carlsbad, CA) and Superscript III reverse transcriptase (200 U/µg of RNA, Invitrogen Life Technologies) at 42C for 1 hr. cDNA was treated with RNAse H to disintegrate dsDNA. Total RNA (1.4 to 1.8 µg) from each Laser capture pool and from a pool of rhesus tissues was transcribed and stored as cDNA at a concentration of 250 ng/µl.
The cDNA (10–20 ng) from each laser capture pool and from the rhesus tissue pool (100ng cDNA) was used in a 50 µl multiplex PCR reaction with Platinum Taq polymerase (Invitrogen Life technology, Carlsbad, CA). Each of the external primer sets (20 µM) was added to the multiplex PCR reaction. Then, 1µl was removed for each of the internal primer sets and the quantitative polymerase chain reaction. qPCR was conducted with Platinum SYBR-Green qPCR Super Mix UDG (Invitrogen Life Technologies). The reaction (final volume 20 µl) contained 1 µl PCR product of external PCR reaction, 10 µM of forward and reverse primers and 1× SYBR-Green ER qPCR mix Invitrogen (11760-500). PCR Mastermix contains a modified Platinum DNA Taq Polymerase, uracil DNA glycosylase (UDG) and SYBR-Green II fluorescent dye, which is specific for double stranded DNA. In addition, 500 nM of ROX was added to control PCR artifacts and to control the baseline of the multiplex reaction. There was a log linear increase in fluorescence detected as the concentration of amplified double stranded product cDNA increased during the reaction. The fluorescence was detected as cycle threshold (Ct) with an ABI 7900 thermal cycler during forty cycles. A standard curve was generated with a pool of RNA from various rhesus tissues and rhesus embryonic stem cells (rhesus standard pool). The PCR product from the multiplex reaction on the rhesus standard pool was diluted to generate a standard curve for each of the internal primer sets. The slope of the curve was used to calculate the relative pg of each transcript in the RNA extracted from the laser captured pools. Then, the ratio of each transcript to GAPDH was calculated.
Validation of Nested PCR
Initially, cold PCR was conducted for all primer sets using cDNA from the rhesus tissue pool to verify that the internal reactions produced one product. To further verify PCR amplification, the product of the internal qPCR reaction was sequenced.
Primer selection
For our genes of interest, the probe set ID was saved as a .txt file. This file was launched to batch query at Affymetrix Netaffix program on the web. This program enabled retrieval of the annotation list of the genes of interest and provides direct access to Genebank sequence at NCBI. Thus, using Netaffix, the oligonucleotide location on each cDNA sequence was identified. Based upon the oligoset distribution, a target area of each gene of interest was selected. The target sequence was then loaded into Primer Express software, which chooses the primers for optimum qRT-PCR. For each gene, two sets of primers were generated; set one was used to generate an external larger PCR product of about 400bp–600bp. The second set was used to amplify internal or nested PCR product of about 250bp–350bp off of the larger product. The primers utilized for qRT-PCR on the RNA extracted from the laser-captured serotonin neurons are shown in Table 1.
Table 1.
Sequence of the external and internal primers for nested qRT-PCR on mRNA from laser captured serotonin neurons.
| Synaptic Plasticity Primers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence | TM | Amplicon size | Name | Sequence | TM | Amplicon size | ||
| RhoA | Profilin | ||||||||
| ExtF | TGGGGCAGGAAGATTATGATCG | 65.3 | 436 | ExtF | GGCGCTGCGGTAAGGAG | 63.05 | 305 | ||
| ExtR | GCAGCTCTCGTAGCCATTTCA | 62.85 | ExtR | AGCTCTGCCGACAGCAACAT | 63.44 | ||||
| IntF | CCCTCTCCTACCCAGATACCG | 62.08 | 251 | IntF | CCGCCGCTGGTTAGTCAG | 62.9 | 251 | ||
| IntR | CCCAAAAAGCACCAATCCTGTT | 64.37 | IntR | CTCTACCAGCTCTGCCGACAG | 62.5 | ||||
| Rock2 | CDC42 | ||||||||
| ExtF | GGTTCGTCATAAGGCATCACA | 60.09 | 548 | ExtF | ACGACCGCTGAGTTATCCACA | 62.94 | 351 | ||
| ExtR | CCCCCACTAGCATCTCAAAAAG | 62.1 | ExtR | TTCTTCGGTTCTGGAGGCTC | 61.81 | ||||
| IntF | ATGCCTGGTGGAGACCTTGTA | 61.5 | 298 | IntF | TTTTCAGTGGTCTCTCCATCTTCAT | 62.4 | 202 | ||
| IntR | AGTAACCATCACCCCCTTGTG | 61.04 | IntR | GGTCACGGGCCAGCTTTT | 63.5 | ||||
| MLCK | IRSp53 | ||||||||
| ExtF | GCCACGTGAACCTGATCCA | 63.16 | 551 | ExtF | GGCTTTATCACCAGGATGTCACA | 63.3 | 463 | ||
| ExtR | CAGCAACCGGGAAACAAAGT | 62.34 | ExtR | CGGCTTTACCTTGAACTGAGGT | 61.85 | ||||
| IntF | GACCTCAAGCCGGAGAACATATT | 62.81 | 251 | IntF | TTTGGAAACATCTGTGGACTCTGTT | 63.2 | 263 | ||
| IntR | TCTGCATCTGTTTCCCCTAGAAAT | 62.4 | IntR | GGTGCACTCTCCTAGCATGCT | 61.9 | ||||
| CRIPAK | WAVE | ||||||||
| ExtF | CTCGCTTTGTGCCAATGTG | 61.43 | 1361 | ExtF | TTCAACTCCTCCCCCTCCA | 62.94 | 456 | ||
| ExtR | CCTCTCCTCTTCCATCTCATCC | 61.2 | ExtR | TTAGCTTCCTGTTCACGCTGC | 62.8 | ||||
| IntF | TCACACAAAGCCCTGGCAT | 62.6 | 154 | IntF | ACCTGGCATTCGACCATCAT | 62.6 | 253 | ||
| IntR | CCTACGTTATCTCGAGGTGCAG | 61.1 | IntR | ACGCTGCTCTTCTACTTTGCG | 62.09 | ||||
| LIMK2 | WASP | ||||||||
| ExtF | AGGTGGAGGATGCAATTAGCC | 62.5 | 356 | ExtF | GACAGAGCAGGCTGGGAGAA | 63.02 | 463 | ||
| ExtR | CAGGACCTCCCCATGGATTA | 62.01 | ExtR | TGCTATCAGGCTTTGAGCCC | 62.6 | ||||
| IntF | GTTGATTGAACATGACCCCGT | 61.9 | 251 | IntF | GGTCTGGGAGGACTGTTCCA | 62.47 | 208 | ||
| IntR | CAACGAAGGGATTCTGAGCG | 63.1 | IntR | GGTTCCTCTGAGGCTCTGGG | 63.56 | ||||
| Cofilin1 | ARP3 | ||||||||
| ExtF | GAAAAGCGCGCGAGATC | 63.5 | 451 | ExtF | TGCTAAGGCAGTAAAGGAGCG | 61.87 | 453 | ||
| ExtR | TGATGGCATCCTTGGAACTG | 62.02 | ExtR | CGCTGCATGTGGTGTGTAATG | 63.48 | ||||
| IntF | GGTGTGGCTGTCTCTGATGGT | 62.5 | 259 | IntF | ATGATACAGATGGGTCAAAATGGATTA | 62.7 | 252 | ||
| IntR | GCATCATAGAGGGCGTAGCG | 63.07 | IntR | TGAACATGGTTGAACCTCCAGA | 62.7 | ||||
| RAC1 | GAPDH | ||||||||
| ExtF | TTGTGGTAGATGGAAAACCGG | 61.9 | 351 | ExtF | CCCTCCAAAATCGAGTGTGGT | 60 | 315 | ||
| ExtR | TGTGAGCACCAAACACTCCAG | 62.3 | ExtR | AGGAAAACTGGCAGTGATGGC | 60 | ||||
| IntF | TGTCCGTGCAAAGTGGTATCC | 63.1 | 182 | IntF | CTGACACTGAGTACGTCGTGGA | 57 | 268 | ||
| IntR | TTTTGCAGCACCAATCTCCTT | 62.03 | IntR | TGGACTGTGGTCATGAGTCCTT | 58 | ||||
| PIP5 | PPIA | ||||||||
| ExtF | TTCAAGCTTTTGAAAGGCCTCAC | 62.93 | 405 | ExtF | TCGACGGCGAGCCCCT | 59 | 451 | ||
| ExtR | GCATTGTCATCCTGCACTTCC | 62.9 | ExtR | GTCCACAGTCAGCAATGGTGAT | 59 | ||||
| IntF | TTCCATCGCCGCCTTAAAAG | 63.3 | 201 | IntF | TCGTGCTCTGAGCACTGGAG | 59 | 301 | ||
| IntR | GAAATTGTGGTCGCCAAGGA | 63.25 | IntR | CATGCCTTCTTTCACTTTGCC | 58 | ||||
| Gelsolin | GUSP | ||||||||
| ExtF | GCCACCCTTCCTCTCCATCT | 62.8 | 454 | ExtF | TGATCGCTCACACCAAAGCTT | 59 | 453 | ||
| ExtR | CTGGAAAGGTAGGACAAGCCC | 62.2 | ExtR | TTTTGGTTGTCTCTGCCGAGT | 58 | ||||
| IntF | TGTGGCTGATGAGAACCCCT | 63 | 251 | IntF | ACTTCGGGCACCTGGAGTT | 58 | |||
| IntR | CCCGCCAGTTCTTGAAGAACT | 62.9 | IntR | TCATGAAATCGGCAAATTCC | 58 | 253 | |||
Statistical analysis
The average signal intensities on the microarrays in the E and E+P groups were examined for 2-fold or greater increases or decreases from the ovx-control group (n=2 animals or chips/group). No further statistical test was performed on the signal intensities. The average relative expression of the 3 groups (n=3 animals/group) from the qRT-PCR assay was first compared with ANOVA followed by Student-Newman-Keuls posthoc pairwise comparison. In a few comparisons, the variance within one group hindered the ANOVA analysis. This was not unusual for this type of preparation, and more animals would likely reduce the variance. However, labor and cost were prohibitive. Therefore, a two-tailed t-test was performed in which each treatment group was compared to the ovx-control group. To confirm the variance was significantly different between groups, an F test was performed. Hence, the mean of a treatment group was significantly different from the ovx-control group with Student-Newman-Keul’s or a t-test; or the variance within the treatment group was significantly different from the ovx-control group with an F test. Either result indicates that the groups are different. Comparisons were considered significant when there was greater than a 95% chance that the groups were different (p<0.05). Prism 5.0 from Graph Pad (San Diego, CA) was used for all comparisons.
Results
Expression Changes Related to Dendritic Spines in Laser Captured Serotonin Neurons
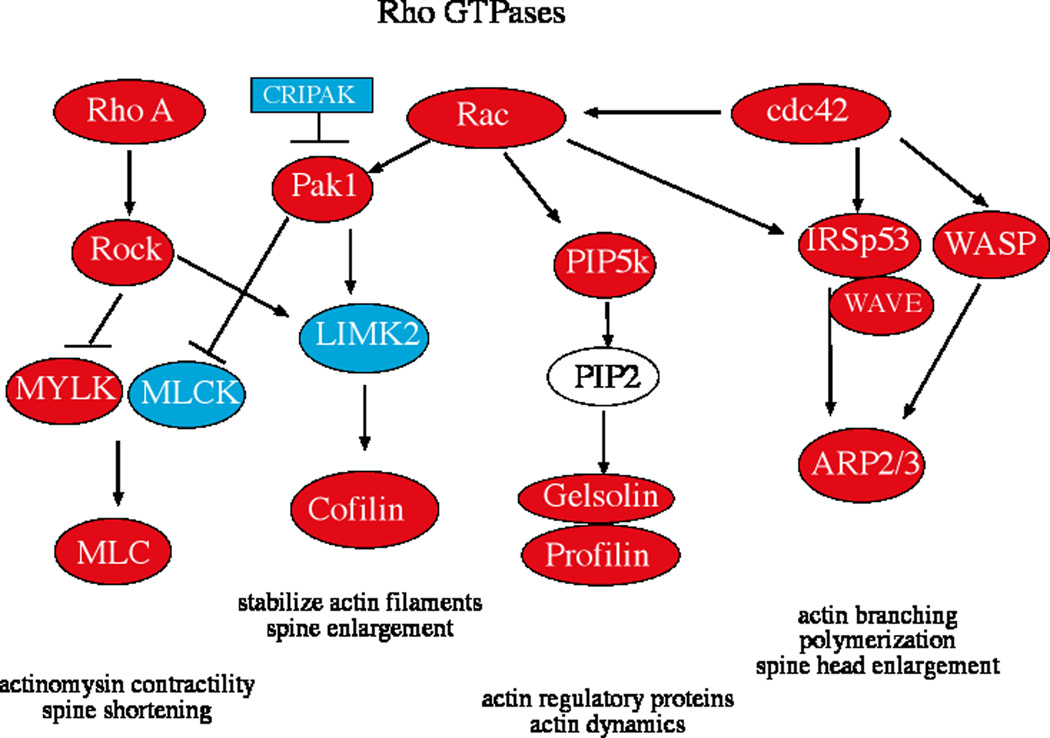
Figure 1 illustrates the 2-fold changes in gene expression that were detected on the microarrays. Genes in red increased 2-fold or greater in animals treated with E or E+P. Genes in blue decreased 2-fold or greater in animals treated with E or E+P. There was increase in expression of RhoA, Rac1, and cdc42 in the E or E+P treated animals. Downstream of CDC42, there was an increase in expression of IRSp53, WASP, WAVE and subunits of actin related protein (ARP2/3), which promotes actin enucleation, branching, polymerization and spine head enlargement. Downstream of Rac1, there was an increase in expression of PIP5K and the actin regulatory proteins, cofilin, gelsolin and profilin, which reduce actin dynamics, stabilize actin filaments and thereby stabilize spines. Also downstream of Rac1, there was a decrease in expression of CRIPAK1, LIMK2 and MLCK, which could maintain activation of cofilin. Downstream of RhoA, there was an increase in expression of ROCK2 and MLC, which code for proteins that facilitate actinomysin contractility and spine shortening. MLCK is also known as MYLK. The microarray results were confounding as there were both increases and decreases in probe sets with these acronyms. We show them in red and blue on the figure and performed qRT-PCR for confirmation below. Table 2 contains the average signal intensities for the probe sets corresponding to each of these genes from the duplicate microarray chips in each group. The results of the 6 Affymetrix microarray chips have been submitted to the Gene Expression Omnibus public database and assigned the GEO accession number GSE16169.
Figure 1.
Chart illustrating the key Rho GTPases and their downstream effectors for actin and myosin activation leading to dendritic spine protrusion and transformation. RNA extracted from laser captured serotonin neurons was labeled and hybridized to the Rhesus Affymetrix GeneChip (n=2 animals/treatment) and the results were analyzed with GeneSifter. The genes in red increased 2-fold or greater and the genes in blue decreased 2-fold or greater with E or E+P treatment compared to ovariectomized placebo treated controls.
Table 2.
Average Signal Intensity of 2 microarray chips per group for selected plasticity probe sets.
| OVX | E | E+P | |
|---|---|---|---|
| Probe Sets | |||
| Rho A | 1057 | 2436 | 1682 |
| Rac (probe set 1) | 130 | 1570 | 687 |
| Rac (probe set 2) | 2099 | 6242 | 3468 |
| cdc42 (probe set 1) | 1876 | 3790 | 2682 |
| cdc42 (probe set 2) | 461 | 1136 | 921 |
| Rock | 475 | 1110 | 723 |
| CRIPAK | 365 | 188 | 150 |
| Pak 1 | 527 | 2049 | 780 |
| PIP5K1B | 143 | 390 | 221 |
| IRSp53 | 292 | 831 | 339 |
| WAVE (WASPIP*) | 568 | 1275 | 881 |
| N-WASP (WASL) | 836 | 3006 | 1619 |
| LIMK2 | 86 | 8 | 44 |
| MLCK | 693 | 43 | 447 |
| MYLK | 549 | 1923 | 730 |
| MLC | 205 | 778 | 534 |
| Cofilin | 487 | 2255 | 1321 |
| Gelsolin | 1245 | 3182 | 1910 |
| Profilin | 2066 | 4932 | 3484 |
| ARP2/3 subunit2 | 154 | 799 | 277 |
| ARP2/3 subunit3 | 380 | 1366 | 809 |
| ARP2/3 subunit5 | 20 | 554 | 268 |
NWASP/WASL interacting protein
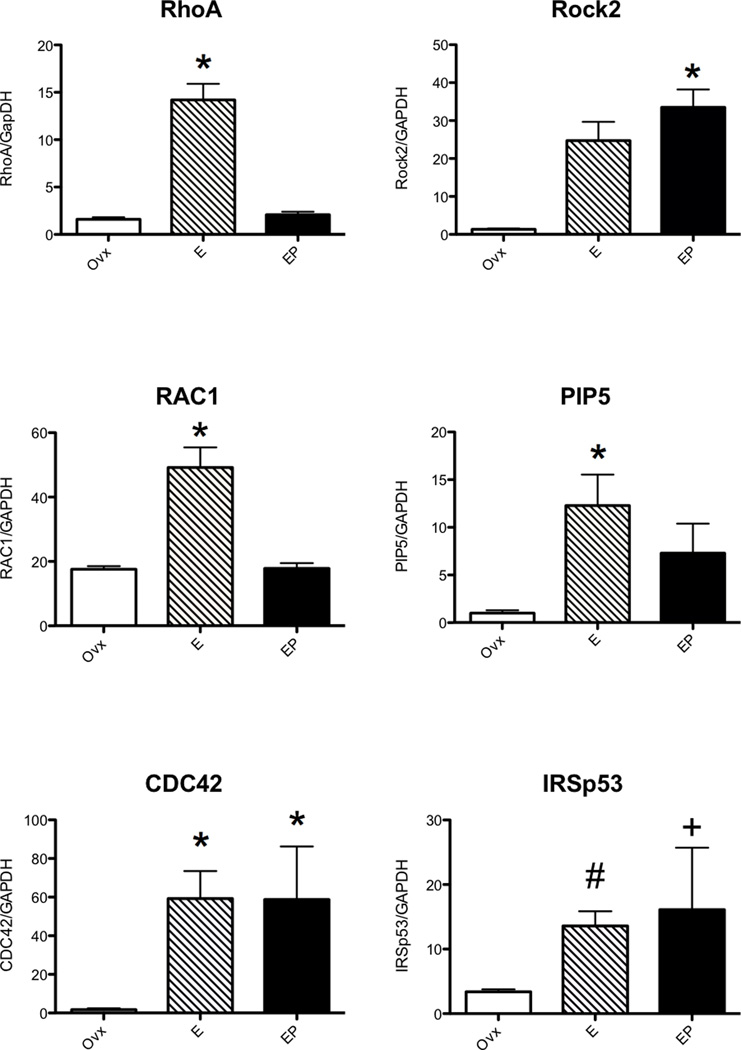
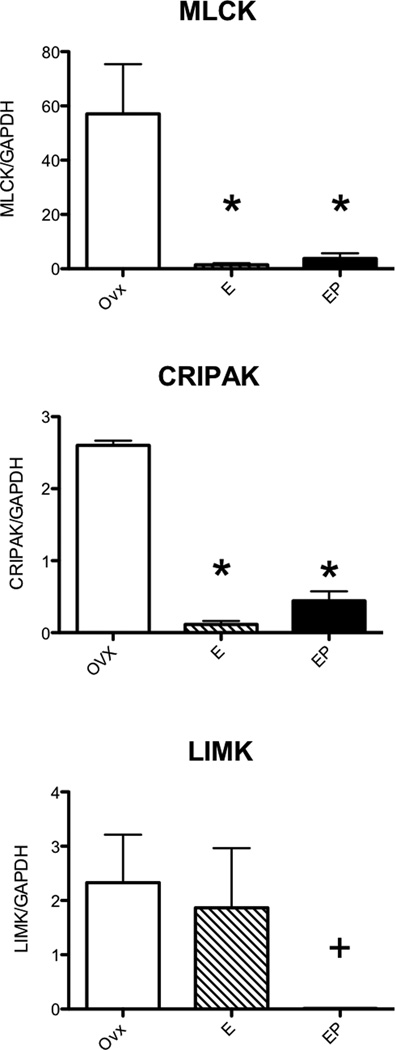
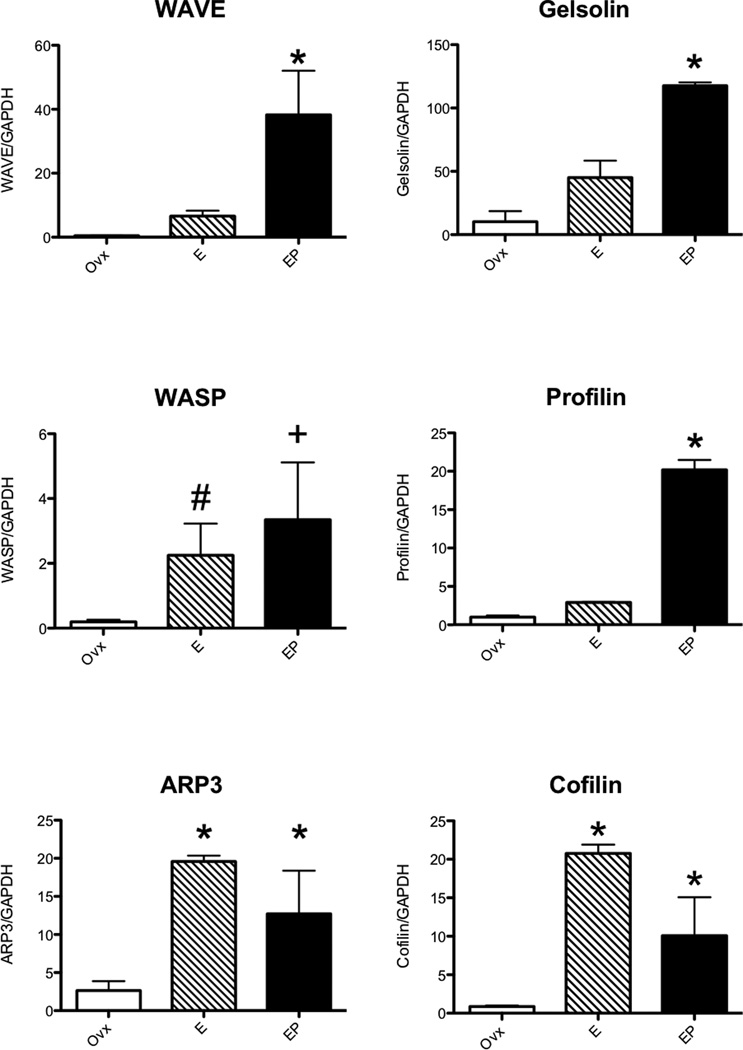
Validation of Expression Changes in Laser Captured Serotonin Neurons
Significant increases in expression of RhoA, Rac1, CDC42, ROCK, PIP5k, IRSp53, WASP, WAVE, cofilin, gelsolin, profilin, and ARP3; and significant decreases in expression of LIMK2, CRIPAK1 and MLCK, were confirmed with nested qRT-PCR and the results are shown in Figures 2–4. In Figures 2 and 3, there were different patterns of increased expression. That is, RhoA, Rac1 and PIP5K increased with E and then returned to placebo levels with supplemental P; whereas CDC42, ROCK2, IRSp53, WASP, ARP3 and cofilin increased with E and remained elevated with supplemental P; and finally WAVE and profilin increased only with E+P. Similarly, MLCK and CRIPAK were decreased with E and E+P whereas LIMK was decreased with E+P only.
Figure 2.
Histograms illustrating changes in gene expression obtained with nested qRT-PCR on RNA extracted from laser captured serotonin neurons (n=3 animals/treatment). The Rho GTPases, RhoA, and Rac1 significantly increased with E alone (ANOVA p<0.0002 and 0.00115, respectively) whereas CDC42 increased with E and E+P (ANOVA p<0.05). Downstream effectors ROCK2 increased with E+P (ANOVA p<0.0047) whereas PIP5K and IRSp53 increased with E alone (ANOVA p<0.05 and #t-test p<0.0114, respectively).
The variance in IRSp53 in the E+P treated group was significantly different from the ovx group (+F test p<0.0028).
* p<0.05 different from ovx control with Student-Newman-Keuls posthoc pairwise comparison after ANOVA.
Figure 4.
Histograms illustrating changes in gene expression obtained with nested qRT-PCR on RNA extracted from laser captured serotonin neurons (n=3 animals/treatment). Downstream of Rac1, MLCK was significantly decreased by E and E+P treatment (ANOVA p<0.0166) whereas LIMK2 was significantly decreased by E+P treatment (#t-test p<0.05; F-test p<0.0001). The endogenous Pak1 inhibitor, CRIPAK, was significantly decreased by E and E+P treatment (ANOVA p<0.0001).
* p<0.05 different from ovx control with Student-Newman-Keuls posthoc pairwise comparison after ANOVA.
Figure 3.
Histograms illustrating changes in gene expression obtained with nested qRT-PCR on RNA extracted from laser captured serotonin neurons (n=3 animals/treatment). Intermediaries WAVE and WASP were significantly increased with E treatment, respectively (ANOVA p< 0.0129 and #t-test p<0.0001). The variance in WASP in the E+P treated group was significantly different from the ovx group (+F test p<0.0027). The actin regulatory proteins ARP3 and cofilin significantly increased with E and E+P treatment (ANOVA p<0.0329 and p<0.0093, respectively). However, gelsolin and profilin increases only with E+P (ANOVA p<0.0029 and p<0.0001, respectively).
* p<0.05 different from ovx control with Student-Newman-Keuls posthoc pairwise comparison after ANOVA.
Steroid Hormone Verification
The concentration of E and P in a serum sample obtained from each animal at necropsy was obtained to verify the efficacy of the Silastic implants. The concentration of E in the serum of the E and E+P-treated animals was 136.5±15 pg/ml and the concentration of P in the serum of the E+P-treated animals was 8.17±1.15 ng/ml. The concentrations of E and P in the serum of the untreated spayed control animals were 11.0 ± 0.1 pg/ml and 0.16±0.13 ng/ml, respectively (significantly different from treated animals by ANOVA, p <0.01).
Discussion
Dendritic spine extrusion and remodeling is a multi-step process that requires actin reorganization. Actin reorganization is a process of enucleation and branching. Filopodia precede spine formation and glutamate promotes filopodia extensions. When filopodia contact an axon, or glutamatergic buton, they may be transformed into spines. This involves decreased motility, shortening of the neck and growth of the head (2–4). The dynamic organization of the actin cytoskeleton provides the force for filopodia extrusion and spine morphogenesis. It is regulated by small GTPases of the Rho family, in particular RhoA, Rac1 and CDC42. These G-Proteins function as molecular switches in signal transduction pathways by cycling between an active GTP-bound and an inactive GDP-bound state. RhoA controls stress fiber formation and the attachment of contractile bundles of actin and myosin filaments to the cell membrane at points of focal adhesion, where integrin clusters are present. Rac1 and CDC42 signal transduction pathways lead to actin rearrangements in the form of filopodia (microspikes). Rac1 and RhoA bind to and specifically activate their downstream effectors, which are either kinases such as ROCK (Rho-Associated Coiled-Coil-Containing Protein Kinase), PAK (p21-Activated Kinase) and PI5K (Phosphatidylinositol-5-OH Kinase) or scaffolding proteins such as GDIA, WASP (Wiskott-Aldrich syndrome protein) and IRSp53. Constitutively active GDIA lacking Rho-binding domains cooperates with activated ROCK to form stress fibers. PAK activates LIM-kinases (LIMK1 and LIMK2) to phosphorylate cofilins. This allows signals flowing through Rho family GTPases to coordinate the initiation of new filaments through WASP and the ARP2/3 complex. GTP-bound RhoA activates ROCK (Rho-associated kinase), which phosphorylates the myosin-binding subunit of MLCK (Myosin Light Chain Kinase, also known as MYLK), inactivating it and thereby preventing MLC dephosphorylation. As a result, RhoA activation leads to an accumulation of the phosphorylated MLC and, subsequently, to the stimulation of actomyosin ATPase activity. CDC42 and IRSp53 activation of WAVE (WASP-family Verprolin-Homologous Protein), another member of the WASP family, also induces actin alterations in response to upstream signals. In addition, activation of Rac1 can activate PI5K, which stimulates biosynthesis of PIP2 (Phosphatidylinositol-4, 5-Bisphosphate) and leads to promotion of actin assembly from profilin and gelsolin (14–16).
We found that E or E+P increased gene expression on the microarray of RhoA, Rac1 and CDC42 and their downstream effectors Pak1, Rock, PIP5K, IrSp53, WASP and WAVE in serotonin neurons. In addition, E or E+P increased gene expression of the actin and myosin regulatory proteins MLC, cofilin, gelsolin, profilin and ARP2/3. MLCK, also known as MYLK, was confounding. Two probe sets increased and three probe sets decreased. We show two probe sets in Table 2, named MLCK and MYLK, but these seem to be the same or similar genes on the microarray. With qRT-PCR primers based upon the decreased probe sets there was a decrease in MLCK expression. Of interest, the decrease in gene expression of CRIPAK and MLCK could act to promote accumulation of phosphorylated MLC and its stimulation of actomyosin ATPase activity. The decrease in gene expression of LIMK2 is puzzling, but it suggests that the Rac/PIP5k/gelsolin:profilin and the CDC42/IRSp53/WASP/WAVE/ARP2/3 pathways may play a more prominent role in dendritic spine proliferation in serotonin neurons. Most of these gene expression changes were confirmed with nested qRT-PCR, which strongly indicates that the microarray reports are accurate. It was noted that some genes responded to E alone, some genes responded similarly to E and E+P and other genes responded to only E+P. These responses indicate that P may have different actions depending on the gene in question. It may reverse the action of E, it may have no effect yielding a similar response in E and E+P treated groups, or it may act solely after E priming. Each of these types of responses has been observed in other systems. However, in order to explain them, a great deal more information is needed such as the steroid receptor response elements in each promoter.
It is noteworthy that 31 other guanidine exchange factor (GEF) probe sets were changed 2-fold or greater with E or E+P treatment including Rap GEFs 2,3,6 and 7; Rho rac GEFs 2,7,9, and 17; GEF-T, Kaliren, spectrin and FYVE (CMT4H, RhoGEF and PH containing domain 4). In addition, 36 probe sets with GTPase activating activity (GAPs) were changed 2-fold or greater with E or E+P treatment including RAB GAP1, RAB GAP150, RAB3 GAP, and Rho GAPs 5, 15, 26.
These changes in gene expression indicate that E and/or E+P promote dendritic spine proliferation on the dendritic arbor of serotonin neurons. This action would play a central role in governing the excitatory input to the serotonin neurons. Dendritic spines express excitatory glutamate receptors, both ionotroic and metabotropic, and the maintenance of a spine requires continuous glutamate signals from afferent neurons (3, 4). Indeed, AMPA receptors can promote spine formation, transformation, stabilization and maintenance. Moreover, inhibition of synaptic transmission in hippocampal slices causes spine loss and atrophy (17, 18).
The ability of E to induce spine proliferation is well documented in the hippocampus (19, 20), hypothalamus (21), cerebellum (22) and cortex (23, 24) so the responses of the serotonin neurons to ovarian hormones may be a general phenomena. However, analysis of gene expression upstream of actin reorganization was lacking in any area. Nonetheless, it is now necessary to demonstrate that the changes observed in gene expression translate to protein expression and spine proliferation on serotonin neurons.
The ability of estrogen to increase dendritic spines in the hippocampus involves NMDA receptor activity (19) and CREB activation (25). Additional work has shown that estrogen and NMDA receptor activation increase dendritic spine number in the developing hypothalamus, which is dependent on an increase in presynaptic glutamate release. In turn, the estrogen-enhanced glutamate release occurs via a nongenomic mechanism and activation of PI3 kinase (21).
In light of these reports, it is important to speculate whether changes in genes related to dendritic spine proliferation in serotonin neurons are due to a direct action of E or P via nuclear steroid receptors or, are due to an indirect action via increased glutamate release, or both. We have shown that serotonin neurons express ERβ and PR (7, 8, 26), and these receptors are gene transcription factors. Thus, it is reasonable to think that the regulation of gene expression is due to the activity of these transcription factors perhaps causing an increase in filopodia. Nonetheless, an increase in extracellular glutamate may also play a pivotal role and contribute to the transformation of filopodia to mature spines. One would predict glutamate to increase with E or E+P, but as noted above, the gene expression patterns varied. For example, E treatment likely increased glutamate, but some genes increased and others did not. Likewise, glutamate may be increased with supplemental P treatment, however some genes increased, others were equivalent with E treatment, and others were suppressed. It will be hard to test this notion in whole monkeys, or even monkey raphe slices, but with our development of embryonic stem cell-derived serotonin neurons in culture (27), we may begin to unravel the mechanisms involved.
Summary
Altogether, the data indicate that administration of ovarian steroids to ovariectomized monkeys for 1 month alters gene expression in serotonin neurons in a manner that would promote dendritic spine protrusion and an increase in excitatory synapses. This action would enhance serotonin neurotransmission and in turn, improve mood and decrease anxiety.
Acknowledgements
We are deeply grateful to the dedicated staff of the Division of Animal Resouces including the staff of the Departments of Surgery and Pathology for their expertise and helpfulness in all aspects of monkey management. The staff of the OHSU Gene Microarray Shared Resource was essential for this study.
Supported by NIH grants: MH62677 to CLB, U54 contraceptive Center Grant HD 18185, and RR000163 for the operation of ONPRC
Footnotes
This data was presented at the Annual Meeting of the American College of Neuropsychopharmacology, December 8–11, 2009.
Literature Cited
- 1.Manji HK, Quiroz JA, Sporn J, Payne JL, Denicoff K, et al. Enhancing neuronal plasticity and cellular resilience to develop novel, improved therapeutics for difficult-to-treat depression. Biol Psychiatry. 2003 Apr 15;53(8):707–742. doi: 10.1016/s0006-3223(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 2.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005 Feb;75(3):161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Butler AK, Uryu K, Chesselet MF. A role for N-methyl-D-aspartate receptors in the regulation of synaptogenesis and expression of the polysialylated form of the neural cell adhesion molecule in the developing striatum. Dev Neurosci. 1998;20(2–3):253–262. doi: 10.1159/000017319. [DOI] [PubMed] [Google Scholar]
- 4.McKinney RA, Capogna M, Durr R, Gahwiler BH, Thompson SM. Miniature synaptic events maintain dendritic spines via AMPA receptor activation. Nat Neurosci. 1999 Jan;2(1):44–49. doi: 10.1038/4548. [DOI] [PubMed] [Google Scholar]
- 5.Cooke BM, Woolley CS. Gonadal hormone modulation of dendrites in the mammalian CNS. J Neurobiol. 2005 Jul;64(1):34–46. doi: 10.1002/neu.20143. [DOI] [PubMed] [Google Scholar]
- 6.Murphy DD, Cole NB, Segal M. Brain-derived neurotrophic factor mediates estradiol-induced dendritic spine formation in hippocampal neurons. Proc Natl Acad Sci U S A. 1998 Sep 15;95(19):11412–11417. doi: 10.1073/pnas.95.19.11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bethea CL. Colocalization of progestin receptors with serotonin in raphe neurons of macaque. Neuroendocrinology. 1993;57:1–6. doi: 10.1159/000126334. [DOI] [PubMed] [Google Scholar]
- 8.Gundlah C, Lu NZ, Mirkes SJ, Bethea CL. Estrogen receptor beta (ERb) mRNA and protein in serotonin neurons of macaques. Molecular Brain Research. 2001;91:14–22. doi: 10.1016/s0169-328x(01)00108-5. [DOI] [PubMed] [Google Scholar]
- 9.Bethea CL, Lu NZ, Gundlah C, Streicher JM. Diverse actions of ovarian steroids in the serotonin neural system. Frontiers in Neuroendocrinology. 2002;23:41–100. doi: 10.1006/frne.2001.0225. [DOI] [PubMed] [Google Scholar]
- 10.Lu NZ, Bethea CL. Ovarian steroid regulation of 5HT1A receptor binding and G protein activation in female monkeys. Neuropsychopharmacology. 2002;27:12–24. doi: 10.1016/S0893-133X(01)00423-7. [DOI] [PubMed] [Google Scholar]
- 11.Lu NZ, Eshleman AJ, Janowsky A, Bethea CL. Ovarian steroid regulation of serotonin reuptake transporter (SERT) binding, distribution and function in female macaques. Molecular Psychiatry. 2003;8:353–360. doi: 10.1038/sj.mp.4001243. [DOI] [PubMed] [Google Scholar]
- 12.Bethea C, Reddy A. Effect of ovarian hormones on survival genes in laser captured serotonin neurons from macaques. J Neurochem. 2008;105:1129–1143. doi: 10.1111/j.1471-4159.2008.05213.x. [DOI] [PubMed] [Google Scholar]
- 13.Tokuyama Y, Reddy A, Bethea C. Neuroprotective actions of ovarian hormones without insult in the raphe region of rhesus macaques. Neuroscience. 2008;154:720–731. doi: 10.1016/j.neuroscience.2008.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci. 2001 Nov;114(Pt 21):3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- 15.Wherlock M, Mellor H. The Rho GTPase family: a Racs to Wrchs story. J Cell Sci. 2002 Jan 15;115(Pt 2):239–240. doi: 10.1242/jcs.115.2.239. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson-Dykstra S, Higgs HN, Harris ES. Actin dynamics: growth from dendritic branches. Curr Biol. 2005 May 10;15(9):R346–R357. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Matus A, Brinkhaus H, Wagner U. Actin dynamics in dendritic spines: a form of regulated plasticity at excitatory synapses. Hippocampus. 2000;10(5):555–560. doi: 10.1002/1098-1063(2000)10:5<555::AID-HIPO5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 18.Fischer M, Kaech S, Wagner U, Brinkhaus H, Matus A. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat Neurosci. 2000 Sep;3(9):887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- 19.Woolley CS, Weiland NG, McEwen BS, Schwartzkroin PA. Estradiol increases the sensitivity of hippocampal CA1 pyramidal cells to NMDA receptor-mediated synaptic input: correlation with dendritic spine density. J Neurosci. 1997 Mar 1;17(5):1848–1859. doi: 10.1523/JNEUROSCI.17-05-01848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao J, Janssen WG, Tang Y, Roberts JA, McKay H, Lasley B, et al. Estrogen increases the number of spinophilin-immunoreactive spines in the hippocampus of young and aged female rhesus monkeys. J Comp Neurol. 2003 Oct 27;465(4):540–550. doi: 10.1002/cne.10837. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz JM, Liang SL, Thompson SM, McCarthy MM. Estradiol induces hypothalamic dendritic spines by enhancing glutamate release: a mechanism for organizational sex differences. Neuron. 2008 May 22;58(4):584–598. doi: 10.1016/j.neuron.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasahara K, Shikimi H, Haraguchi S, Sakamoto H, Honda S, Harada N, et al. Mode of action and functional significance of estrogen-inducing dendritic growth, spinogenesis, and synaptogenesis in the developing Purkinje cell. J Neurosci. 2007 Jul 11;27(28):7408–7417. doi: 10.1523/JNEUROSCI.0710-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao J, Rapp PR, Leffler AE, Leffler SR, Janssen WG, Lou W, et al. Estrogen alters spine number and morphology in prefrontal cortex of aged female rhesus monkeys. J Neurosci. 2006 Mar 1;26(9):2571–2578. doi: 10.1523/JNEUROSCI.3440-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava DP, Woolfrey KM, Jones KA, Shum CY, Lash LL, Swanson GT, et al. Rapid enhancement of two-step wiring plasticity by estrogen and NMDA receptor activity. Proc Natl Acad Sci U S A. 2008 Sep 23;105(38):14650–14655. doi: 10.1073/pnas.0801581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy DD, Segal M. Morphological plasticity of dendritic spines in central neurons is mediated by activation of cAMP response element binding protein. Proc Natl Acad Sci U S A. 1997 Feb 18;94(4):1482–1487. doi: 10.1073/pnas.94.4.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bethea CL. Regulation of progestin receptors in raphe neurons of steroid-treated monkeys. Neuroendocrinology. 1994;60:50–61. doi: 10.1159/000126719. [DOI] [PubMed] [Google Scholar]
- 27.Salli U, Reddy AP, Salli N, Lu NZ, Kuo H-C, Pau FK-Y, et al. Serotonin neurons derived from rhesus monkey embryonic stem cells: similarities to CNS serotonin neurons. Experimental Neurology. 2004;188:351–364. doi: 10.1016/j.expneurol.2004.04.015. [DOI] [PubMed] [Google Scholar]