Abstract
Pancreatic cancer is a genetic disease in which somatic mutations in the KRAS proto-oncogene are detected in a majority of tumors. KRAS mutations represent an early event during pancreatic tumorigenesis, crucial for cancer initiation and progression. Recent studies, including comprehensive sequencing of the pancreatic cancer exome, have implicated the involvement of a number of additional core signaling pathways during pancreatic tumorigenesis. Improving our understanding of genetic interactions between KRAS and these additional pathways represents a critical challenge, as these interactions may provide novel opportunities for diagnosis and treatment. However, studying these interactions requires the expression of multiple transgenes in relevant cell types, an effort that has proven very difficult to achieve using gene targeted mice and is also technically challenging in zebrafish. Based on the ability of the Gal4 transcriptional activator to drive the expression of multiple transgenes under regulation of UAS (upstream activator sequence) regulatory elements, the Gal4/UAS system represents an attractive strategy for the study of genetic interactions. In this chapter, we review our experience using the Gal4/UAS system to model KRAS-initiated pancreatic cancer in zebrafish, as well as our early efforts using this system to study the influence of other cooperating oncogenes. We also describe techniques used to identify and characterize pancreatic tumors in adult transgenic fish.
I. Introduction
Pancreatic cancer is a deadly disease, with a predicted 43,140 new cases and 36,800 deaths occurring in US in 2010 (Jemal et al., 2010). The 5-year survival rate for this type of cancer remains around 5% (Altekruse et al., 2010). The poor outcome of the disease is largely due to the lack of effective early-stage diagnostic tools as well as intrinsic resistance to current therapeutic modalities.
The vast majority of pancreatic cancers are categorized as pancreatic ductal adenocarcinomas (PDACs). Over 90% of PDACs carry point mutations in the KRAS gene, resulting in constitutive activation of the KRAS protein and enhanced activity of downstream signaling cascades, including MAPK, PI3K/AKT, and PLCε/PKC pathways. Abnormal KRAS activation represents an early event during pancreatic tumorigenesis, as KRAS mutations have been detected in pancreatic intraepithelial neoplasia (PanIN), a common PDAC precursor. Evidence from mouse models indicates that mutant KRAS plays a critical role in the initiation of PDAC. (Reviewed in (Karreth and Tuveson, 2009)).
In addition to KRAS mutations, recent advances in genome sequencing have documented a complex picture of genetic interactions in the initiation and progression of pancreatic cancer (Jones et al., 2008). This analysis has demonstrated frequent genetic alterations in a wide variety of core signaling pathways, including TGFβ, Wnt, Notch, and Hedgehog. While these core pathways were altered in the majority of analyzed tumors, the specific pathway components found to be mutated in any individual tumor varied widely, and the functional significance of the many identified mutations remains unclear. Further understanding of these changes at a functional level, especially related to genetic interactions between mutant KRAS and other observed mutations, will doubtlessly be important in the identification of new genetic targets for both early diagnosis and more effective treatment.
The zebrafish pancreas shares significant anatomical similarity with its mammalian counterparts. More importantly, the underlying regulatory molecular mechanisms for pancreas development are well-conserved between zebrafish and mammals (Reviewed in (Kinkel and Prince, 2009; Tiso et al., 2009)). To study KRAS-initiated pancreatic tumorigenesis, our lab generated a transgenic zebrafish model that specifically expresses the human KRASG12V mutant fused with an N-terminal eGFP tag in the exocrine pancreas (Park et al., 2008). The transgene was placed under control of zebrafish ptf1a regulatory elements through BAC recombineering. Ptf1a (pancreas specific transcription factor 1 a) is a basic-helix-basic transcription factor that plays a crucial role in pancreas development both in mammals and zebrafish (Kawaguchi et al., 2002; Lin et al., 2004; Zecchin et al., 2004). In the mouse model, constitutive activation of a mutant human KRAS protein in Ptf1a-expressing cells led to the development of PanIN and eventual PDAC, fully recapitulating the progression sequence observed in humans (Hingorani et al., 2003). ptf1a:eGFP-KRASG12V transgenic fish developed pancreatic tumors that displayed some histo-logical resemblance to human pancreatic cancer, albeit with a strong bias towards acinar cell carcinomas as opposed to classical PDAC (Park et al., 2008). Further analysis of the tumors also showed abnormal activation of Hedgehog signaling pathway, a trait commonly observed in human tumors.
These results indicated that the zebrafish indeed represents a potentially useful organism for studying the biology of KRAS-initiated pancreatic cancer. Furthermore, based on the relative low cost associated with zebrafish transgenesis, the fish may represent the most attractive platform for functionally annotating the emerging pancreatic cancer genome, especially the evaluation of novel dominant candidate oncogenes. However, the generation of multiple BAC transgenic lines in which candidate oncogenes are expressed under the control of ptf1a regulatory elements represents a daunting task, which led us to develop a novel Gal4/UAS approach that allows the simultaneous expression of multiple candidate oncogenes in ptf1a-expressing pancreatic cell types.
A. Rationale
In this chapter, we describe the utility of a dyad Gal4/UAS system for establishing either stable or transient transgenic models that consistently produce tumors in adult fish. The Gal4/UAS system has been successfully adapted to zebrafish, thus providing precise temporal and spatial control of transgene expression (Reviewed in (Halpern et al., 2008)). In the absence of a Gal4 driver gene, transgenes placed downstream of UAS (upstream activator sequence) elements are typically not expressed, thus avoiding the possible toxic effect of oncogene overexpression that can hinder generation of stable transgenic models.
The Gal4/UAS system allows quick and effective generation of transgenic models that can be used to interrogate interactions among multiple genes of interest in a tissue-specific manner. Multiple transgenes can be easily cloned downstream of UAS elements in Tol2-based vectors via conventional cloning methods and then co-injected into an established Gal4 driver line to generate simultaneous transient and/or stable expression of the transgenes within the same tissue. Utilization of Tol2 transposon-mediated transgenesis thereby provides the simultaneous advantages of both high frequency somatic expression as well as high germline transmission efficiency, especially when compared to the lower efficiency associated with BAC transgenesis.
We also describe techniques used to identify and characterize pancreatic tumors in adult transgenic fish. To study the molecular mechanism underlying pancreatic tumor-igenesis, it is essential to be able to detect and document distinct histologic features as well as associated molecular changes during cancer initiation and progression.
II. Transgenic Zebrafish with Gal4/UAS-Mediated eGFP-KRASG12V Expression in the Exocrine Pancreas
A. Generation of UAS Regulated eGFP-KRASG12V Transgene
The fusion eGFP-KRASG12V transgene was cloned downstream of a 14xUAS element in a Tol2-based vector backbone (Davison et al., 2007). A single amino acid change at codon 12 of human KRAS protein, from glycine to valine, leads to membrane localization and constitutive activation of the mutant protein. To avoid the insertion site position effects observed commonly among UAS transgenes, multiple founders need to be identified and examined for phenotypes (Urasaki and Kawakami, 2009).
The UAS construct was co-injected with Tol2 transposase into wildtype AB zebrafish embryos at the one- to two-cell stage. To identify fish carrying the UAS transgene, we routinely genotype fish when they reach reproductive maturation. The Tol2-mediated microinjection protocol and purification of genomic DNA for genotyping PCR have been described elsewhere (Fisher et al., 2006; Westfield, 2007).
Mosaicism and silencing of transgene expression are two major issues commonly encountered when using the Gal4/UAS system (Davison et al., 2007; Halpern et al., 2008). In the case of functional analysis of candidate oncogenes, mosaic transgene expression may be advantageous, because the cells not expressing the oncogene can be used as neighboring controls for comparative analysis against oncogene-expressing cells. However, the silencing issue, together with oncogene toxicity, does present an obstacle to establishing stable transgenic models that provide consistent tumor formation across multiple generations. In our experience, maintaining UAS transgenic fish in the absence of a Gal4-VP16 driver seems to minimize these issues.
B. Gal4/UAS-Mediated Expression of eGFP-KRASG12V Transgene in the Exocrine Pancreas
The UAS:eGFP-KRASG12V transgenic fish are crossed to the ptf1a:Gal4-VP16 transgenic fish, to direct eGFP-KRASG12V expression in the exocrine pancreas (Fig. 1) (Halpern et al., 2008; Pisharath and Parsons, 2009). In zebrafish, the earliest ptf1a-driven expression is observed in the hindbrain at 22 hpf (Lin et al., 2004). The expression in the exocrine pancreas begins around 34 hpf, and continues through adulthood. The embryos are cultured in embryo medium (Westfield, 2007) containing 0.003% phenylthiourea, and screened for pancreatic expression based on eGFP fluorescence (Fig. 1B). The eGFP-KRASG12V protein displays a membrane-bound subcellular localization, as expected based upon normal activated KRAS localization (Fig. 1C).
Fig. 1. Targeted expression of eGFP-KRASG12Vtransgene in zebrafish pancreas.
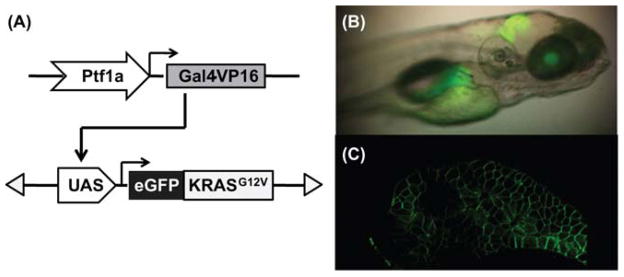
(A) Schematic drawing of the dyad Gal4/UAS system used to drive eGFP-KRASG12V transgene expression in the Ptf1a domain. (B) Lateral view under transmitted and fluorescent illumination of a larval fish at 5 days post fertilization (dpf), showing expression pattern of eGFP-KRASG12V transgene in the Ptf1a domain, including the retina, the hindbrain, and the exocrine pancreas. (C) Two-photon confocal image of the pancreas from a live 5 dpf larval fish, revealing the membrane localization of eGFP-KRASG12V protein. (For color version of this figure, the reader is referred to the web version of this book.)
In our previous model, where the eGFP-KRASG12V transgene was placed directly under ptf1a regulatory elements through BAC recombineering, exocrine pancreatic progenitor cells expressing eGFP-KRASG12V failed to undergo primary exocrine differentiation, leading to an accumulation of undifferentiated progenitor cells expressing oncogenic KRAS (Park et al., 2008). This embryonic phenotype was associated with eventual loss of eGFP fluorescence as early as 96 hpf. The Gal4/UAS-mediated model does not display this type of embryonic phenotype – transgene expression of eGFP-kRASG12V was observed uninterrupted from larval stages through adulthood. We suspect that the difference observed between the two models may be explained by the levels and timing of Gal4 onset, as delayed Gal4 onset in early developmental stages has been observed by others (Zhan and Gong, 2010).
III. Identification and Characterization of Pancreatic Tumors
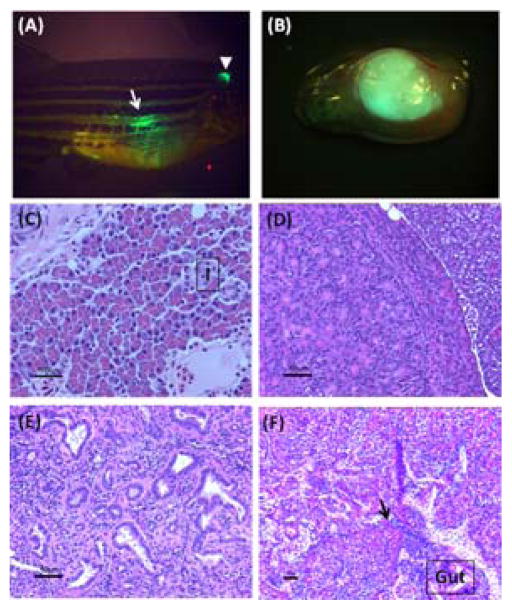
Tagging the oncogenic KRAS mutant with eGFP allows tracking of pancreatic tumor formation by monitoring visceral expression of GFP fluorescence in transgenic fish (Fig. 2A.B). Tumors developing in transgenic fish can be easily distinguished from normal adult pancreas histologically, based upon their size, distribution, and disrupted pattern of epithelial organization. Typically, 50% of ptf1a:Gal4-VP16; UAS:eGFP-KRASG12V fish are found to develop pancreatic tumors by 5 months of age, with detection of tumors as early as 2 months. These tumors display features of acinar cell and/or ductal cell differentiation and invasion into neighboring organs, all demonstrating significant similarity to the human disease (Fig. 2C–F) (Hruban et al., 2007). Transient expression of eGFP-KRASG12V, through injection of the Tol2-based UAS:eGFP-KRASG12V construct into ptf1a: Gal4-VP16 embryos, displayed a mosaic transgene expression pattern in ptf1a domains and did not result in tumor formation in adult fish. Functional analysis of potential oncogenes can thus be carried out in both stable and transient models to address their role in both cancer initiation and/or progression. The stable model of ptf1a:Gal4-VP16; UAS:eGFP-KRASG12V may be more suitable for the analysis of oncogene interactions during cancer progression, whereas the transient approach may be more informative for studies of tumor initiation.
Fig. 2.
Identification and histological evaluation of KRAS-initiated pancreatic tumors in transgenic zebrafish.
(A) Lateral view under transmitted and fluorescent illumination of an adult transgenic fish carrying a tumor in the pancreas. Expression of eGFP-KRASG12V transgene remains strong in the cerebellum (white arrowhead) of the adult fish. Formation of a tumor is indicated by the presence of intense eGFP fluorescence (white arrow) detected transcutaneously. (B) Lateral view under transmitted and fluorescent illumination, of an entire viscera with an eGFP-positive tumor. (C) Normal zebrafish adult pancreas, showing well-organized acinar cells in the exocrine pancreas (I: islet). (D) A tumor with acinar cell differentiation. The periphery of acinar cell carcinoma is generally circumscribed with minimal stroma within the tumor. Acinar cell differentiation of the tumor can be further confirmed by the presence of pancreatic exocrine enzymes, such as trypsin and elastase. (E) Part of a tumor showing ductal differentiation. The haphazard growth pattern of the ducts associated with stromal desmoplastia is a prominent histological feature of tumors with ductal differentiation. (F) A malignant tumor invading neighboring intestinal tube (gut). Infiltrating tumor cells (black arrow) disrupt normal borders between pancreas and adjacent organs. (See color plate.)
In both BAC and Gal4/UAS transgenic models, expression of the eGFP-KRASG12V transgene in the hindbrain is observed during embryonic stages and later restricted to the cerebellum in adult fish (Fig. 1B, 2A). We have not identified any tumor developing in the hindbrain/cerebellum region of transgenic fish expressing eGFP-KRASG12V. However, when overexpression of mutant KRAS is combined with transient expression of an additional oncogene – for example, a constitutively active mutant form of human YAP1 protein – rapid formation of tumors was observed in the hindbrain/cerebellum region by 1 month of age. Many oncogenes are involved in tumorigenesis only in selected tissues, thus the hindbrain/cerebellum region presents an additional organ site for evaluating a target gene’s oncogenic function.
To best preserve anatomical details of tumors and surrounding organs, we embed tissues in paraffin and then evaluate tumor histology after hematoxylin and eosin staining. Immunohistochemical and immunofluorescent staining are performed on paraffin-embedded and/or frozen sections to further confirm eGFP-KRASG12V trans-gene expression and to determine the expression patterns of other genes that may be involved in pancreatic tumorigenesis. Together, these techniques allow the initial detection and characterization of oncogene-initiated pancreatic cancer in zebrafish.
A. Identifying Pancreatic Tumors Expressing eGFP-KRASG12V in Living Fish
In adult zebrafish, the anatomical structure of the pancreas evolves from the distinct head–neck–tail morphology observed during larval and early juvenile stages to organization of four linked lobes surrounding the intestinal tube (Chen et al., 2007). Prior to tumor formation, diffuse pancreatic eGFP expression in ptf1a:Gal4-VP16; UAS:eGFP-KRASG12V transgenic fish is indistinguishable from autofluorescence from the intestinal tube when the whole fish is visualized under a fluorescent dissecting microscope. In contrast, when tumors arise in the pancreas, the intensity of the eGFP expression from most of the tumor mass is usually strong enough to be detected transcutaneously, except in the few cases where very small tumor masses are nested in between folds of intestinal tube.
To check for pancreatic tumor formation in ptf1a:Gal4-VP16; UAS:eGFP-KRASG12V transgenic fish, anesthetize fish in a 250 ml beaker containing 100 ml of system water and 4–5 ml of 400 mg/ml tricaine stock solution prepared with dH2O. Lay fish sideways on a petri dish lid. Confirm transgene expression by viewing eGFP fluorescence in the cerebellum with a fluorescence dissecting microscope (Fig. 2A). Then look for discernible eGFP expression in the abdominal area (Fig. 2A). After examination, return fish promptly to fresh system water for recovery.
B. Dissection and Fixation of Transgenic Fish Bearing KRAS-Initiated Pancreatic Tumors
1. Dissection of Visceral Organs
A major concern when obtaining pancreatic tumor samples from transgenic fish is the prevention of autolysis, because pancreatic tissue carries high levels of both protease and ribonuclease activity. To minimize degradation of tissues during the process, we recommend using a rapid cooling method for euthanasia (Wilson et al.,2009). Dissection should be carried out as soon as fish are euthanized. Normal adult pancreas is heavily infiltrated with fatty tissues, giving an opaque “slimy” appearance when visualized under brightfield illumination. Tumors are densely packed and present themselves as solid white masses under brightfield with detectable eGFP fluorescence (Fig. 2B).
Place euthanized fish on a petri dish lid, with right side of the fish facing up. Use microdissection scissors to make an incision near the cloaca, and gently open up the right abdominal wall from posterior to anterior, to expose internal organs.
Under a dissecting microscope, use sharp tipped forceps to separate the skin from the viscera. Once separated, cut the skin with scissors to remove the right abdominal wall in its entirety.
Confirm tumor location via bright field and/or fluorescence microscopy.
Remove the two swim bladders with forceps. Make a final cut right below the heart to dissect out the entire viscera. Segments of tumor can be removed with scissors at this stage if further RNA/protein purification is desired. Rinse the remaining tissue briefly in PBS. Blot dry tissue and proceed to fixation.
2. Fixation
Fixation solutions should be at least 10 times the volume of tissue.
Fixation for paraffin-embedded sections: Incubate dissected tissue in 10% buffered formalin solution at room temperature overnight. If sample will be used solely for paraffin-embedded sections, whole fish can be fixed prior to dissection. The fixation hardens tissue and facilitates organ separation during dissection. For whole-fish fixation, first inject 100–200 μl of 10% buffered formalin intraperitonially into euthanized fish with a 1-ml syringe fitted with a 26-gauge needle. This step helps to minimize autolysis of the pancreas during fixation process. The whole fish can then be incubated in 10% buffered formalin at room temperature overnight.
Fixation for frozen sections: Incubate dissected tissue in 4% paraformaldehyde (PFA) prepared in PBS overnight at 4 °C. Our experience suggests that frozen sections are not suitable with whole fish.
C. Preparing Paraffin-Embedded Sections of Tumors
Paraffin-embedded sections best preserve the overall structure of tumors and their surrounding organs and therefore are optimal for evaluating tumor histology. However, paraffin-embedded sections are not compatible for labeling with certain antibodies and are especially problematic when observed by immunofluorescence microscopy.
1. Decalcification (optional)
Decalcification is recommended if an entire adult fish is embedded in paraffin.
Incubate fish in 0.5 M EDTA, pH 7.8, for 7 days at room temperature.
Wash three times with dH2O for 20 min each.
2. Preliminary Embedding in Agar (Optional)
Embedding tissues in agar prior to paraffin embedding is very beneficial when working with small and friable samples (Lund et al., 1961). It minimizes the loss of tissue during vigorous processing procedures, allows placement of multiple small tissues within one block, and provides orientation for sectioning.
Prepare agar solution per manufacturer’s instruction. Sterilize agar solution and let cool to −50 °C.
Blot dry tissues briefly. Arrange tissues on a glass slide, with the region of interest in contact with the slide surface.
Slowly drip agar solution on top of tissues. Let solidify. If the tissue does not stay flat on its own, use forceps to hold the tissue in place during agar solidification.
Use a razor blade to trim agar block to desired size for paraffin embedding.
3. Paraffin Embedding and Sectioning
Place tissue in an embedding cassette (FisherScientific, 22-272-417). Use working solution at least 10 times the volume of tissue embedded for each step. When processing small sized tissue samples, the length of incubation time in the initial dehydration steps may be shortened to avoid over-drying of samples and resulting tissue tearing during sectioning.
Incubate tissue in two washes of 70% ethanol for 15 min each at room temperature with gentle agitation.
Incubate tissue in 85% ethanol for 30 min at room temperature with gentle agitation.
Incubate tissue in 95% ethanol for 30 min at room temperature with gentle agitation.
Incubate tissue in two washes of 100% ethanol for 30 min each at room temperature with gentle agitation.
Incubate tissue in Histoclear (National Diagnostics: HS-200) for 15–30 min at room temperature with gentle agitation.
Incubate tissue in Histoclear for 15–30 min at 60 °C.
Incubate tissue in Histoclear/paraffin (V/V = 1:1) for 30 min at 60 °C.
Incubate tissue in two washes of paraffin for 30–60 min each at 60 °C.
Incubate tissue in paraffin for 30–60 min at 60 °C. Alternatively, tissue can be left in paraffin overnight at 60 °C.
Fill the bottom of a stainless steel base mold (Sakura Finetek, 4163) with paraffin at 60 °C. If tissues have been embedded in agar, place the agar block in the middle of the mold, then proceed to STEP k. If tissue has not been preliminarily embedded in agar, place tissue in the middle of mold, with region of interest facing down, then place the mold on cold plate briefly to immobilize tissue in the mold while leaving most of the paraffin in liquid form.
Fill the mold with paraffin at 60 °C. Leave mold on cold plate until paraffin solidifies.
Process paraffin blocks on Leica RM2135 microtome or equivalent. Sections are cut at 5 μm thickness and then placed on superfrost plus-charged microscope slides (FisherScientific, 12-550–15). Microtome sectioning techniques will not be discussed here.
Heat slides at 60 °C for 20 min, or at 42 °C overnight, to ensure complete dehydration and flattening of the paraffin. The slides can then be stored at room temperature away from light.
D. Preparing Frozen Sections of Dissected Tumors
Although some histologic detail is lost with frozen sections, this is typically the embedding method of choice for immunofluorescent labeling due to better epitope preservation. In addition, native eGFP fluorescence expressed from the transgene is preserved when tissue is fixed in 4% PFA and embedded in O.C.T. compound. This allows rapid assessment of transgene expression in frozen sections.
1. Sucrose Treatment
Rinse PFA fixed tissue twice with PBS. Blot dry tissue.
Incubate tissue in 30% sucrose prepared in PBS at 4 °C overnight.
2. O.C.T Compound Treatment
Place a plastic cryomold (Sakura Finetek, 4557) on dry ice. Cover bottom of the cryomold with a layer of O.C.T. Compound (Sakura Finetek, 4583). Let the compound freeze.
Rinse tissue briefly in PBS. Blot dry the tissue, place on top of the layer of O.C.T. compound, and orient the tissue for sectioning.
Add O.C.T. compound to cover tissue in the cryomold. Allow compound to freeze at −80 °C (dry ice) or by placing in liquid nitrogen. Store frozen block at −80 °C.
3. Sectioning
Frozen blocks are sectioned using a Leica CM1850 cryostat or equivalent. Sections are cut at 5–10 μm thickness and then placed on superfrost plus charged microscope slides. Air dry slides briefly and store slides at −80 °C.
4. Rapid Evaluation of eGFP-KRASG12V Expression
Wash slides three times in PBS for 5 min each.
Remove excess PBS from slides. Add Vectashield mounting media (Vector Laboratories, H-1200) to slides for fluorescence microscopy. Gently place coverslip on slides to avoid air bubbles. Slides can be sealed with nail polish around coverslip edges.
Check slides for eGFP expression under an inverted fluorescence microscope.
E. Hematoxylin and Eosin (H&E) Staining
H&E staining provides optimal histologic detail for characterizing tumor differentiation, invasion, and/or metastatic behavior. Staining of paraffin-embedded sections requires deparaffinization/rehydration. For frozen sections, thaw slides to room temperature and start from STEP 2.
1. Deparaffinization/Rehydration
Deparaffinize slides in two washes of Histoclear for 5 min each.
Hydrate slides by two washes of 100% ethanol for 2 min each, then two washes of 95% ethanol for 2 min each.
Wash slides twice in dH2O for 5 min each.
2. H&E staining
The intensity of H&E staining can be modified by adjusting the incubation time in hematoxylin and/or eosin solution.
Incubate slides in Harris hematoxylin staining solution (Sigma–Aldrich, HHS16) for 30 s to 2 min.
Rinse in running tap water for 5 min.
Incubate slides in 95% ethanol for 1 min.
Incubate in eosin Y solution (Sigma–Aldrich, HT1101128), prepared per manufacturer’s instruction, for 15 s to 2 min.
Dehydrate slides in 95% ethanol for 2 min then wash twice with 100% ethanol (2 min each).
Air dry slides briefly. g. Add Histomount (Invitrogen, 00-8030) to cover sections. Gently place coverslip on slides to avoid air bubbles.
F. Immunohistochemical (IHC) Staining for Paraffin-Embedded Sections
We usually test new antibodies using the following protocol as starting point. All steps are performed at room temperature unless specified.
Deparaffinize and Rehydrate Sections as Described in Section II.E.
Antigen Unmasking (Optional)
Our lab routinely uses the sodium citrate buffer method for antigen unmasking. For most antibodies we tested, the addition of this step prior to staining leads to more intense signals. However, there are cases where antigen unmasking does not help or actually weakens staining signals.
Place the slides in a slide rack and immerse them in a plastic staining dish with 250 ml of antigen unmasking solution (Vector Laboratories, H-3300). Bring solution to boiling in water bath or microwave. Maintain temperature at 95–99 °C for 10 min
Place the plastic staining dish with solution and slides on ice. Allow slides to cool for 30 min.
Wash slides three times in dH2O for 5 min each.
1. IHC Staining Using ABC Avidin/Biotin Method
Quench endogenous peroxidase activity by incubating slides in 3% hydrogen peroxide diluted in dH2O for 10 min.
Wash slides twice in dH2O for 5 min each.
Incubate slides in blocking buffer (PBS-0.1%Tween-20 (PBST) containing 10% normal serum from the same species as the secondary antibody) for 60 min.
Prepare primary antibody dilution in blocking buffer. Remove blocking buffer from slides. Add antibody solution to slides, and incubate overnight at 4 °C.
Wash slides three times in PBST for 5 min each.
Incubate slides in biotinylated secondary antibody diluted in blocking buffer for 1 h.
Wash slides three times with PBST for 5 min each.
During STEPs f and g, prepare ABC reagent (Vector Laboratories, PK-6100) per manufacturer’s instructions and incubate solution for 30 min.
Add ACB reagent to cover sections and incubate for 30 min.
Wash slides three times with PBST for 5 min each.
Add DAB substrate (Vector Laboratories, SK-4100) to each section. Monitor the sections develop staining. When the staining reaches desired intensity, immerse slides immediately into dH2O.
Follow Section II.E.2a to counterstain slides in hematoxylin (Optional).
Wash slides twice with dH2O for 5 min each.
Dehydrate slides.
Incubate slides in 95% ethanol for 2 min, then in two washes of 100% ethanol for 2 min each, and finally in two washes of Histoclear for 2 min each.
Add Histomount to slides. Gently place coverslip on slides to avoid air bubbles. Slides should be stored away from light at room temperature.
G. Immunofluorescent (IF) Staining
IF staining can be performed on paraffin-embedded or frozen sections depending on the characteristics of the antibody tested. IF staining allows detection of multiple antigens on the same slide. However, the utility of this method is limited by the availability and quality of antibodies.
Paraffin-embedded sections require deparaffinization/rehydration and antigen unmasking (optional) described in Sections II.E and II.F prior to the IF staining procedure. All steps are performed at room temperature unless specified. Place slides in a humidified chamber during the procedure to prevent drying.
1. Blocking
Rinse slides in PBS for 5 min.
Incubate slides in blocking buffer (PBST containing 5% normal serum from the same species as the secondary antibody) for 60 min.
2. Incubation in Primary Antibody
Prepare primary antibody dilution in blocking buffer.
Remove blocking buffer. Add primary antibody solution to slides, and incubate overnight at 4 °C.
Wash slides three times in PBST for 5 min each.
3. Detection
Prepare fluorochrome-conjugated secondary antibody dilution in PBST.
Incubate slides in secondary antibody solution for 1–2 h in dark.
Wash slides three times in PBST for 5 min each.
Add Vectashield mounting media to slides. Gently place coverslip on slides to avoid air bubbles. Slides can be sealed with nail polish around coverslip edges. IF stained slides should be stored flat at 4 °C in dark.
IV. Conclusions
The Gal4/UASA-mediated zebrafish pancreatic cancer model is a useful tool for examining the biology of KRAS-initiated pancreatic neoplasia, especially for delineating crosstalk between KRAS and other signaling pathways involved in the initiation and progression of pancreatic tumor formation. The techniques reviewed in this chapter are suitable for generating and characterizing any trans-genic zebrafish cancer model involving tissue-specific single or multiple candidate oncogenes.
Acknowledgments
We thank Michael J. Parsons for providing ptf1a:Gal4-VP16 transgenic zebrafish for the study. This work is supported by NIH Grant P01 CA134292 (to SDL).
References
- Altekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK, editors. SEER Cancer Statistics Review, 1975–2007. National Cancer Institute; Bethesda, MD: 2010. http://seer.cancer.gov/csr/1975_2007/, based on November 2009 SEER data submission, posted to the SEER web site. [Google Scholar]
- Chen S, Li C, Yuan G, Xie F. Anatomical and histological observation on the pancreas in adult zebrafish. Pancreas. 2007;34:120–125. doi: 10.1097/01.mpa.0000246661.23128.8c. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Pitman MB, Klimstra DS. AFIPAtlas of Tumor Pathology. Tumors of the pancreas. American Registry of Pathology Press; Washington, DC: 2007. [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tuveson DA. Modelling oncogenic Ras/Raf signalling in the mouse. Curr Opin Genet Dev. 2009;19:4–11. doi: 10.1016/j.gde.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Prince VE. On the diabetic menu: zebrafish as a model for pancreas development and function. Bioessays. 2009;31:139–152. doi: 10.1002/bies.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Lund HZ, Forrest WW, Harris JR, Brown EH. Preliminary embedding in agar-agar. Its value in the processing of multiple separately identified specimens, and in the orientation of small biopsy specimens. Tech Bull Regist Med Technol. 1961;31:192–194. [PubMed] [Google Scholar]
- Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisharath H, Parsons MJ. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol Biol. 2009;546:133–143. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- Tiso N, Moro E, Argenton F. Zebrafish pancreas development. Mol Cell Endocrinol. 2009;312:24–30. doi: 10.1016/j.mce.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Kawakami K. Analysis of genes and genome by the tol2-mediated gene and enhancer trap methods. Methods Mol Biol. 2009;546:85–102. doi: 10.1007/978-1-60327-977-2_6. [DOI] [PubMed] [Google Scholar]
- Westfield M. A guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene: 2007. The Zebrafish Book. [Google Scholar]
- Wilson JM, Bunte RM, Carty AJ. Evaluation of rapid cooling and tricaine methane-sulfonate (MS222) as methods of euthanasia in zebrafish (Danio rerio) J Am Assoc Lab Anim Sci. 2009;48 :785–789. [PMC free article] [PubMed] [Google Scholar]
- Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, Peers B, Bortolussi M, Argenton F. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Zhan H, Gong Z. Delayed and restricted expression of UAS-regulated GFP gene in early transgenic zebrafish embryos by using the GAL4/UAS system. Mar Biotechnol (NY) 2010;12:1–7. doi: 10.1007/s10126-009-9217-y. [DOI] [PubMed] [Google Scholar]