Abstract
Osteosarcoma is the most common primary bone malignancy of adolescents and young adults. In order to better understand the genetic etiology of osteosarcoma, we performed a multi-stage genome-wide association study (GWAS) consisting of 941 cases and 3,291 cancer-free adult controls of European ancestry. Two loci achieved genome-wide significance: rs1906953 at 6p21.3, in the glutamate receptor metabotropic 4 [GRM4] gene (P = 8.1 ×10-9), and rs7591996 and rs10208273 in a gene desert on 2p25.2 (P = 1.0 ×10-8 and 2.9 ×10-7). These two susceptibility loci warrant further exploration to uncover the biological mechanisms underlying susceptibility to osteosarcoma.
Main Text
Osteosarcoma is the most common primary malignant bone tumor in children and young adults, affecting approximately four persons per million per year in the U.S.1. The peak incidence correlates with the pubertal growth spurt, occurring earlier in females than in males. It is more common at sites of rapid bone growth. Tall stature and high birth weight are proven risk factors2, and osteosarcoma is a syndrome-associated malignancy in the Li-Fraumeni syndrome, Rothmund-Thomson syndrome, and hereditary retinoblastoma cancer susceptibility syndromes. Several small case-control studies have reported preliminary associations of common genetic variants with osteosarcoma risk in biologically-plausible pathways 3-11, e.g., growth and DNA repair, but statistical power has been limited by small sample sizes12.
We developed an international, multi-institutional collaborative study of osteosarcoma to conduct a GWAS (Supplementary Table 1). Germline genomic DNA was extracted from either blood or buccal cells drawn from osteosarcoma case series using standard methods. Participating subjects provided informed consent under the auspices of local Institutional Review Boards. Control subjects were selected from previously-scanned cancer-free adults over the age of 55 years drawn from large case control and cohort studies as per an approach successfully used for Ewing sarcoma13 and pediatric acute lymphoblastic leukemia (ALL)14.
Genotyping of all cases was conducted using the Illumina OmniExpress SNP microarray over time resulting in two parts, known as Stage 1A and 1B, due to the staggered receipt of samples in this multi-institutional study design (Supplementary Figure 1, Online Methods). Stage 1A genotyped 910 available osteosarcoma cases from five studies. Based on quality control filtering (including locus or sample missing rates, sample heterozygosity, Hardy-Weinberg equilibrium, and sex verification) and assessment of underlying population substructure (STRUCTURE and principal components analyses), 596 cases of European ancestry were advanced to the primary GWAS analysis (Supplementary Figures 2 and 3); 97 African American and 99 Hispanic cases were identified and excluded from the present analysis. A later download of samples, known as Stage 1B consisting of 218 osteosarcoma cases, was also scanned with the OmniExpress SNP microarray (Supplemental Table 1). The same quality filtering and population substructure analyses were applied, resulting in an additional 98 cases of European ancestry for the final analysis. The choice of SNPs for follow-up was generated based on the first set (Stage 1A) but for the final analysis, we present the results of the full set of 694 cases (namely, the combined Stages 1A and 1B).
Overall, 698,968 SNPs passed quality control metrics for the full set of 694 cases for final analysis. The number of SNPs overlapping with pooled controls was 510,856, 510,856, 310,384 and 304,092 for the USA, Spain, Italy and UK components, respectively, and were used in the association analyses (see Online Methods).
We combined already scanned controls drawn from two large cohort studies, Prostate, Lung, Colon and Ovarian Cancer Prevention Trial [PLCO]15 and the American Cancer Society Cancer Prevention Study-II [CPSII]16 scanned on Illumina Omni 2.5M array and three European studies (i.e., Spanish Bladder Cancer Study17 on the HumanHap 1M SNP microarray, Environment and Genetics Lung Cancer Etiology Study [EAGLE]18/on the HumanHap 550 SNP microarray and the Wellcome Trust Case Control Consortium [WTCCC]19 with the HumanHap 550 SNP microarray). We selected 2,703 controls based on comparable quality control metrics for comparison with the scanned cases. A systematic assessment of the underlying population substructure was conducted as described above to remove highly admixed individuals (non-European admixture of >20%) from a model to investigate cases of European background (Supplementary Figures 2 and 3).
Association analyses were performed using a 1-degree of freedom trend test adjusted by sex and eigenvectors. Optimized TaqMan assays were designed based on SNP associations observed in the first set of cases in the scan, known as Stage 1A (Supplementary Table 2). This set of 30 SNPs was genotyped in a follow-up set of 247 osteosarcoma cases (Supplemental Table 1) and 588 controls drawn from PLCO and NHS. A fixed effect meta-analysis was applied to the combined results of the two scanned sets comprising Stage 1 (596 cases in Stage 1A and 98 cases in Stage 1B yielding a total of 694 total, unique cases) with the replication stage of 30 SNPs with P <1.0 × 10-4.
In a fixed effect meta-analysis of all cases and controls of European ancestry, 941 cases and 3,291 cancer-free adult controls, we observed two regions that achieved genome-wide significance on chromosomes 6p21.3 and 2p25.2 (Table 1 and Supplementary Table 2).
Table 1. Summary of combined association results for 941 cases and 3,291 controls.
| dbSNP locus | Positiona | Alleleb | Stage | Cases | Controls | P | OR (95% CI) |
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| EAF | EAF | ||||||
| rs1906953 | 34144424 | C/T | Discovery | 0.19 | 0.15 | 4.0 × 10-05 | 1.45 (1.21-1.73) |
| 6p21.31 | Replication | 0.21 | 0.12 | 6.5 × 10-06 | 2.00 (1.47-2.72) | ||
| Combined | 8.0 × 10-09 | 1.57 (1.35-1.83) | |||||
| rs7591996 | 6378872 | A/C | Discovery | 0.50 | 0.45 | 4.8 × 10-05 | 1.30 (1.15-1.47) |
| 2p25.2 | Replication | 0.55 | 0.43 | 5.7 × 10-06 | 1.69 (1.35-2.13) | ||
| Combined | 1.0 × 10-08 | 1.39 (1.23-1.54) | |||||
| rs10208273 | 6441445 | G/A | Discovery | 0.36 | 0.30 | 3.5 × 10-06 | 1.37 (1.2-1.56) |
| 2p25.2 | Replication | 0.34 | 0.29 | 2.7 × 10-02 | 1.31 (1.03-1.67) | ||
| Combined | 2.9 × 10-07 | 1.35 (1.21-1.52) | |||||
| rs17206779 | 64483533 | T/C | Discovery | 0.53 | 0.47 | 2.2 × 10-05 | 1.32 (1.16-1.49) |
| 5q12.3 | Replication | 0.54 | 0.46 | 6.7 × 10-03 | 1.37 (1.09-1.72) | ||
| Combined | 5.1 × 10-07 | 1.33 (1.19-1.47) |
The discovery stage consists of the combined scans of Stage 1A (596 cases) and Stage 1B (98 cases) for a total of 694 scanned osteosarcoma cases and 2,703 cancer-free adult controls, all of European ancestry. The independent replication set consisted of 247 osteosarcoma cases genotyped by TaqMan and 588 controls previously scanned on Illumina Omni2.5 or OmniExpress. The combined results report the data from the meta analysis of Stage 1and the TaqMan replication (941 cases and 3,291 controls).
Abbreviations: EAF, effect (coded) allele frequency; OR, odds ratio; CI, confidence interval.
Build 36 position;
Reference/Effect alleles.
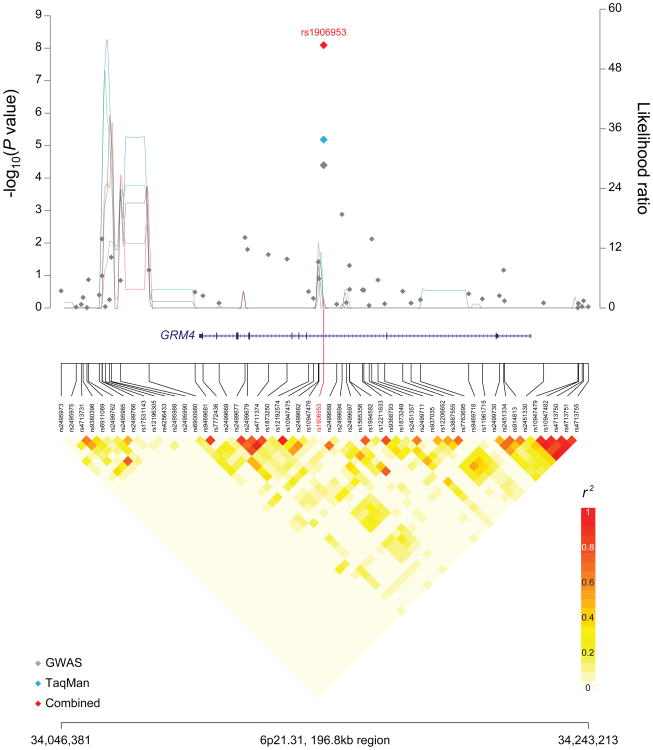
The locus on 6p21.3, marked by rs1906953 is associated with susceptibility to osteosarcoma (P=8.0×10-9, odds ratio (OR) 1.57, 95% confidence interval (CI) 1.35-1.83) (Table 1). The SNP, rs1906953, is located in intron 7 (IVS 7-32066 C>T) of the glutamate receptor metabotropic 4 (GRM4) gene (Figure 1A). Our most significant SNP resides within the gene, GRM4, which is within a distinct haplotype block from the HLA Class II region, which is more than 1Mb telomeric on 6p21.3. It is also notable that between the HLA Class II region and our signal for OS is a distinct region containing BAK1, which in published GWAS studies has been conclusively associated with platelet counts20-22, chronic lymphocytic leukemia23, and testicular germ cell tumors24.
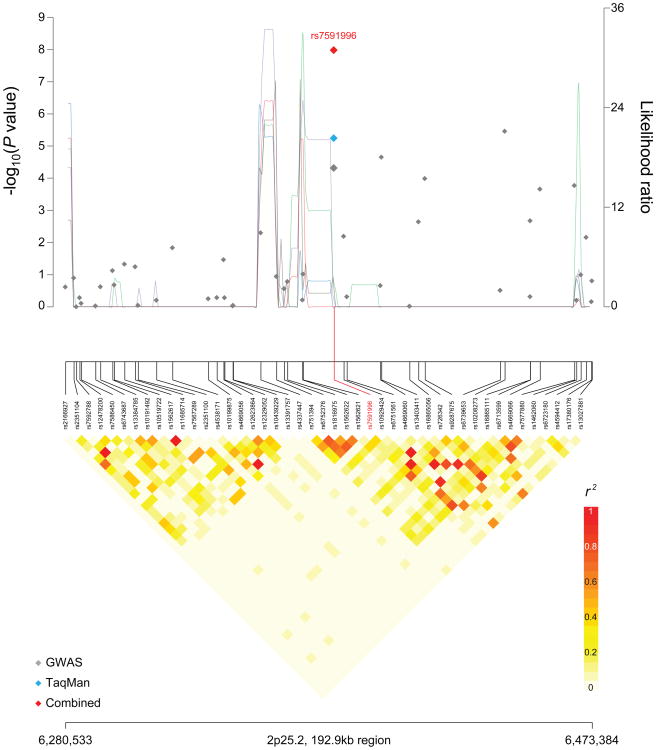
Figure 1. Regional plots of loci associated with osteosarcoma.
Regional plots of association results, recombination hotspots and linkage disequilibrium for the (a) 6p21.31:34,046,381–34,243,213 and (b) 2p25.2:6,280,533–6,473,384 osteosarcoma susceptibility loci. Association results from a trend test in –log10P values (y axis, left; gray diamonds, combined GWAS result; sky blue diamonds, TaqMan result; red diamonds, combined result) of the SNPs are shown according to their chromosomal positions (x axis). Linkage disequilibrium structure based on controls (n=2,703) was visualized by snp.plotter software. The line graph shows likelihood ratio statistics (y axis, right) for recombination hotspot by SequenceLDhot software and 5 different colors represent 5 tests of 100 controls without resampling. Physical locations are based on NCBI human genome build 36. Gene annotation was based on the NCBI RefSeq genes from the UCSC Genome Browser.
The reference allele (C) of GRM4 is highly conserved and its allele frequency is higher in more ancient populations; the global minor allele frequency (MAF) is 0.29 in the 1000 Genomes Project (Phase 1 genotype data from 1094 individuals25) data, with a greater frequency in individuals of Asian or African ancestry (MAF 0.46 and 0.59, respectively). While there are four surrogates in individuals of European background (r2>0.6 and within ±500kb) in the 1000 Genomes data, the index SNP, rs1906953, maps to a DNaseI hypersensitivity region in the Encyclopedia of DNA Elements (ENCODE)26 data set, raising the distinct possibility that the variant is within a region that is accessible to open chromatin (Supplementary Table 3) and could manifest active regulatory elements. One intronic surrogate SNP (rs73410895) is predicted to alter known regulatory motifs (Supplementary Table 3).
GRM4 is a plausible candidate gene that has been implicated in intracellular signaling and inhibition of the cyclic AMP (cAMP) signaling cascade. In mice, a cAMP-dependent protein kinase (Prkar1α) is an osteosarcoma tumor suppressor gene involved in tumorigenesis27,28, suggesting the cAMP pathway is important in osteosarcoma. Although, glutamate signaling is best characterized in the central nervous system (CNS), where it is known to be involved in the excitability of gonadotropin releasing hormone neurons, it also occurs in bone29. The GRM4 receptor is expressed in bone osteoblast (bone-building) and osteoclast (bone-resorbing) cells, suggesting that glutamate signaling is involved in cell differentiation and regulation of bone formation and resorption30. GRM4 is expressed in human osteosarcoma cells31 and associated with poor prognosis in colorectal cancer32, pediatric CNS tumors33, rhabdomyosarcoma, and multiple myeloma34, as well as cancer cell proliferation in vitro35.
A second signal was observed in an intergenic region on chromosome 2p25.2 (Figure 1B); rs7591996 (OR 1.39, 95% CI 1.23-1.54, P=1.0 × 10-8) achieved genome-wide significance. A second SNP, rs10208273, was moderately correlated with the first signal (r2=0.32 in HapMap 3 release 2) and approached genome-wide significance (OR 1.35, 95% CI 1.21-1.52, P=2.9 × 10-7) (Figure 1B and Table 1). The risk allele frequency for rs7591996 in our cases of European ancestry was 0.50 and 0.45 for the controls. rs7591996 has a global MAF of 0.38 in the 1000 Genomes data; the risk allele is the minor allele in CEU individuals and the major allele in individuals of Asian or African ancestry (MAF 0.35 and 0.14, respectively). Both rs7591996 and rs10208273 occur in a region of relatively low linkage disequilibrium (Figure 1B). The 1000 Genomes data for European populations documents at least 27 surrogate SNPs (r2>0.6 and within ±500kb). Based on ENCODE26 data, the two index SNPs do not localize to a region of active regulatory elements or transcription factor binding sites. However, several of the surrogate SNPs alter known regulatory motifs and transcription factor binding sites (Supplementary Table 3) and may affect regulation. These findings suggest that further sequencing and fine-mapping will be required to determine which variants will be optimal for functional studies needed to explain the direct association.
Finally, we report a third locus that is promising but does not yet achieve genome-wide significance, rs17206779, (OR 0.75, 95% CI 0.68 to 0.84, P=5.1×10-7) located in the ADAM metallopeptidase with thrombospondin type 1 motif, 6 (ADAMTS6) gene (g.64447777C>T) on chromosome 5q12.3. It is notable that variation in genes encoding members of the ADAMTS protein family have been associated with height36, a known risk factor for osteosarcoma2. Further data are required to confirm this locus, and then fine-map the region prior to conducting functional studies.
To further explore the two significant regions reported in this manuscript, we imputed SNPs based on the 1000 Genomes data (March 2012 release) together with the DCEG Imputation Reference Set version 137 using the IMPUTE2 program38 across a 1 Mb region centered on each index SNP (see Online Methods). A subsequent association analysis did not identify new signals that are substantially stronger than the genotyped SNPs for both 2p25.2 and 5q12.3 regions (Supplementary Figure 4). Although there appear to be stronger signals in the imputed data for the HLA Class II region (Supplementary Figure 4a), there is a strong recombination hotspot separating it from the region marked by the index SNP rs1906953. In this regard, it will be critical to pursue more detailed mapping and genotyping of the HLA class II region to determine if there is an independent signal in addition to that which resides in the GRM4 gene.
An assessment of loci previously reported in candidate gene association studies3-12 did not reveal that the reported loci approached genome-wide significance (Supplementary Table 4). It is notable that the SNPs in the 8q24.3 locus as well as the multi-cancer 8q24.21 region appear to be promising candidates10; the strongest signal of a set of highly correlated SNPs on 8q24.21, rs11777807, demonstrated a P= 4.6 × 10-5, and rs369051 on 8q24.3 had a P = 1.16 × 10-4. Further sample sizes are needed to determine if these are stable or perhaps due to chance.
We have conclusively identified two susceptibility regions, on chromosome 2p25.2 and 6p21.3, the latter harboring a plausible candidate gene, GRM4, in a multistage GWAS of osteosarcoma. It is noteworthy that the loci we have identified demonstrate estimated odds ratios greater than 1.5, values greater than those observed for most adult cancer GWAS. Our findings are consistent with the high risk estimates reported for Ewing's sarcoma13 and ALL14, other rare pediatric tumors. Further investigation of these associated loci is warranted to uncover the biological mechanisms underlying susceptibility to osteosarcoma.
Online Methods
Genome-wide SNP Genotyping
Genome-wide SNP genotyping of osteosarcoma cases was conducted using the Illumina OmniExpress Beadchip at the NCI Cancer Genomics Research Laboratory (CGR) in the Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Bethesda, USA. Genotype analysis occurred in two stages because of the sequential receipt of samples. Stage 1A consisted of 910 unique cases from the Children's Oncology Group (COG), Toronto Study, PamplonaCUN, Rizzoli (Italy), Ankara (Turkey) and University College London (UCL). Adult, cancer-free controls were drawn from previously scanned studies in the US (add references), namely, the Prostate, Lung, Colon and Ovary Prevention Trial (PLCO) and the Cancer Prevention Study-II (CPS-II) of the American Cancer Society, both scanned on the Omni 2.5 M SNP microarray; European controls were drawn from the EAGLE (Italy) scanned on the HumanHap550, the Welcome Trust Case-Control Cohort-UK (WTCCC-UK) scanned on the Human Hap550 and the NCI-Spanish Bladder Cancer Study, scanned on the Human Hap 1 M SNP microarray chip. Stage 1B consisted of unique 218 cases scanned on the Omni Express Beadchip drawn from UCL, Rizzoli, PamplonaCUN studies plus two additional studies from the PeterMac (Australia) and Brazil. Cancer-free adult controls were drawn from the PLCO and the Nurses Health Study (NHS), scanned on the Illumina 2.5 M Omni and the OmniExpress, respectively.
Each participating study obtained informed consent from study participants and approval from its Institutional Review Board (IRB) for this study and obtained IRB certification permitting data sharing in accordance with the NIH Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies (GWAS). The CGEMS data portal provides access to individual level data from the NCI scan ONLY to investigators from certified scientific institutions after approval of their submitted Data Access Request.
Quality Control Assessment
Systematic quality control was conducted which included QC steps specific for the performance of different arrays at distinct times. For SNP assays, exclusions included those with less than 90% of completion rate, and SNPs with extreme deviation from fitness for Hardy-Weinberg proportion (P <1×10-7). There were 29 duplicated cases in Stages 1A and 1B; concordance rates were 99.96%.
Analysis of Stage 1A
In the QC analysis of stage 1A, samples were excluded based on: 1) completion rates lower than 94% (n=28 samples); 2) abnormal heterozygosity values of less than 20% or greater than 31% (n=8); 3) expected duplicates (n=23 pairs); 4) abnormal X chromosome heterozygosity (n=1); 5) phenotype exclusions (due to ineligibility or incomplete information) (n=57). Genotypes for all subject pairs were also computed for close relationships (1-2nd degree) using GLU qc.ibds module with an IBD0 threshold of 0.70; no first-degree relatives were identified in the cases scanned. Utilizing a set of 12,000 un-linked SNPs (pair-wise r2 <0.004) common to the GWAS chips used herein33, 264 subjects with less than 80% European ancestry were excluded based on STRUCTURE analysis34 and PCA35; majority of these represent 97 cases of African American ancestry and 99 of Hispanic ancestry. The final participant count for the association analysis for Stage 1A was 596 cases and 2,703 controls of European ancestry. 698, 968 SNPs were available after the QC filtering. The number of SNPs overlapping with pooled controls are follows, 510,856, 510,856, 310,384 and 304,092 for the USA, Spain, Italy and UK components, respectively, were used in the downstream association analyses. PCA analyses per components did not reveal significant differences between cases and controls using the same 12,000 SNPs described above.
Analysis of Stage 1B
Similar quality control metrics were applied to the second set of additional scanned cases, known as Stage 1B. For the current analysis, an additional 98 cases were added to the association analysis of individuals of European ancestry.
Quality control metrics for previously scanned cancer-free adult controls
The pooled PLCO controls were scanned on the Illumina Human Omni2.5 array, we excluded samples with missing rate > 6%; excluded SNPs with missing rate > 10%; excluded samples with mean heterozygosity > 21% or < 16%; excluded one from each of cryptic related pair; excluded non-CEU admixed individuals based on the STRUCTURE analysis. The pooled SPBC controls were scanned on Illumina 1M SNP array and the genotype QC were previously described17; the pooled EAGLE controls were scanned on Illumina 550K SNP array and the genotype QC were previously published18; the pooled WTCCC controls were previously described39.
Replication and TaqMan genotyping
A follow-up analysis of the 30 SNPs selected for replication (based on Stage 1A) was conducted using the TaqMan genotyping assays (ABI, Foster City, CA) that were optimized in the CGR pipeline. A total of 247 cases and 550 controls were analyzed; the studies included 99 histologically confirmed osteosarcoma cases and 65 hospital-based cancer-free pediatric controls from the National Osteosarcoma Etiology Study Group plus 148 cases drawn from the studies used in the scan in which there was insufficient DNA for scanning. Cancer-free controls were also selected from the PLCO and NHS studies (previously analyzed on the Omni2.5M and OmniExpress, respectively).
Statistical analysis
Associations between the SNPs and the risk of osteosarcoma were estimated using unconditional logistic regression by the odds ratio (OR) and 95% confidence interval (CI) using the multivariate unconditional logistic regression assuming a trend genetic model (the effect of the variant by log-additive model with 1 degree of freedom). PCA analysis revealed four significant (p<0.05) eigenvectors when included in the NULL model. The main effect model was adjusted by sex, country, four eigenvectors, based on significance with p<0.05 observed in the NULL model. For the replication studies, the analysis was adjusted for sex only.
For stage 1 (A + B), in addition to the joint analysis detailed as above, a fixed effect meta-analysis was performed to combine results for each country (component) to facilitate filtering out artifacts by checking for the heterogeneity across countries. Similar meta-analysis was also performed to combine scan (discovery) association results with taqman (replication) association results for those 30 SNPs.
The estimated inflation factors, λ and adjusted λ1000 of the test statistic for the combined Stage 1A and 1B were 1.036 and 1.033.
Recombination hotspots were identified in the vicinity of osteosarcoma associated loci on 6p21.31 and 2p25.2 using SequenceLDhot40, a program that uses approximate marginal likelihood method41 and calculates likelihood ratio statistics at a set of possible hotspots. We tested 5 unique sets of 100 control samples drawn from controls. PHASE v2.1 program was used to calculate background recombination rates42,43 and LD heatmap was visualized in r2 using snp.plotter program44.
Imputation
In order to further interrogate the loci associated with osteosarcoma, we imputed additional SNPs within 1 Mb on either side of the implicated SNPs using IMPUTE2 software and the reference data from both the 1000 Genomes project13 and the DCEG Imputation Reference Set version 137. SNPTEST was used to analyze the posterior SNP dosages from IMPUTE2, adjusting for sex, country, and four eigenvectors, as described above45.
Data Analysis
Data analysis and management was performed with GLU (Genotyping Library and Utilities version 1.0), PLINK, Eigenstrat, IMPUTE2, and SNPTEST.
URLs
CGEMS portal: http://cgems.cancer.gov/
GLU: http://code.google.com/p/glu-genetics/
EIGENSTRAT: http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm
STRUCTURE: http://pritch.bsd.uchicago.edu/structure.html
PLINK: http://pngu.mgh.harvard.edu/∼purcell/plink/
SequenceLDhot: http://www.maths.lancs.ac.uk/∼fearnhea/Hotspot/
snp.plotter: http://cbdb.nimh.nih.gov/∼kristin/snp.plotter.html
PHASE v2.1: http://www.stat.washington.edu/stephens/phase/download.html
IMPUTE2: http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
SNPTEST: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
Supplementary Material
Acknowledgments
We thank Dr. Giovanna Maganoli for tissue banking, Dr. Marilù Fanelli for DNA isolation, and Dr. Cristina Ferrari for updating of clinicopathologic data at the Orthopaedic Rizzoli Institute. We thank Anthony Griffin and Diana Marsilio for data collection and Teresa Selander and the Biospecimen Repository staff at Mount Sinai Hospital, Toronto. We acknowledge the advice of Francisco Real at the Spanish National Cancer Research Centre, CNIO. We thank Francine Tesser Gamba at the Pediatric Oncology Institute, GRAACC-UNIFESP, São Paulo SP Brazil. We also thank the International Sarcoma Kindred Study, East Melbourne, Victoria, Australia.
Funding Support: This study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health; Bone Cancer Research Trust UK.
Research is supported by the Chair's Grant U10 CA98543 and Human Specimen Banking Grant U24 CA114766 of the Children's Oncology Group from the National Cancer Institute, National Institutes of Health, Bethesda, MD, USA. Additional support for research is provided by a grant from the WWWW (QuadW) Foundation, Inc. to the Children's Oncology Group.
This work was supported by grants to I.L.A. and J.S.W. from the Ontario Research Fund, and Canadian Foundation for Innovation.
This study was also supported by biobank grants from the Regione Emilia-Romagna, by the infrastructure and personnel of the Royal National Orthopaedic Hospital Musculoskeletal Research Programme and Biobank. Support was also provided to A.M.F. (UCL) by the National Institute for Health Research UCLH Biomedical Research Centre, and UCL Experimental Cancer Centre, funding from PI10/01580, FIS, ISCIII and CAN Programme “Tú eliges, tú decides” to Dr. Patino-Garcia and Dr. L. Sierrasesúmaga, and AECC project to F.L.
Footnotes
Author Contributions: Project design was carried out by S.A.S., S.J.C.
Sample collection and clinical characterization was performed by J.M.G.F, R.G., C.K., A.M.F., R.T., I.L.A, J.S.W., N.G., L.S., D.A.B., N.M., A.P.G., L.S., F.L., M.S., C.H., P.P., N.K., I.E.I., N.S., S.R.C.T., S.P., M.F.A., D.H., D.M.T., C.D., P.M., S.I.B., M.P.P., N.E.C., M.T., N.R., M.T.L., D.T.S., P.K., D.J.H., N.M., M.K., S.W., R.T., L.H., J.F.F, R.N.H
Genotyping was performed by K.J., C.C.C, M.Y., Z.W.
Statistical analysis was performed by Z.W., L.M.
The manuscript was written by S.A.S., L.M., Z.W, S.J.C., and reviewed by all co-authors.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Mirabello L, Troisi R, Savage S. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, et al. Height at diagnosis and birth-weight as risk factors for osteosarcoma. Cancer Causes Control. 2011;22:899–908. doi: 10.1007/s10552-011-9763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patio-Garcia A, Sotillo-Pieiro E, Modesto C, Sierrases-Maga L. Analysis of the human tumour necrosis factor-alpha (TNFalpha) gene promoter polymorphisms in children with bone cancer. J Med Genet. 2000;37:789–792. doi: 10.1136/jmg.37.10.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruza E, Sotillo E, Sierrasesúmaga L, Azcona C, Patiño-Garcia A. Analysis of polymorphisms of the vitamin D receptor, estrogen receptor, and collagen Iα1 genes and their relationship with height in children with bone cancer. J Pediatr Hematol Oncol. 2003;25:780–786. doi: 10.1097/00043426-200310000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Savage SA, et al. Germ-line genetic variation of TP53 in osteosarcoma. Pediatr Blood Cancer. 2007;49:28–33. doi: 10.1002/pbc.21077. [DOI] [PubMed] [Google Scholar]
- 6.Savage SA, et al. Analysis of genes critical for growth regulation identifies Insulin-like Growth Factor 2 Receptor variations with possible functional significance as risk factors for osteosarcoma. Cancer Epidemiol Biomarkers Prev. 2007;16:1667–1674. doi: 10.1158/1055-9965.EPI-07-0214. [DOI] [PubMed] [Google Scholar]
- 7.Koshkina N, et al. Exploratory analysis of Fas gene polymorphisms in pediatric osteosarcoma patients. J Pediatr Hematol Oncol. 2007;29:815–21. doi: 10.1097/MPH.0b013e3181581506. [DOI] [PubMed] [Google Scholar]
- 8.Toffoli G, et al. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin Cancer Res. 2009;15:3550–6. doi: 10.1158/1078-0432.CCR-08-2249. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, et al. Association between TGFBR1*6A and osteosarcoma: A Chinese case-control study. BMC Cancer. 2010;10:169. doi: 10.1186/1471-2407-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirabello L, et al. Genetic variation at chromosome 8q24 in osteosarcoma cases and controls. Carcinogenesis. 2010;31:1400–4. doi: 10.1093/carcin/bgq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirabello L, et al. A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer. 2011;11:209. doi: 10.1186/1471-2407-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage S, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postel-Vinay S, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 14.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–10. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 16.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothman N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landi M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium., W.T.C.C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum Mol Genet. 2013;22:1457–64. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gieger C, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qayyum R, et al. A Meta-Analysis and Genome-Wide Association Study of Platelet Count and Mean Platelet Volume in African Americans. PLoS Genet. 2012;8:e1002491. doi: 10.1371/journal.pgen.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slager S, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120:843–6. doi: 10.1182/blood-2012-03-413591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molyneux S, et al. Prkar1a is an osteosarcoma tumor suppressor that defines a molecular subclass in mice. J Clin Invest. 2010;120:3310–3325. doi: 10.1172/JCI42391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin K, et al. A transgenic mouse bearing an antisense construct of regulatory subunit type 1A of protein kinase A develops endocrine and other tumours: comparison with Carney complex and other PRKAR1A induced lesions. J Med Genet. 2004;41:923–31. doi: 10.1136/jmg.2004.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowan RW, Seidlitz EP, Singh G. Glutamate Signaling in Healthy and Diseased Bone. Front Endocrinol (Lausanne) 2012;3:89. doi: 10.3389/fendo.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skerry T. The role of glutamate in the regulation of bone mass and architecture. J Musculoskelet Neuronal Interact. 2008;8:166–73. [PubMed] [Google Scholar]
- 31.Kalariti N, Lembessis P, Koutsilieris M. Characterization of the glutametergic system in MG-63 osteoblast-like osteosarcoma cells. Anticancer Res. 2004;24:3923–9. [PubMed] [Google Scholar]
- 32.Chang H, et al. Metabotropic glutamate receptor 4 expression in colorectal carcinoma and its prognostic significance. Clin Cancer Res. 2005;11:3288–95. doi: 10.1158/1078-0432.CCR-04-1912. [DOI] [PubMed] [Google Scholar]
- 33.Brocke K, et al. Glutamate receptors in pediatric tumors of the central nervous system. Cancer Biol Ther. 2010;9:455–68. doi: 10.4161/cbt.9.6.10898. [DOI] [PubMed] [Google Scholar]
- 34.Stepulak A, et al. Expression of glutamate receptor subunits in human cancers. Histochem Cell Biol. 2009;132:435–45. doi: 10.1007/s00418-009-0613-1. [DOI] [PubMed] [Google Scholar]
- 35.Luksch H, et al. Silencing of selected glutamate receptor subunits modulates cancer growth. Anticancer Res. 2011;31:3181–92. [PubMed] [Google Scholar]
- 36.Hirschhorn J, Lettre G. Progress in genome-wide association studies of human height. Horm Res. 2009;71(2):5–13. doi: 10.1159/000192430. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet. 2011;44:6–7. doi: 10.1038/ng.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howie B, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purdue MP, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43:60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 41.Fearnhead P, Donnelly P. Approximate likelihood methods for estimating local recombination rates. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2002;64:657–680. [Google Scholar]
- 42.Crawford DC, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 43.Li N, Stephens M. Modeling Linkage Disequilibrium and Identifying Recombination Hotspots Using Single-Nucleotide Polymorphism Data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23:774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- 45.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.