Abstract
Background & Aims
As in other tumor types, progression of pancreatic cancer may require a functionally unique population of cancer stem cells. Although such cells have been identified in many invasive cancers, is not clear whether they emerge during early or late stages of tumorigenesis. Using mouse models and human pancreatic cancer cell lines, we investigated whether pre-invasive pancreatic neoplasia contains a subpopulation of cells with distinct morphologies and cancer stem cell-like properties.
Methods
Pancreatic tissue samples were collected from the KCPdx1, KPCPdx1, and KCiMist1 mouse models of pancreatic intraepithelial neoplasia (PanIN) and analyzed by confocal and electron microscopy, lineage tracing, and fluorescence-activated cell sorting. Subpopulations of human PDAC cells were similarly analyzed and also used in cDNA microarray analyses.
Results
The microtubule regulator DCLK1 marked a morphologically distinct and functionally unique population of pancreatic cancer-initiating cells. These cells displayed morphologic and molecular features of gastrointestinal tuft cells. Cells that expressed DCLK1 also expressed high levels of ATAT1, HES1, HEY1, IGF1R, and ABL1, and manipulation of these pathways in PDAC cell lines inhibited their clonogenic potential. Pharmacologic inhibition of γ–secretase activity reduced the abundance of these cells in murine PanIN, in a manner that correlated with inhibition of PanIN progression.
Conclusions
Human PDAC cells and pancreatic neoplasms in mice contain morphologically and functionally distinct subpopulations that have cancer stem cell-like properties. These populations can be identified at the earliest stages of pancreatic tumorigenesis, and provide new cellular and molecular targets for pancreatic cancer treatment and/or chemoprevention.
Keywords: PanIN, Kras, Notch, acetylated tubulin
INTRODUCTION
Normal human tissues are characterized by hierarchical precursor/progeny relationships between resident stem cells and their differentiated offspring. Similar relationships may also exist in malignant human tumors, with unique subpopulations of “cancer stem cells” or “tumor-initiating cells” thought to be responsible for tumor maintenance. Several recent studies involving formal in vivo lineage tracing have confirmed the critical role played by cancer stem cells in multiple primary tumor types1–3. With respect to pancreatic cancer, subpopulations of cells with tumor-initiating capacities have been identified in human pancreatic cancer cell lines as well as in primary xenografts of human pancreatic ductal adenocarcinoma (PDAC)4–7. However, the role of stem cell populations in the maintenance and progression of pre-invasive pancreatic cancer (pancreatic intra-epithelial neoplasia; PanIN) remains unknown. In addition, while cancer stem cell populations have typically been distinguished based on unique patterns of cell surface marker expression, no information is available regarding whether or not these cells can be morphologically distinguished from their non-stem cell neighbors. To address these issues, we have examined the temporal onset of cellular and functional heterogeneity in early pancreatic cancer. These studies have revealed a novel and morphologically distinct tumor-initiating pancreatic cancer cell type, marked by expression of Doublecortin and Ca2+/Calmodulin-Dependent Kinase-Like 1 (Dclk1). These findings suggest that cellular heterogeneity and functional diversity represent defining features of both invasive and pre-invasive pancreatic cancer.
MATERIALS AND METHODS
All animal experiments described herein were approved by Johns Hopkins University Institutional Animal Care and Use Committees.
Mouse lines
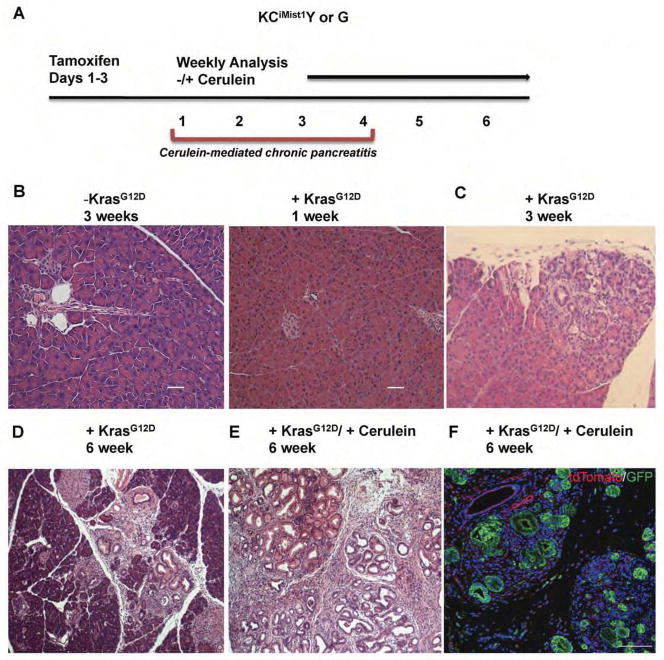
The following murine models of pancreatic intraepithelial neoplasia (mPanIN) and invasive cancer were utilized: KCPdx1, KPC Pdx1 and KCiMist1. Each model utilizes Cre recombinase (C) to activate oncogenic KrasG12D (K), either during development or in adulthood. The KCPdx1 and KPC Pdx1 models utilize a Pdx1:Cre allele to activate oncogenic Kras (KCPdx1) in embryonic pancreatic progenitor cells, either alone (KCPdx1)8 or in combination with inactivation of a floxed p53 allele (KPC Pdx1)9. In contrast, the KCiMist1 model uses an inducible Mist1:CreERT2 driver line to activate oncogenic Kras in adult acinar cells10. Both models lead to the induction of pancreatic “ductal” neoplasia, with the progressive accumulation of mPanIN occurring over several months. For the KCiMist1 model, mPanIN formation was further accelerated by the induction of associated chronic pancreatitis using cerulein (Figure 1A–F). For experiments requiring either lineage tracing or fluorescence-activated cell sorting (FACS), selected KCiMist1 mice were also crossed onto either the either the Rosa26:LSL-YFP Cre reporter line (Y) or the Rosa26:loxP-membrane tdTomato-loxP-membrane GFP (mTmG) Cre reporter line (G), generating KCiMist1Y mice and KCiMist1G mice, respectively (Figure 1F).
Figure 1. Histological analysis of mPanIN progression model after activation of oncogenic Kras in the acinar cell compartment.
(A) Schematic illustrating tamoxifen induction of CreERT2 activity with and without concomitant cerulein-induced chronic pancreatitis in Mist1:CreERT2; LSL-Kras; LSL-YFP (KCiMist1Y) and Mist1:CreERT2; LSL-Kras; mTmG (KCiMist1G) mice. (B–E) Progressive PanIN formation with and without concomitant chronic pancreatitis. (B) No PanIN are detected in either the absence of KrasG12D activation or 1 week following KrasG12D activation. (C) Representative section depicting mPanIN three weeks after oncogenic Kras expression, at which point mPanINs typically occupy ~5% of cross sectional area. (D) Increased PanIN density 6 weeks following KrasG12D activation, at which point mPanINs typically occupy ~10–15% of cross sectional area. (E) Accelerated PanIn formation following KrasG12D activation in combination with cerulein-mediated chronic pancreatitis; PanIN lesions occupy greater than 70% of the pancreas. (F) Antibody labeling for GFP and tdTomato in pancreatic tissue harvested from KCiMist1G mice confirms acinar cell origin of ADM and PanIN.
Microarray analysis
AcTubHI and AcTubLO human PDAC cells were FACS sorted and RNA was isolated using the Qiagen RNeasy Isolation Kit. cDNA microarrays were performed using Agilent Human GE 4x44K arrays, with analysis of differentially expressed genes performed as previously described11.
Clonogenic assays
FACS-sorted murine PanIN and human PDAC cell populations were subjected to clonogenic sphere-forming assays as previously described12.
Additional detailed descriptions of all materials and experimental methods are provided in supplementary material.
RESULTS
A subpopulation of cells containing high levels of Dclk1 and acetylated-αTubulin are present in murine PanIN
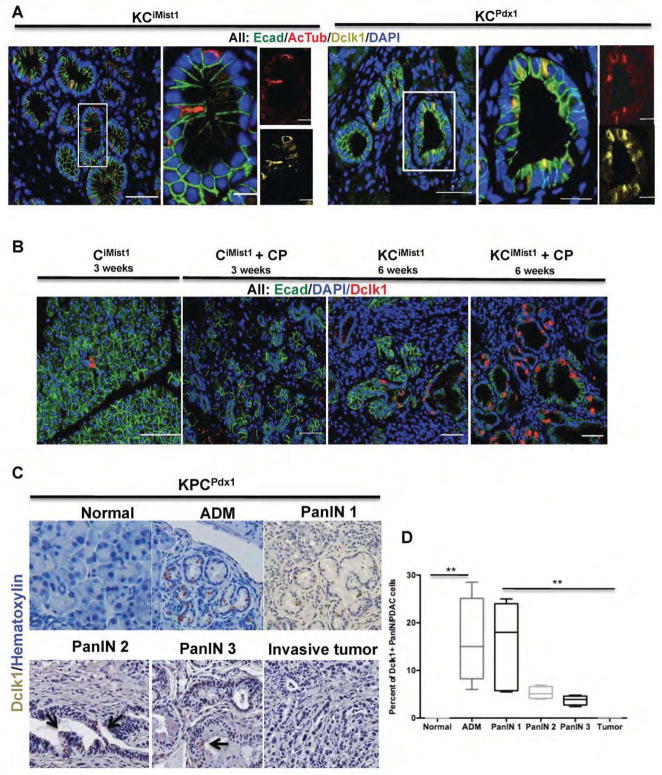
In a survey of markers identifying unique PanIN cell subpopulations, we examined expression of acetylated α-Tubulin (AcTub) in mPanIN. Using immunofluorescent labeling, we characterized the prevalence and distribution of AcTub expressing cells in both KCiMist1 and KCPdx1 mouse pancreas. While AcTub expression was observed in association with primary cilia in normal ductal epithelium and in areas of acinar-to-ductal metaplasia (ADM), we found no evidence of AcTub labeling of primary cilia in mPanIN epithelium (Figure S1A), consistent with the previously reported absence of primary cilia in these lesions13. However, we did observe a low abundance cell population in mPanIN epithelium containing high levels of both cytoplasmic and apical AcTub (Figure 2A), reminiscent of gastrointestinal tuft cells14–17. To further clarify the identity of AcTub positive PanIN epithelial cells, we performed double-labeling for AcTub and Doublecortin and Ca2+/ Calmodulin-Dependent Kinase-Like 1 (Dclk1), a tuft cell marker previously reported to regulate epithelial-mesenchymal transitions in pancreatic cancer cells18. Using immunofluorescenct confocal imaging, we observed a high degree of overlap between Dclk1 and AcTub expression in ADM and early mPanIN lesions from both KCiMist1 and KCPdx1 mice (Figure 2A).
Figure 2. Dclk1 and AcTub label a distinct cell type in mPanIN.
(A) Expression of E-cadherin (green), Dclk1 (yellow) and AcTub (red) in KCiMist1+CP (6 weeks post tamoxifen) and KCPdx1 (18 weeks old) mice. White boxes outline representative areas depicted at higher magnification in adjacent images. Note overlapping labeling for Dclk1 (yellow) and AcTub (red) in distinct subset of mPanIN epithelial cells. (B) Cerulein treatment increases the abundance of Dclk1-expressing cells within mPanIN. (C) Immunohistochemical analysis of Dclk1 expressing cells in KPCPdx1 mice. (D) Dclk1 expressing cells are significantly increased in ADM, mPanIN and invasive PDAC relative to normal pancreatic ductal epithelium. Compared to PanIN1, however, the relative abundance of Dclk1-expressing cells progressively decreases in advanced PanINs and invasive PDAC (**p<0.01).
In order to determine how the number of Dclk1-positive cells was dynamically regulated during early pancreatic neoplasia, we quantified their relative abundance in normal pancreatic epithelium as well as in mPanIN and PDAC. In CiMist1 mice lacking oncogenic Kras, we observed minimal non-ciliary labeling for AcTub, but did identify Dclk1 expression in a small fraction (1–2 cells per field) of normal ductal epithelial cells. In contrast, we found that 10–20% of epithelial cells within ADM lesions and mPanIN1 expressed Dclk1 (Figure 2B–D). Although still much more frequent than in normal ductal epithelium, Dclk1-expressing cells became progressively less abundant in mPanIN2 and mPanIN3 lesions from KPCPdx1 mice, and were rarely identified in invasive murine PDAC (Figure 2C,D). Combined analysis of Dclk1 and AcTub expression confirmed a high correlation between these two markers in ADM and early mPanIN, with progressive divergence in advanced mPanIN or invasive murine PDAC (Figure 3A). Notably, at both the late PanIN and PDAC stages we also observed increases in the abundance of stromal cells characterized by high levels of AcTub (Figure 3A).
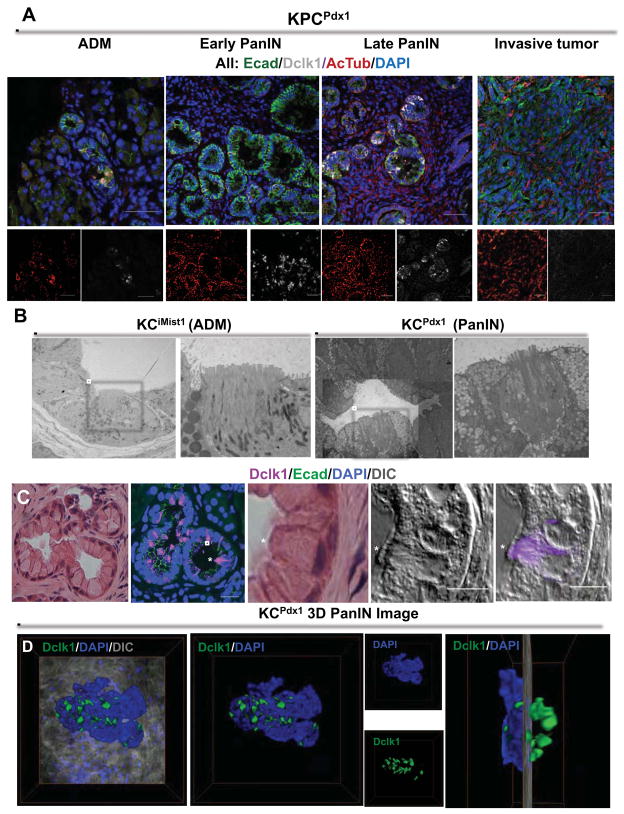
Figure 3. Dclk1- and AcTub-expressing PanIN cells display a tuft cell phenotype.
(A) Immunoflourescent analysis of E-cadherin (green) Dclk1 (white) and AcTub (red) in ADM, early and late PanIN and invasive tumors from KPCPdx1 mice. (B) Transmission electron micrograph depicting ADM (left) and mPanIN (right) from the KCiMist1 model. White boxes indicate cells with pronounced microvilli, shown at higher magnification in adjacent images.. (C) H&E staining on mPanIN from a 9 month old KCPdx1 mouse. Confocal microscopy showing overlay of Dclk1 (purple) and E-cadherin (green). H&E and high-resolution confocal analysis of PanIN tuft cell (marked by *), displaying prominent apical microvilli and expressing Dclk1 (purple). (D) Three dimensional PanIN image from a 9 month old KCPdx1 mouse showing spatial orientation of Dclk1 (green) positive cells. Right most image depicts 90 degree rotation of three-dimensional image, revealing clusters of Dclk1 expressing PanIN cells.
To identify the cellular origin of AcTub+/ Dclk1+ epithelial cells in mPanIN, we performed lineage tracing experiments using KCiMist1Y mice. This analysis demonstrated that virtually all AcTub-positive mPanIN epithelial cells were also marked by YFP (Figure S1A), confirming that the AcTub+/ Dclk1+ cell subpopulation within KCiMist1 ADM and PanIN epithelium is acinar cell-derived.
AcTub+/DCLK1+ PanIN epithelial cells display characteristic tuft cell morphology
To determine whether AcTub+/Dclk1+ epithelial cells displayed classical tuft cell morphology, we performed additional analysis on pancreatic tissue from ADM- and PanIN-bearing KCiMist1 and KCPdx1 mice, using both high-resolution DIC/confocal and transmission electron microscopy (TEM). As recently reviewed19, tuft cells are specialized epithelial cells exhibiting abundant microtubule bundles in association with prominent apical microvilli. In ADM lesions identified in KCiMist1 mice, TEM confirmed the presence of a subpopulation of cells with apical microvilli (Figure 3B). These cells were interspersed among another minority cell population with acinar cell features (i.e. zymogen granules) and a more abundant population with duct cell features. TEM further confirmed the presence of low-abundance, morphologically discernible tuft cells in mPanIN lesions isolated from both KCiMist1 and KCPdx1 mice (Figure 3B). In addition to prominent apical microvilli, the paucity of mucin granules in mPanIN tuft cells further distinguished them from the more predominant mPanIN cell type, in which these granules filled the entire apical cytoplasm. Combining high resolution DIC imaging with confocal microscopy, we were further able to confirm that tuft cell morphology and Dclk1 expression were tightly correlated (Figure 3C). Three-dimensional confocal imaging provided additional perspective on how these cells were spatially organized within mPanIN lesions, with areas of tuft cell clustering suggestive of specialized zones within each PanIN (Figure 3D; Supplemental Movie M1).
A subpopulation of cells in human PanIN and PDAC also display tuft cell features
In parallel to our analysis of murine PanIN and PDAC, we also analyzed the possible presence of a tuft cell-like subpopulation in human chronic pancreatitis, PanIN, and invasive PDAC. Immunostaining in human tissue was limited to AcTub, as multiple Dclk1 antibodies failed to generate reliable and reproducible labeling patterns. As in the case of murine PanIN and PDAC, human PanIN and PDAC lesions contained a minority cell population characterized by high level, cytoplasmic AcTub labeling (Figure S2A–C). To objectively determine whether this pattern of labeling was associated with distinct tuft cell morphology, we surveyed multiple TEM fields from human PDAC and quantified maximal microvillus length for individual human PDAC cells. For comparison, a similar analysis was performed on murine PanIN. This analysis confirmed a dramatic bimodal distribution in maximal apical microvillus length in mPanIN, consistent with a subset of cells displaying classical tuft cell morphology. While median microvillus length in murine PanIN cells was less than 0.2 microns, mPanIN tuft cells displayed apical microvilli up to 4 microns in length (Figure S2F). In contrast, classical tuft cells were less evident on TEM imaging of invasive human cancer. Nevertheless, human PDAC cells also displayed a bimodal distribution in maximal microvillus length, with a subpopulation of cells displaying microvillus lengths in excess of 1 micron (Figure S2E,F). These findings suggest that while cells displaying classical tuft cell morphology are lost as human PanIN lesions progress to invasive cancer, some element of morphologic specialization may be retained.
AcTub+/DCLK1+ mPanIN cells display unique functional capacities
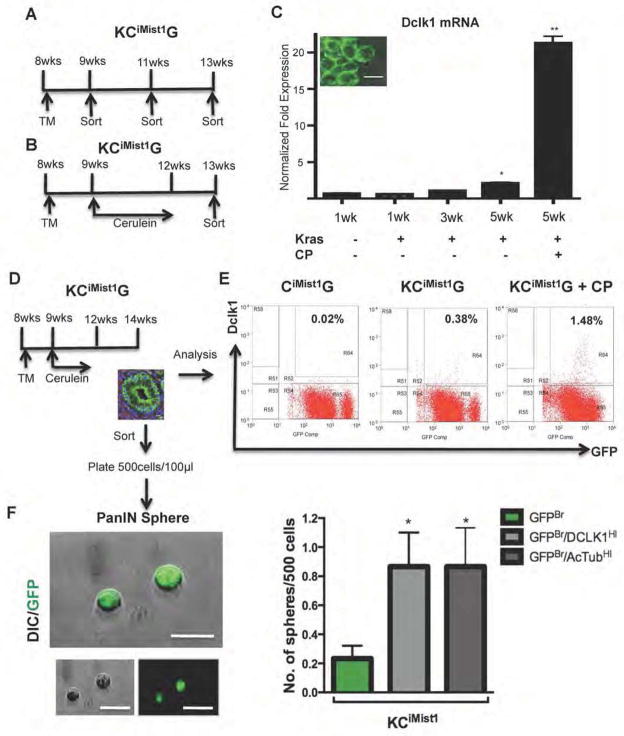
As a means to isolate and functionally characterize AcTub+/Dclk1+ PanIN tuft cells, we crossed KCiMist1 mice onto mTmG Cre reporter line, thereby generating KCiMist1G mice and CiMist1G control mice (Figure 4A,B). Using qRT-PCR to quantify Dclk1 gene expression among FACS sorted mGFP+ cells, we confirmed a progressive increase in Dclk1 expression during KrasG12D-induced mPanIN formation (Figure 4C). Using GFP in combination with an antibody recognizing cell surface Dclk120, we further quantified the number of Dclk1+ cells during mPanIN formation by flow cytometry (Figure 4D, E). This revealed a 19-fold increase in the relative abundance of these cells in KCiMist1G mice compared to CiMist1G controls; this difference was further increased to 70-fold when PanIN formation was accelerated by the induction of chronic pancreatitis (Figure 4E).
Figure 4. Dclk1 and AcTub cells show progenitor capability in preinvasive lesions.
(A,B) Schematic outline of the sorting strategy to isolate eGFP+ cells at different time points after KCiMist1G mice were treated with either tamoxifen alone (A) or tamoxifen and cerulein (B). (C) Dclk1 mRNA levels significantly increase in FACS sorted GFP+ cells during mPanIN progression (**p<0.01). Inset shows imaging confirming membrane eGFP expression in isolated cells. (D) Sorting strategy for FACS-based isolation of eGFP+ cells from PanIN bearing KCiMist1G + CP mice. To generate PanIN spheres, KCiMist1G mice were treated with tamoxifen (TM) and cerulein, then left untreated for two weeks before the isolation of eGFP+ cells. (E) FACS analysis of KCiMist1G + CP demonstrating dramatic increase in Dclk1-expressing cells. Percentages indicate fraction of all pancreatic cells comprising the Dclk1HI/GFPBr subpopulation. (F) PanIN-sphere formation by Dclk1HI/GFPBr, AcTubHI/GFPBr, and GFPBr subpopulations. Dclk1HI/GFPBr, AcTubHI/GFPBr, and GFPBr cells were FACS-sorted, plated at clonal density and cultured for seven days in low attachment plates. Graph indicates relative sphere-forming efficiency for different cell populations (*p<0.05 vs. GFPBr control).
In order to determine the functional correlates of cell surface Dclk1 expression, we isolated GFPBr/Dclk1HI and GFPBr/Dclk1LO cells from KCiMist1G mouse pancreas. Surprisingly, we also found that a fraction of GFPBr cells isolated from KCiMist1G mice also had AcTub epitopes detectable on their cell surface (discussed further, below), correlating with their observed high levels of apical and cytoplasmic AcTub. In addition, the GFPBr/Dclk1HI and GFPBr/AcTubHI populations were largely overlapping, as assessed by flow cytometry. Based on the previous identification of Dclk1 as a marker of tumor stem cells16, 21, we assayed FACS-isolated GFPBr/Dclk1LO, GFPBr/Dclk1HI and GFPBr/AcTubHI cells for clonogenic activity in a suspension culture system. In the absence of oncogenic Kras, the numbers of identified GFPBr/Dclk1HI and GFPBr/AcTubHI were extremely low, and no clonogenic activity was observed. In contrast, in the presence of oncogenic Kras, we observed an increased “PanIN sphere”-forming capacity in cells derived from either the GFPBr/Dclk1HI or the GFPBr/AcTubHI subpopulation relative to the GFPBr/DCLK1LO/AcTubLO fraction alone (Figure 4F and Table S1). Based on these findings, we conclude that morphologically distinct GFPBr/DCLK1HI/AcTubHI mPanIN epithelial cells display unique sphere-forming capacities similar to those displayed by cancer stem cells.
Cell surface labeling for AcTub and DCLK1 in human PDAC cells
Based on the identification of a functionally unique subpopulation of GFPBr/DCLK1HI/AcTubHI in murine PanIN, we subjected human pancreatic cancer cell lines as well as primary human tumor xenografts to cell surface labeling for AcTub and DCLK1, and in each case detected low-abundance (2–4% for cell lines; 3–9% for xenografts) DCLK1HI and AcTubHI cell populations. In these analyses, we found that greater than 90% of AcTubHI cells were also positive for DCLK1 (Supplemental Figure S3A). As in the case of AcTubHI cells identified in mPanIN, it was somewhat surprising to identify AcTub epitopes on the cell surface of human pancreatic cancer cells. We therefore employed a number of techniques to confirm this finding, including immunofluorescent labeling and confocal imaging of fixed and unfixed cells, immunogold labeling followed by TEM, and cell surface protein biotinylation followed by immunoblot analysis. Each of these analyses confirmed the presence of enriched AcTub epitopes on the cell surface of AcTubHI cells. In addition, AcTubHI cells displayed increased expression of α-tubulin acetyltransferase (ATAT1; Figure S4A), and siRNA knockdown of ATAT1 reduced their relative abundance (Supplemental Figure S4B). Together, these findings strongly support the presence of cell surface AcTub epitopes on a subset of human pancreatic cancer cells.
The DCLK1HI/AcTubHI phenotype is shared by other pancreatic cancer stem cell populations
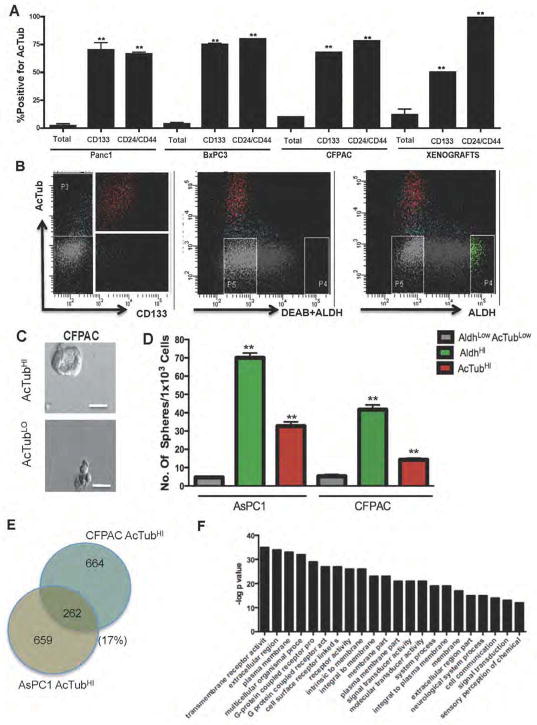
Based on our finding of enhanced clonogenic activity in the AcTub- and Dclk1-expressing mPanIN subpopulation, we hypothesized that AcTubHI/DCLK1HI PDAC cells might also display unique functional capacities. We used FACS analysis to compare the frequency of AcTubHI labeling in cell populations expressing a series of previously published PDAC cancer stem cell markers, including CD1337, CD24/CD44/ESA4, and ALDH5. In these analyses, we found that cell populations labeling positive for either CD133 or CD24/CD44/ESA demonstrated up to 30-fold enrichment for AcTubHI cells (Figure 5A,B). In contrast, we observed no enrichment and minimal overlap between the AcTubHI and ALDHHI populations (Figure 5B), suggesting that these two markers define independent PDAC cell subpopulations.
Figure 5. DCLKHI/AcTubHI cells are pancreatic cancer initiating cells.
(A) Enrichment for cell surface AcTub labeling among CD133 or CD24/CD44/ESA positive cells in Panc1, BxPC3 and CFPAC cell lines and in two harvested human xenografts. Cells expressing CD133 or CD24/CD44/ESA are also enriched for expression of cell surface AcTub compared to the bulk cell population (**p<0.001). (B) FACS plots showing overlap of AcTub cell surface labeling with other published markers of pancreatic cancer stem cells, CD133 and ALDH. Significant overlap is observed for AcTub and CD133, but not for AcTub and ALDH (C) Tumor sphere derived from FACS sorted AcTubHI cells from the CFPAC cell line (scale bar 50μm). (D) Graphical representation of in vitro tumor initiating capacity of AcTubHI/DCLK1HI subpopulations isolated from two different pancreatic cancer cell lines. The non-overlapping AcTubHI/DCLK1HI and ALDHHI subpopulations formed significantly more spheres than the control AcTubLO/ALDHLO fraction (**p<0.01). (E) Whole transcriptome analysis on FACS-sorted AcTubHI cells from both the CFPAC and AsPC1 cell lines. In these analyses, 926 genes were upregulated in AcTubHI vs. AcTubLO CFPAC cells and 921 genes were upregulated in AcTubHI ASPC1 vs AcTubLO cells. A total of 262 genes were upregulated in both CFPAC AcTubHI and ASPC1 AcTubHI cells. (F) Gene set enrichment analysis of 262 upregulated genes. The GO category “transmembrane receptor activity” was the most highly enriched functional group (p=1.322e-35), and many other groups related to cell surface receptors were similarly enriched. A complete listing of differentially expressed genes is provided in Table S2.
DCLK1HI/AcTubHI PDAC cells have tumor- and tumorsphere-initiating capabilities
We next determined whether DCLK1HI/AcTubHI PDAC cells were capable of clonogenic growth, similar to their mPanIN counterparts. Knowing that the DCLK1HI/AcTubHI and ALDHHI phenotypes defined distinct and non-overlapping cell populations, and because ALDHHI cells have been previously shown to display clonogenic and tumor-initiating capabilities, we directly compared DCLK1HI/AcTubHI and ALDHHI for tumorsphere- and tumor xenograft-initiating capabilities. In these studies, only AcTub was used as a marker for DCLK1HI/AcTubHI cells, given the near-universal co-expression of DCLK1HI within the AcTubHI cell population. In tumorsphere-forming assays, AcTubHI cells isolated from two different PDAC cell lines displayed an approximately 10-fold increase in sphere-forming efficiency compared to the more abundant AcTubLO/ALDHLO population, although they were ~50% less efficient than ALDHHI cells (Figure 5C,D).
To determine the tumor initiating potential and growth potential of the AcTubHI population in vivo, we sorted AcTubHI and ALDHHI cells from two human xenografts and implanted limiting dilutions of cells into immunodeficient mice. We injected either 100, 500, 1000 or 5000 cells per mouse and monitored tumor initiation over a series of 20 weeks. In these experiments, there was no tumor formation observed following injection of AcTubLO/ALDHLO cells. AcTubHI cells formed tumors with a calculated tumor-initiating cell frequency of 1/1454 (95% confidence limits 1/3523 to 1/600), compared to 1/540 (95% confidence limits 1/1248-1/234) for ALDHHI cells (Table 1). These findings suggest that that AcTub- and Dclk1-expressing human PDAC cells have cancer stem cell-like functional capabilities.
Table 1.
Xenograft initiating capacity of FACS sorted AcTubHI and ALDHHI vs AcTubLO/ALDHLO cell populations.
| Sorted Cell Population | No. Cells per injection | No. of tumors/No. of injections | TIC Frequency | P-value (vs. bulk) |
|---|---|---|---|---|
| Bulk | 100 | 0/4 | ND | ND |
| 500 | 0/4 | |||
| 1000 | 0/4 | |||
| 5000 | 0/4 | |||
| AcTubHI | 100 | 0/4 | 1/1454 (1/3523-1/600) | 0.0000539 |
| 500 | 1/4 | |||
| 1000 | 2/4 | |||
| 5000 | 4/4 | |||
| ALDHHI | 100 | 0/4 | 1/540 (1/1248-1/234) | 0.00000045 |
| 500 | 2/4 | |||
| 1000 | 4/4 | |||
| 5000 | 4/4 |
Both AcTubHI and ALDHHI isolated cells had significantly increased xenograft initiating frequency compared to control cells (**p<0.001). TIC frequency was calculated using the Extreme Limiting Dilution Analysis software (Hu and Smyth, 2009).
Tubulin acetylation and polymerization both regulate the clonogenic potential of DCLK1HI/AcTubHI PDAC cells
ATAT1 is the primary enzyme responsible for the acetylation of Tubulin on Lysine 4022, while DCLK1 is a known regulator of tubulin polymerization23. Using quantitative RT-PCR, we determined that FACS-sorted AcTubHI/DCLK1HI and AcTubLO/DCLK1HI cells both had significant increases in DCLK1 and ATAT1 transcript abundance relative to AcTubLO/DCLK1LO cells (Supplemental Figure S4A). To determine whether tubulin acetylation was required for tumor sphere initiation, we performed siRNA knockdown of ATAT1 in two different human PDAC cell lines (AsPC1 and CFPAC). This resulted in a 3- to 7-fold reduction in sphere forming capacity compared to cells transfected with control siRNA (**p<0.01 for both AsPC1 and CFPAC) (Figure S4C,E). As an additional means to interrogate the apparent contribution of tubulin acetylation and polymerization to the clonogenic activity of AcTubHI/DCLK1HI cells, we also performed siRNA knockdown of both DCLK1 and Histone deacetylase 6 (HDAC6), the primary enzyme responsible for tubulin deacetylation 24–26. For these experiments, we used bulk (unsorted) CFPAC cells and were able to achieve 75–80% reductions in target gene expression (Figure S4D). Knockdown of HDAC6 led to a 1.5 fold increase in sphere formation (*p<0.05), consistent with tubulin acetylation exerting a positive influence on the clonogenic activity of PDAC cells (Supplemental Figure S4C). As in the case of ATAT1, DCLK1 gene knockdown also resulted in a significant reduction in clonogenicity (*p<0.05) (Figure S4C).
Whole transcriptome analysis reveals gene and pathway enrichment in DCLK1HI/AcTubHI PDAC cells
As a means to identify genes and pathways potentially regulating the tumor-initiating capacities of DCLK1HI/AcTubHI PDAC cells, we performed whole transcriptome analysis on FACS-sorted cells from both the CFPAC and AsPC1 cell lines (Figure 5E,F). A complete listing of differentially expressed genes is provided in Table S2. Among 926 genes upregulated in DCLK1HI/AcTubHI cells from the CFPAC cell line and 921 genes similarly upregulated in DCLK1HI/AcTubHI cells from the AsPC1 cell line, 262 genes were upregulated in both cell lines (Figure 5E). Gene Ontology (GO) functional group enrichment analysis on these 262 genes defined transmembrane receptor activity as the most highly enriched functional group, with many other categories related to cell surface receptors also enriched (Figure 5F). Among genes found to be upregulated in DCLK1HI/AcTubHI cells were the tuft cell markers TAS2R31, OR5A2 and DCLK1 itself, as well as the Notch response genes HES1, HES7 and HEY1. Additional genes found to be upregulated in DCLK1HI/AcTubHI cells included ATAT1, ABL1 and IGF1R. We therefore performed additional functional studies investigating the role of these signaling pathways in regulating the relative abundance and/or the clonogenic capacities of DCLK1HI/AcTubHI PanIN and PDAC cells.
AcTubHI/DCLK1HI cell abundance is regulated by Notch signaling in both human PDAC and murine PanIN
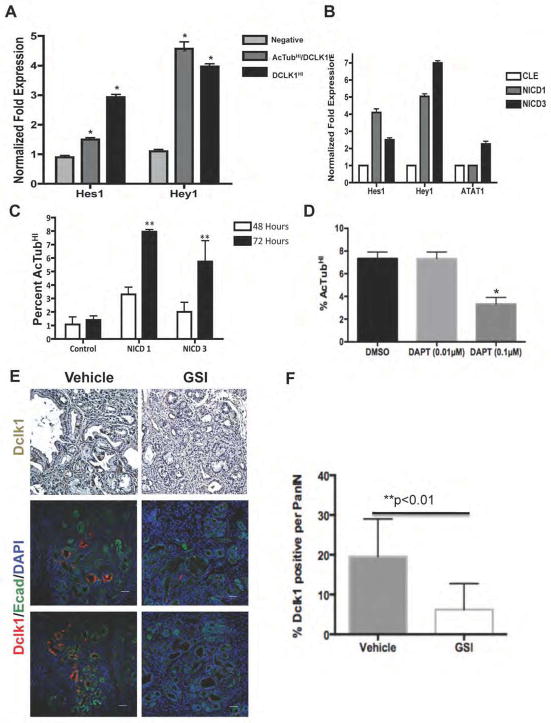
Prior studies have implicated the Notch signaling pathway in the pathogenesis of pancreatic cancer27, 28. Using qRT-PCR, we confirmed upregulated expression of both HES1 and HEY1 in AcTubHI/DCLK1HI PDAC cells (Figure 6A). In order to determine the functional significance of Notch signaling in these cells, we forced Notch pathway activation in PDAC cells using retroviral vectors encoding the active, intracellular domain of either NOTCH1 (NICD1) or NOTCH3 (NICD3). Infection of PDAC cells with these retroviral vectors resulted in upregulated expression of HES1 and HEY1, confirming effective pathway activation (Figure 6B). Notch pathway activation induced by ectopic expression of NICD3 also increased the expression of ATAT1. Expression of either NICD1 or NICD3 also dramatically increased the fraction of AcTubHI PDAC cells (Figure 6C), while inhibition of Notch signaling using the γ-secretase inhibitor DAPT decreased the relative abundance of AcTubHI cells (Figure 6D). Thus Notch signaling appears to be a potent regulator of AcTubHI/DCLK1HI cell abundance in human PDAC.
Figure 6. AcTubHI/DCLK1HI PDAC stem cell function is regulated by Notch.
(A) Quantitative Real Time PCR confirming increased expression of Hes1 and Hey1 in FACS sorted AcTubHI/DCLK1HI cells. (B) Expression of NICD1 or NICD3 increases the expression of Hes1, Hey1 and ATAT1 mRNA levels. (C) Expression of NICD1 or NICD3 significantly increases the percentage of cells with detectable cell surface AcTub (**p<0.01). (D) Treatment of the CFPAC cell line with DAPT decreases the percentage of cells with cell surface AcTub labeling (*p<0.05). (E) Immunohistochemistry and immunoflourescent analysis of pancreatic tissue from vehicle-treated and γ-secretase inhibitor (MRK-300)-treated KPCPdx1 mice. (F) Quantification of Dclk1 labeling shown in (E). In vivo inhibition of Notch signaling significantly reduces the fraction of mPanIN epithelial cells expressing Dclk1 (**p<0.01).
We next sought to determine whether the ability of Notch to regulate the abundance of AcTubHI/DCLK1HI cells in vitro might be correlated with a therapeutically relevant in vivo effect. To achieve this, we compared the density of cells expressing Dclk1 in KPCPdx1 mice treated with either vehicle or with the γ-secretase inhibitor MRK-003. In these studies, MRK-003 was delivered using an intermittent dosing regimen that effectively blocked PanIN progression29. In examining these tissues, in vivo γ-secretase inhibition was associated with a significant reduction in the number of Dclk1-expressing cells, expressed as the percentage of Dclk1-positive cells per PanIN (Figure 6E,F).
Inhibition of ABL and IGF1R reduce the sphere forming capacity of CFPAC cells
Microarray analysis also suggested upregulated expression of ABL1 and IGF1R in Dclk1HI/AcTubHI PDAC cells, a result further confirmed in by qRT-PCR (Figure S6A). We further confirmed a significant increase in ABL1 transcript levels from FACS sorted GFPBr/ DCLK1HI relative to GFPBr/DCLK1LO mPanIN cells from the KCiMist1G mouse model (Figure S6B). We therefore examined the effect of pharmacologic inhibition ABL1 on the clonogenic potential of bulk CFPAC cells. Using nilotinib to target ABL1, we observed a potent and dose-dependent reduction in tumorsphere formation mediated through a pro-apoptotic mechanism (Figure S6C–D) Similar effects were observed following inhibition of IGF1R signaling using linsitinib (data not shown).
CONCLUSIONS
In addition to identifying a novel and morphologically distinct subpopulation of tumor-initiating cells in human PDAC, these studies emphasize both morphologic and functional heterogeneity among epithelial cells within both murine and human PanIN. The current results complement a recent report demonstrating that distinct epithelial and mesenchymal subpopulations are generated following oncogenic Kras activation in adult acinar cells30 and emphasize the need to consider PanIN lesions as a complex, multicellular tissue.
Prior studies have suggested that Dclk1 may mark tumor-initiating cells in a variety of tumor types21, 31, 32. In a recent report using the ApcMin model, formal Cre/lox-based lineage tracing was employed to demonstrate that Dclk1-expressing cells within intestinal adenomas are responsible for the continuous production of tumor cell progeny, and that diphtheria toxin-mediated ablation of these cells results in tumor regression33. DCLK1 expression has previously been empirically demonstrated in both murine and human PanIN and PDAC. DCLK1 is also expressed by isolated cells within the pancreatic duct and islets of normal mouse pancreas, and prior studies have suggested that these non-neoplastic Dclk1-expressing pancreatic cells may also be associated with progenitor-like function20.
While the cancer stem cell hypothesis has been most forcefully proposed as a feature of invasive malignancy, a number of recent studies have confirmed this principle to also be operative in several forms of pre-invasive neoplasia, including intestinal adenomas and skin papillomas34. We have demonstrated that DCLK1HI/AcTubHI mPanIN cells display specialized morphology, unique patterns of gene expression and enhanced “PanIN sphere” forming ability. The ability to more formally implicate DCLK1HI/AcTubHI mPanIN cells as PanIN stem cells is currently hindered by the fact that our current methodologies for lineage tracing and oncogenic Kras activation both require Cre/lox activity. Future progress in this area will depend on either the development of lineage tracing techniques using alternate DNA recombinases (e.g. Dre, Flp), or the development of non Cre/lox-based methods for Kras activation in murine pancreas.
Using whole transcriptome analysis, we identified multiple pathways that contribute to the cancer stem cell-like properties of DCLK1HI/AcTubHI pancreatic cancer cells. Among these, tubulin acetylation itself appears to be required for optimal clonogenic function, establishing inhibition of ATAT1 or activation of HDAC6 as potential new therapeutic strategies. The role of Notch signaling in pancreatic cancer is well established27, 29, 35, 36, and recent studies have further implicated Notch in regulating pancreatic cancer stem cell function37; our studies suggest that this effect may in part be due to an influence on the relative size of the DCLK1HI/AcTubHI subpopulation. Our studies also identified ABL1 and IGF1R to be highly expressed in DCLK1HI/AcTubHI cells, complementing ongoing preclinical and clinical evaluation of ABL1 and IGF1R pathway inhibition as new forms of targeted therapy for pancreatic cancer38, 39.
In summary, these studies suggest that both preinvasive and invasive pancreatic cancer may depend upon a unique subpopulation of DCLK1-expressing cells with cancer stem cell capabilities. As in the case of intestinal tumorigenesis21, targeting this cell population may have therapeutic potential in the treatment and/or chemoprevention of pancreatic cancer.
Supplementary Material
Acknowledgments
The authors wish to thank Danielle Blake, Mara Swaim, Jeffrey Roeser, Anzer Habibulla, Dr. Hao Ho and Dr. Xiaogang Zhong for expert technical assistance and Dr. James Goldenring for many helpful discussions. We also thank Barbara Smith for expertise in TEM. TMA arrays were provided by the Hopkins Pathology Core. JMB is supported by the 2011 Pancreatic Action Network-AACR Pathway to Leadership Award and NCI 5F32 CA157044. JA is supported through the 5T32 CA126607 and the immixGroup Foundation Fellowship. ZR is supported by the Pancreatic Cancer Action Network-AACR Pathway to Leadership award. FM is supported by NIGMS T32GMO66691, 2012 Pancreatic Cancer Action Network-AACR Fellowship, in memory of Samuel Stroum, Grant Number 12-40-25-MCAL and Conquer Cancer Foundation Young Investigator Award 2012. WM is supported by R01 CA150142. SDL and AM were supported by NCI P01 CA134292. SDL was further supported by the Paul K. Neumann Professorship in Pancreatic Cancer at Johns Hopkins University.
Footnotes
Jennifer M. Bailey has no Conflict of interest
Janivett Alsina has no Conflict of Interest
Zeshaan A. Rasheed has no Conflict of Interest
Ya-Yuan Fu has no Conflict of Interest
Ruben Pletz has no conflict of Interest
Florencia McAllister has no Conflict of Interest
Hao Zhang has no Conflict of Interest
Nabeel Bardeesy has no conflict of interest
Pankaj Pasricha has no Conflict of Interest
William Matsui has no Conflict of Interest
Anirban Maitra has no Conflict of Interest
Steven Leach has no Conflict of Interest
Supplemental Information includes supplemental experimental procedures, five figures, two tables and one supplemental movie.
Author Contributions: J.M.B. participated in study concept, study design, data acquisition, data interpretation and manuscript preparation. J.A., Z.R., Y.F., F.M.M. and H.Z. participated in the acquisition of data. P.J.P., N.B. and R.P. provided material support; W.M. and A.M. provided important intellectual support, S.D.L. participated in study concept and design, study supervision, data interpretation and manuscript preparation. J.M.B. and S.D.L. obtained funding.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 5.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong SM, Koorstra JB, Rajeshkumar NV, He X, Goggins M, Iacobuzio-Donahue C, Berman DM, Laheru D, Jimeno A, Hidalgo M, Maitra A, Matsui W. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–51. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olempska M, Eisenach PA, Ammerpohl O, Ungefroren H, Fandrich F, Kalthoff H. Detection of tumor stem cell markers in pancreatic carcinoma cell lines. Hepatobiliary Pancreat Dis Int. 2007;6:92–7. [PubMed] [Google Scholar]
- 7.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 9.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, Depinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–8. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provost E, Wehner KA, Zhong X, Ashar F, Nguyen E, Green R, Parsons MJ, Leach SD. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development. 2012;139:3232–41. doi: 10.1242/dev.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YJ, Bailey JM, Rovira M, Leach SD. Sphere-forming assays for assessment of benign and malignant pancreatic stem cells. Methods Mol Biol. 2013;980:281–90. doi: 10.1007/978-1-62703-287-2_15. [DOI] [PubMed] [Google Scholar]
- 13.Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 2009;69:422–30. doi: 10.1158/0008-5472.CAN-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saqui-Salces M, Keeley TM, Grosse AS, Qiao XT, El-Zaatari M, Gumucio DL, Samuelson LC, Merchant JL. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol. 2011;136:191–204. doi: 10.1007/s00418-011-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam KT, O’Neal R, Lee YS, Lee YC, Coffey RJ, Goldenring JR. Gastric tumor development in Smad3-deficient mice initiates from forestomach/glandular transition zone along the lesser curvature. Lab Invest. 2012;92:883–95. doi: 10.1038/labinvest.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–7. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 17.Hofer D, Drenckhahn D. Cytoskeletal markers allowing discrimination between brush cells and other epithelial cells of the gut including enteroendocrine cells. Histochem Cell Biol. 1996;105:405–12. doi: 10.1007/BF01463662. [DOI] [PubMed] [Google Scholar]
- 18.Sureban SM, May R, Lightfoot SA, Hoskins AB, Lerner M, Brackett DJ, Postier RG, Ramanujam R, Mohammed A, Rao CV, Wyche JH, Anant S, Houchen CW. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 2011;71:2328–38. doi: 10.1158/0008-5472.CAN-10-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerbe F, Legraverend C, Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69:2907–17. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May R, Sureban SM, Lightfoot SA, Hoskins AB, Brackett DJ, Postier RG, Ramanujam R, Rao CV, Wyche JH, Anant S, Houchen CW. Identification of a novel putative pancreatic stem/progenitor cell marker DCAMKL-1 in normal mouse pancreas. Am J Physiol Gastrointest Liver Physiol. 2010;299:G303–10. doi: 10.1152/ajpgi.00146.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, Isomura A, Kawada K, Sakai Y, Yanagita M, Kageyama R, Kawaguchi Y, Taketo MM, Yonehara S, Chiba T. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2012;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 22.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A. 2010;107:21517–22. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–61. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro-Castro A, Janke C, Montagnac G, Paul-Gilloteaux P, Chavrier P. ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of alpha-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur J Cell Biol. 2012;91:950–60. doi: 10.1016/j.ejcb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Bobrowska A, Paganetti P, Matthias P, Bates GP. Hdac6 knock-out increases tubulin acetylation but does not modify disease progression in the R6/2 mouse model of Huntington’s disease. PLoS One. 2011;6:e20696. doi: 10.1371/journal.pone.0020696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Jiang H, Chang M, Xie H, Hu L. HDAC6 alpha-tubulin deacetylase: a potential therapeutic target in neurodegenerative diseases. J Neurol Sci. 2011;304:1–8. doi: 10.1016/j.jns.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, Schmid RM, Leach SD. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 28.Doucas H, Mann CD, Sutton CD, Garcea G, Neal CP, Berry DP, Manson MM. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol. 2008;97:63–8. doi: 10.1002/jso.20894. [DOI] [PubMed] [Google Scholar]
- 29.Plentz R, Park JS, Rhim AD, Abravanel D, Hezel AF, Sharma SV, Gurumurthy S, Deshpande V, Kenific C, Settleman J, Majumder PK, Stanger BZ, Bardeesy N. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2009;136:1741–9. e6. doi: 10.1053/j.gastro.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, Leach SD, Stanger BZ. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vega KJ, May R, Sureban SM, Lightfoot SA, Qu D, Reed A, Weygant N, Ramanujam R, Souza R, Madhoun M, Whorton J, Anant S, Meltzer SJ, Houchen CW. Identification of the putative intestinal stem cell marker doublecortin and CaM kinase-like-1 in Barrett’s esophagus and esophageal adenocarcinoma. J Gastroenterol Hepatol. 2012;27:773–80. doi: 10.1111/j.1440-1746.2011.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sureban SM, May R, Ramalingam S, Subramaniam D, Natarajan G, Anant S, Houchen CW. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology. 2009;137:649–59. 659 e1–2. doi: 10.1053/j.gastro.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, Isomura A, Kawada K, Sakai Y, Yanagita M, Kageyama R, Kawaguchi Y, Taketo MM, Yonehara S, Chiba T. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 34.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–12. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazur PK, Einwachter H, Lee M, Sipos B, Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Kloppel G, Schmid RM, Siveke JT. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2010;107:13438–43. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, Banerjee S, Azmi AS, Miele L, Sarkar FH. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307:26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Ali Y, Lin Y, Gharibo MM, Gounder MK, Stein MN, Lagattuta TF, Egorin MJ, Rubin EH, Poplin EA. Phase I and pharmacokinetic study of imatinib mesylate (Gleevec) and gemcitabine in patients with refractory solid tumors. Clin Cancer Res. 2007;13:5876–82. doi: 10.1158/1078-0432.CCR-07-0883. [DOI] [PubMed] [Google Scholar]
- 39.Rieder S, Michalski CW, Friess H, Kleeff J. Insulin-like growth factor signaling as a therapeutic target in pancreatic cancer. Anticancer Agents Med Chem. 2011;11:427–33. doi: 10.2174/187152011795677454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.