Abstract
Objective
Hyperactivity of the type I interferon (IFN) pathway is involved in the pathogenesis of systemic lupus erythematosus (SLE). Immunoglobulin like transcript (ILT3) is an immunohibitory transmembrane molecule which is induced by type I IFNs. ILT3 is expressed by plasmacytoid dendritic cells (PDCs), monocytoid dendritic cells (MDCs), and monocytes/macrophages. Given the pathogenic role of IFN in SLE, we hypothesised that the IFN-induced immunosuppressive ILT3 receptor may be dysfunctional in human SLE.
Methods
132 European-derived and 79 Hispanic-American SLE patients were genotyped for two coding-change single nucleotide polymorphisms (SNPs) predicted to interfere with protein folding in ILT3 (rs11540761 and rs1048801). 116 control DNA samples and sera from healthy controls were also studied. We detected associations between ILT3 genotype and serum cytokine profiles. ILT3 expression levels on PDCs and MDCs from 18 patients and 10 controls were studied by flow cytometry.
Results
The rs11540761 SNP in the extracellular region was associated with decreased cell surface expression of ILT3 on circulating MDCs and to a lesser extent PDCs in SLE patients. The cytoplasmically located rs1048801 SNP was not associated with a change in dendritic cells expression of ILT3. Both SNPs were significantly and independently associated with increased levels of serum type I IFN activity in SLE patients. The rs1048801 SNP was also associated with increased serum levels of TNF-α.
Conclusions
Loss-of-function polymorphisms in ILT3 are associated with increased inflammatory cytokine levels in SLE, supporting a biological role for ILT3 in SLE.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a severe systemic autoimmune disease characterised by significant morbidity and mortality, and peak incidence in early adulthood.1 The immunopathogenesis of SLE is poorly understood, but work from our group and others suggests that dysregulation of the type I interferon (IFN) pathway predisposes to the loss of immunological self-tolerance.2–4 Type I IFN is a pleiotropic cytokine that can break tolerance to self by activating antigen presenting cells causing them to present self-antigens.2,5 Serum type I IFN is elevated in many SLE patients and correlates with disease activity and severity.6–9 Numerous genetic variations that associate with SLE susceptibility impact the type I IFN pathway in SLE patients, supporting the idea that gain-of-function polymorphisms in the type I IFN pathway result in risk of SLE.10–14 TNF-α may also have a pathogenic role in SLE since circulating levels are high in many patients.15
While the studies above focus on amplifiers of the IFN-α pathway, negative regulators of this pathway are also important. Immunoglobulin-like transcript 3 receptor (ILT3) is an immunosuppressive receptor inducible by type I IFNs that could represent one possible negative feedback pathway for regulating type I IFNs in vivo. Although the ligand for ILT3 has not been identified, studies show that ILT3 functions primarily to inhibit immune responses. The cytoplasmic tail of ILT3 contains immunoreceptor tyrosine-based inhibition motifs (ITIMs) through which it recruits and activates SH2 containing tyrosine phosphatases. Co-ligation of ILT3 with immunoreceptor tyrosine-based activation motif (ITAM) containing receptors on dendritic cells (DCs), such as FcγRI, and FcRIIa, blocks their activation and downstream signalling.16–18 Monocytoid dendritic cells (MDCs) with high ILT3 levels suppress T cell activation and are tolerogenic.16,17,19,20 In addition, we have shown that ILT3 levels are higher in multiple sclerosis patients during remission or in those who are being treated with type I IFN, supporting the concept that IFN-induced ILT3 expression is immunosuppressive.21
To examine a potential role for ILT3 in SLE, we examined coding change genetic polymorphisms in ILT3 in lupus patients. We determined if polymorphisms which would be predicted to disrupt protein folding would impact upon surface expression of ILT3 and/or pathogenic cytokine levels in SLE patients.
MATERIALS AND METHODS
Patients
Serum and genomic DNA from 132 European-American and 79 Hispanic-American SLE patients, recruited from The University of Chicago Translational Research in the Department of Medicine (TRIDOM) registry, were used to detect association between ILT3 genotype and serum cytokine profiles. Blood from 18 patients and 10 controls were examined with flow cytometry. All patients met the revised 1982 American College of Rheumatology criteria for SLE.22 One hundred and sixteen control DNA samples were obtained from the TRIDOM registry and each screened by medical record review for absence of autoimmune disease. All subjects gave informed consent. The University of Chicago Institutional Review Board approved this study.
Genotyping
Patients and controls were genotyped at the rs11540761 and rs1048801 single nucleotide polymorphisms (SNPs) in ILT3 using custom designed Applied Biosystems Taqman primers and probes on an Applied Biosystems 7900HT PCR machine with greater than 98% genotyping success (for primer and probe sequences see online supplementary table S1). Each genotyping scatter plot was reviewed for quality. Genotype frequencies did not deviate significantly from expected Hardy-Weinberg proportions in controls (p>0.01).
Type I IFN bioassay
A previously described type I IFN bioassay, shown to be highly informative in multiple human autoimmune diseases, was used.4,9,23–26 Briefly, WISH cells (ATCC #CCL-25) were cultured with 50% sera or supernatant for 6 h and lysed. cDNA, made from cellular mRNA, was quantified by real-time PCR using primers for the genes IFN-induced protein with tetratricopeptide repeats 1, myxovirus resistance 1 and dsRNA-activated protein kinase.4 PCR gene products were normalised to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase. Fold increase in expression levels of the IFN-induced genes of cells cultured with samples over cells without samples was determined. A standard curve showing expression of these genes following stimulation with recombinant IFN-α is shown in the online supplementary figure S1. Results were standardised to a healthy multi-ancestral reference population and type I IFN activity was calculated based upon the mean and SD of the reference population.4 SLE patients with activity scores greater than 2 SDs above the mean of healthy controls were considered ‘high IFN’, and others ‘low IFN’.
Tumour necrosis factor α (TNF-α) measurement
TNF-α was measured using a commercially available ELISA kit (Pierce, Rockford, Illinois, USA), according to the manufacturer instructions.
Cells
Peripheral blood mononuclear cells (PBMCs) were isolated on Ficoll-Paque gradients (Cederlane, Burlington, North Carolina, USA). Plasmacytoid dendritic cells (PDCs) (>95% pure) were isolated using a negative selection kit (Miltenyi Biotec, Auburn, California, USA) except that the bead bound cells were not centrifuged but deposited into a 0.5 ml Kimble tube, placed against a magnet for 10 min, and then added to an LS column.
Flow cytometry
PBMC (7.5×105 cells in phosphate buffered saline plus 2% goat serum plus 0.1% NaN3) were incubated with anti-lineage 1-fluorescein isothiocynate (Biolegend, San Diego, California, USA), anti-HLADR-eFluor 450 (LN3; Ebioscience, San Diego, California, USA) and either anti-CD123-PE-Cy7 (6H6) or anti-CD11c-PE-Cy7 (3.9) antibodies for 30 min at 4°C. Cells were co-incubated with anti-ILT3-PE (ZM4.1) and with IgG1-PE (MOPC-21; all from Biolegend). Cells were washed, suspended in 1% formalin, and analysed within 24 h using a BD LSR II cytometer with FACSDiva software (BD Biosciences, San Jose, California, USA). Non-specific fluorescence was determined using fluorochrome conjugated controls. Phycoerythrin (PE) fluorescence of PDCs (lin−CD123+HLADR+) and of MDCs (lin−CD11c+HLADR+) was analysed for cells incubated with anti-ILT3-PE and with IgG1-PE. Minimally 4×105 PBMC were recorded per sample. Geometric mean fluorescent intensities (GMFI) of PE fluorescence for each DC subset were determined (FlowJo software). Specific fluorescent intensities (SFI)=(GMFI of cells incubated with anti-ILT3-PE)/(GMFI of cells incubated with IgG1-PE) are shown.
Plasmacytoid dendritic cells
PDCs were incubated 1 h with anti-ILT3 ZM4.1 and anti-immunoglobulin like transcript 7 (ILT7) 17G10.2 (10 μg/ml; Biolegend) at 4°C. Cells were also incubated with control IgG1 MOPC-21 since crosslinking FcRs on PDCs inhibits IFN secretion.27 Cells were washed, suspended in medium (2.5×105 cells/ml), and added (80 μl) to duplicate wells of enzyme immunoassay (EIA) plates (Costar #3361) previously coated with goat F(ab′)2 anti-mouse IgG (10 μg/ml; Pierce, Rockford, Illinois, USA). IL-3 enhances PDC survival, and 1 ng in 10 μl was added per well.28 Plates were centrifuged (5 min at 150×G) with a slow brake. After a 1 h incubation at 37°C, 0.01 ml/well of oligodeoxy-nucleotide (ODN) 2216 (5 μm) (Invivogen, San Diego, California, USA) was added and supernatants collected 18 h later.
Statistics
For normally distributed data, mean and SD are shown, and columns of data were compared by unpaired t-test. Non-parametric Mann-Whitney U was used to compare the non-normally distributed quantitative IFN-α data in patients between genotype subgroups, with median and IR shown. p Values given are uncorrected for multiple comparisons. To account for multiple comparisons, we used a threshold p value of 0.025 or smaller to control the family-wise type I error rate at 0.05 using a Bonferroni correction for the number of polymorphisms tested in this study. Comparisons reported as significant meet or exceed this threshold p value.
RESULTS
Informatic assessment of common coding change SNPs in ILT3 for potential functional impact
The ILT3 gene is highly polymorphic.29 We examined coding-change ILT3 SNPs reported in dbSNP build 134, including SNPs with a reported minor allele frequency greater than 5%. Figure 1 shows the location of 5 SNPs in the ILT3 gene with the coding changes that occur. These SNPs were examined in the PolyPhen2 software program (http://genetics.bwh.harvard.edu/pph2/index.shtml) to determine if they were likely to be damaging coding changes, with results summarised in table 1.
Figure 1.

Diagram of immunoglobulin-like transcript 3 gene with single nucleotide polymorphism (SNP) locations indicated by coloured lines above the gene, yellow indicates an intronic SNP, green indicates a synonymous SNP, and red indicates a coding change SNP. Ig 1 and Ig 2, immunoglobulin-like domains 1 and 2; Stalk, short peptide sequence between TM and Ig 2; TM, transmembrane sequence.
Table 1.
Coding change single nucleotide polymorphisms in immunoglobulin-like transcript 3 with a reported minor allele frequency >5% and corresponding output from PolyPhen2 prediction algorithm
| SNP rs# | aa Position | aa Change | Score | Sensitivity | Specificity |
|---|---|---|---|---|---|
| rs28366008 | 5 | Leu-Phe | 0.000 | 1.00 | 0.00 |
| rs11540761 | 18 | Ser-Arg | 0.953 | 0.72 | 0.93 |
| rs731170 | 223 | Gly-Asp | 0.000 | 1.00 | 0.00 |
| rs11574576 | 335 | Asp-Asn | 0.002 | 0.99 | 0.28 |
| rs1048801 | 413 | Arg-Gln | 0.820 | 0.79 | 0.91 |
aa Change, the aa change that occurs with the aa encoded from the reference allele listed first; aa Position, position in the amino acid (aa) sequence of ILT3; SNP rs#, polymorphism unique number designation; Score, Sensitivity, and Specificity, statistics derived from the PolyPhen2 algorithm to predict the likelihood that the aa change disrupts protein structure, with higher numbers indicating a higher likelihood (Scores range from 0 to 1.0, indicating a very low likelihood and a very high likelihood of a damaging change, respectively).
The rs11540761 (G/T, coding change from Ser to Arg at position 18) and rs1048801 (A/G, coding change from Arg to Gln at position 413) SNPs were predicted to be probably and possibly damaging respectively, and were chosen for genotyping studies.
ILT3 Rs11540761 (Ser18Arg) associates with lower ILT3 levels on DCs
We used flow cytometry to quantitate ILT3 expression levels on blood-derived DCs of 18 SLE patients and 10 controls (see supplementary figure S2). There was a non-significant trend toward greater ILT3 expression in SLE patients (figure 2A,B). We had expected to see a larger difference, as ILT3 is induced by type I IFN, and many patients had high serum type I IFN activity at the time of blood draw (figure 2C). To determine if serum levels of IFN-α impacted ILT3 expression on DCs, we compared ILT3 levels on DCs of SLE patients with high versus low serum type I IFN activity (see Methods) at time of blood sampling but found no difference (figure 2A,B). We genotyped the SLE patients for the two ILT3 polymorphisms noted above, and found that the rs11540761 allele was associated with reduced ILT3 levels on MDCs (p=0.021) and a similar non-significant trend was observed in PDCs (figure 2D). The rs1048801 SNP did not influence ILT3 surface expression (figure 2E). ILT3 levels on DCs did not differ with SLE clinical characteristics, gender, age, or ancestral background (data not shown).
Figure 2.
Immunoglobulin-like transcript 3 (ILT3) expression levels on dendritic cells of healthy controls and systemic lupus erythematosus (SLE) patients. ILT3 expression levels on plasmacytoid dendritic cells (PDCs) and monocytoid dendritic cells (MDCs) was determined by flow cytometry as described in Materials and Methods. ILT3 levels on (A) PDCs and (B) MDCs are similar between healthy controls, SLE patients, and in SLE patient subgroups with high or low serum type I IFN activity. (C) Serum type I IFN activity of patients sampled for data in this figure. ILT3 levels on (D) PDCs and (E) MDCs vary between SLE patients when data is stratified by ILT3 genotype with the rs11540761 single nucleotide polymorphism associating with significantly lower ILT3 levels on MDCs (coding change affecting amino acid position 18, G(Ser) or T(Arg)). Genotypes are shown as allelic combinations, for ex. GG, homozygous for G allele; GT, heterozygous, etc. No subjects were homozygous for rs11540761 in the flow cytometry experiments. Specific fluorescent intensities (SFI) values equal the fold increase in geometric mean fluorescence intensities of cells incubated with anti-ILT3 IgG1-PE to cells incubated with IgG1-PE. Mean+SD are shown. SFI values were compared by Student’s unpaired t-test. N, number of subjects.
ILT3 coding-change polymorphisms associate with altered circulating cytokine profiles in SLE
We found that the ILT3 rs11540761 polymorphism was more frequently present in SLE patients with high serum type I IFN activity compared with SLE with low serum type I IFN activity (table 2).
Table 2.
rs11540761 and rs1048801 allele frequencies in systemic lupus erythematosus patients and controls, stratified by ancestral background and high versus low serum type I IFN activity
| Subject group | rs11540761 MAF | High versus low IFN | Meta-analysis high versus low IFN | rs1048801 MAF | High versus low IFN | Meta-analysis high versus low IFN |
|---|---|---|---|---|---|---|
| EA Controls n=116 | 0.181 | – | – | 0.431 | – | – |
| EA cases n=132 | 0.155 | – | – | 0.333 | – | – |
| High IFN EA cases n=56 | 0.212 | p=0.031 OR=2.05 | p=0.016 | 0.384 | p=0.13 OR=1.48 | p=0.14 |
| Low IFN EA cases n=76 | 0.116 | 0.296 | ||||
| High IFN HA cases n=44 | 0.350 | p=0.052 OR=2.26 | 0.432 | p=0.25 OR=1.46 | ||
| Low IFN HA cases n=35 | 0.192 | 0.343 |
EA, European-American ancestry; HA, Hispanic-American ancestry; MAF, minor allele frequency.
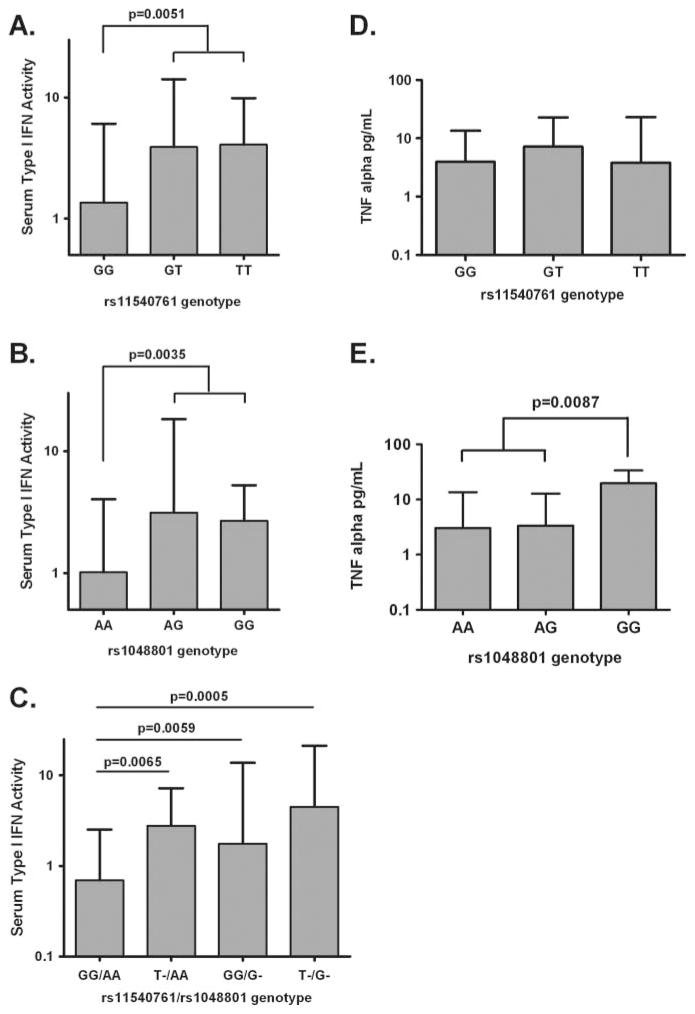
This finding may explain the lack of correlation between serum type I IFN activity levels and ILT3 expression in our SLE cohort, as the low expression allele is more common in SLE patients with high serum type I IFN. A similar non-significant trend was observed with the rs1048801 polymorphism, in which high IFN patients were more likely to carry the allele predicted to disrupt protein folding (table 2). When we analysed quantitative serum type I IFN activity in SLE patients, both functional ILT3 polymorphisms were independently associated with an increase in circulating type I IFN (figure 3). SLE patients homozygous for the rs1048801 polymorphism had higher serum TNF-α than other SLE patients (figure 3). The rs11540761 polymorphism was not associated with serum TNF-α levels.
Figure 3.
Serum type I IFN activity and TNF-α levels in systemic lupus erythematosus (SLE) patients in relation to immunoglobulin-like transcript 3 (ILT3) genotype. SLE patients were stratified by ILT3 genotype at rs11540761 (coding change affecting amino acid position 18, G(Ser) or T(Arg)) and by ILT3 rs1048801 (coding change affecting amino acid position 413, A(Arg) or G(Gln)). Genotypes are shown as allelic combinations, for ex. GG, homozygous for G allele, GT, heterozygous, etc. p Values calculated by using the Mann-Whitney U test. Cytokine data are shown as median+ interquartile range. N, number of subjects.
Crosslinking ILT3 does not inhibit PDC secretion of IFN-α
To determine if crosslinking ILT3 blocks PDC IFN-α secretion, we incubated PDCs from controls with anti-ILT3 Ab, anti-ILT7 Ab, and control IgG1, plated them on F(ab′)2 anti-mouse IgG coated wells and activated them with the TLR9 agonist, ODN 2216. ODN 2216 contains unmethylated cytosine-phosphate-guanine (CpG) motifs, mimics viral DNA, and potently induces PDC IFN secretion.30 In addition to ILT3, we studied the effects of crosslinking PDC-expressed ILT7. ILT7 belongs to the ILT family but unlike ILT3 contains ITAMs within its cytoplasmic tail. Crosslinking ILT7 inhibits TLR9-induced IFN secretion by PDCs.31,32 Crosslinking ILT7, but not ILT3 blocked PDC secretion of IFN (figure 4).
Figure 4.
IFN-α production by TLR9-activated plasmacytoid dendritic cells (PDCs) treated with immunoglobulin-like transcript 3 (ILT3) and immunoglobulin like transcript 7 (ILT7) crosslinking Abs. PDCs were treated with Abs to ILT3 and ILT7, or with control IgG, activated with CpG Type A DNA plus IL-3, and IFN-α activity was assayed in supernatants collected 18 h later. Crosslinking ILT7, but not ILT3, inhibits IFN-α secretion by PDCs. Mean±SD of four experiments is shown.
DISCUSSION
We demonstrate one coding change polymorphism in the extra-cellular portion of ILT3 (rs11540761) which reduces surface expression on MDCs in humans. This polymorphism may interfere with protein folding sufficiently so that the protein exits the endoplasmic reticulum at a reduced rate. The rs11540761 SNP would not be expected to alter gene transcription or mRNA abundance. This polymorphism may interfere with receptor-signalling, either by decreasing the receptor affinity for its ligand, or by interfering with receptor dimerisation. The fact that we also observe increased circulating type I IFN in the context of the low expression allele supports the biological relevance of this ILT3 allele in human SLE.
We found a second coding change polymorphism in the cytoplasmic domain of ILT3 (rs1048801) that is associated with increases in both type I IFN and TNF-α in SLE patients. This polymorphism does not change a known conserved phosphorylated amino acid nor was it associated with altered ILT3 levels on DCs, but might instead impact interactions between the receptor and downstream signalling proteins. It is interesting that the rs1048801 SNP associated with both serum type I IFN activity and serum levels of TNF-α, since we find that levels of both cytokines are high and correlated in SLE patients.15 While a heterozygous change was sufficient to result in a difference in type I IFN, only patients homozygous for this polymorphism demonstrated a difference in TNF-α. This may reflect differential regulation of the two cytokines via ILT3 in cells of the innate immune system. A caveat regarding our results is that we do not include a comprehensive evaluation of all genetic variants in the ILT3 locus, and it is possible that other untyped causal elements are contributing to our findings.
DCs which express heightened levels of ILT3 negatively influence immune responses through multiple pathways. Type I IFN and 1,25-dyhdroxyvitamin D3 treated MDCs upregulate ILT3 expression and are tolerogenic.20,21 MDCs that express high ILT3 levels also induce regulatory T cells.16,33 The ILT3 genotypic dependent differences we observe in serum cytokines of SLE patients may result from the convergence of several immunoregulatory pathways that fail to function normally in these individuals.
Our data suggest that ILT3 does not function as a simple feedback regulator of type I IFN production in PDCs suggesting that the influence of the polymorphisms we observe are likely complex and involve multiple cell types. The modulatory effects of ILT3 on PDC function may be best explored in conjunction with the activation of other receptors expressed by PDCs such as FcγRIIa. Based upon the results of our study, it appears that deficiency of normal negative regulation by ILT3 is involved in the propagation of human lupus. Our findings identify ILT3 has a target gene for the development of therapeutic approaches for the treatment of SLE.
Supplementary Material
Acknowledgments
Funding Supported by NIH Grants R01 AR060861, K08 AI083790, P30 DK42086, NIAID Clinical Research Loan Repayment AI071651, NIH CTSA Core Subsidy Grant, CTSA Pilot Grants from UL1 RR024999, Lupus Research Institute Novel Research Grant, and an Alliance for Lupus Research Target Identification in Lupus Grant.
Footnotes
Contributors All authors contributed to the collection and analysis of data, writing of the manuscript and approved the final submitted work.
Competing interest None.
Ethics approval University of Chicago Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement The authors would be happy to share data regarding our experimental technique or the results presented in this paper with other investigators upon request.
Additional supplementary files are published online only. To view these files please visit the journal online (http://dx.doi.org/10.1136/annrheumdis-2012-202024).
References
- 1.Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40:42–9. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco P, Palucka AK, Gill M, et al. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–3. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 3.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–15. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niewold TB, Hua J, Lehman TJ, et al. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niewold TB, Clark DN, Salloum R, et al. Interferon alpha in systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:948364. doi: 10.1155/2010/948364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko K, Franek BS, Marion M, et al. Genetic ancestry, serum interferon-alpha activity, and autoantibodies in systemic lupus erythematosus. J Rheumatol. 2012;39:1238–40. doi: 10.3899/jrheum.111467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim T, Kanayama Y, Negoro N, et al. Serum levels of interferons in patients with systemic lupus erythematosus. Clin Exp Immunol. 1987;70:562–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Kirou KA, Lee C, George S, et al. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 9.Weckerle CE, Franek BS, Kelly JA, et al. Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum. 2011;63:1044–53. doi: 10.1002/art.30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31:887–92. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson T, Kariuki SN, Franek BS, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kariuki SN, Franek BS, Kumar AA, et al. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R151. doi: 10.1186/ar3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niewold TB, Kelly JA, Kariuki SN, et al. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:463–8. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agik S, Franek BS, Kumar AA, et al. The autoimmune disease risk allele of UBE2L3 in African American patients with systemic lupus erythematosus: a recessive effect upon subphenotypes. J Rheumatol. 2012;39:73–8. doi: 10.3899/jrheum.110590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weckerle CE, Imbuka D, Franek BS, et al. Large scale analysis of tumor necrosis factor alpha levels in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2947–52. doi: 10.1002/art.34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, Ciubotariu R, Manavalan JS, et al. Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat Immunol. 2002;3:237–43. doi: 10.1038/ni760. [DOI] [PubMed] [Google Scholar]
- 17.Cella M, Dohring C, Samaridis J, et al. A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J Exp Med. 1997;185:1743–51. doi: 10.1084/jem.185.10.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu HK, Rentero C, Raftery MJ, et al. Leukocyte Ig-like receptor B4 (LILRB4) is a potent inhibitor of FcgammaRI-mediated monocyte activation via dephosphorylation of multiple kinases. J Biol Chem. 2009;284:34839–48. doi: 10.1074/jbc.M109.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suciu-Foca N, Feirt N, Zhang QY, et al. Soluble Ig-like transcript 3 inhibits tumor allograft rejection in humanized SCID mice and T cell responses in cancer patients. J Immunol. 2007;178:7432–41. doi: 10.4049/jimmunol.178.11.7432. [DOI] [PubMed] [Google Scholar]
- 20.Manavalan JS, Rossi PC, Vlad G, et al. High expression of ILT3 and ILT4 is a general feature of tolerogenic dendritic cells. Transpl Immunol. 2003;11:245–58. doi: 10.1016/S0966-3274(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 21.Jensen MA, Yanowitch RN, Reder AT, et al. Immunoglobulin-like transcript 3, an inhibitor of T cell activation, is reduced on blood monocytes during multiple sclerosis relapses and is induced by interferon beta-1b. Mult Scler. 2010;16:30–8. doi: 10.1177/1352458509352794. [DOI] [PubMed] [Google Scholar]
- 22.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 23.Hua J, Kirou K, Lee C, et al. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54:1906–16. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 24.Niewold TB, Rivera TL, Buyon JP, et al. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 2008;58:541–6. doi: 10.1002/art.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niewold TB, Kariuki SN, Morgan GA, et al. Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum. 2009;60:1815–24. doi: 10.1002/art.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X, Reder NP, Yanamandala M, et al. Type I interferon signature is high in lupus and neuromyelitis optica but low in multiple sclerosis. J Neurol Sci. 2012;313:48–53. doi: 10.1016/j.jns.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bave U, Magnusson M, Eloranta ML, et al. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J Immunol. 2003;171:3296–302. doi: 10.4049/jimmunol.171.6.3296. [DOI] [PubMed] [Google Scholar]
- 28.Grouard G, Rissoan MC, Filgueira L, et al. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–11. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang CC, Silvia EA, Ho EK, et al. Polymorphism and linkage disequilibrium of immunoglobulin-like transcript 3 gene. Hum Immunol. 2008;69:284–90. doi: 10.1016/j.humimm.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Kerkmann M, Rothenfusser S, Hornung V, et al. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol. 2003;170:4465–74. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 31.Cao W, Rosen DB, Ito T, et al. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J Exp Med. 2006;203:1399–405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao W, Bover L, Cho M, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206:1603–14. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenk M, Scheler M, Koch S, et al. Tryptophan deprivation induces inhibitory receptors ILT3 and ILT4 on dendritic cells favoring the induction of human CD4+ CD25+ Foxp3+ T regulatory cells. J Immunol. 2009;183:145–54. doi: 10.4049/jimmunol.0803277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.