Abstract
We describe the landscape of somatic genomic alterations based on multi-dimensional and comprehensive characterization of more than 500 glioblastoma tumors (GBMs). We identify several novel mutated genes as well as complex rearrangements of signature receptors including EGFR and PDGFRA. TERT promoter mutations are shown to correlate with elevated mRNA expression, supporting a role in telomerase reactivation. Correlative analyses confirm that the survival advantage of the proneural subtype is conferred by the G-CIMP phenotype, and MGMT DNA methylation may be a predictive biomarker for treatment response only in classical subtype GBM. Integrative analysis of genomic and proteomic profiles challenges the notion of therapeutic inhibition of a pathway as an alternative to inhibition of the target itself. These data will facilitate the discovery of therapeutic and diagnostic target candidates, the validation of research and clinical observations and the generation of unanticipated hypotheses that can advance our molecular understanding of this lethal cancer.
INTRODUCTION
Glioblastoma (GBM) was the first cancer type to be systematically studied by The Cancer Genome Atlas Research Network (TCGA). The initial publication (TCGA, 2008) presented the results of genomic and transcriptomic analysis of 206 GBMs, including mutation sequencing of 600 genes in 91 of the samples. The observations provided a proof-of-concept demonstration that systematic genomic analyses in a statistically powered cohort can define core biological pathways, substantiate anecdotal observations and generate unanticipated insights.
The initial publication reported biologically relevant alterations in three core pathways, namely p53, Rb, and receptor tyrosine kinase (RTK)/Ras/phosphoinositide 3-kinase (PI3K) signaling (TCGA, 2008). Efforts to link the alterations found in these pathways to the distinct molecular and epigenetic subtypes of glioblastoma revealed that coordinated combinations were enriched in different molecular subtypes, which may affect clinical outcome and the sensitivity of individual tumors to therapy (Noushmehr et al., 2010; Verhaak et al., 2010).
Above and beyond these observations, it has become evident that GBM growth is driven by a signaling network with functional redundancy that permits adaptation in response to targeted molecular treatments. Thus, a comprehensive catalogue of molecular alterations in GBM, based on multidimensional high-resolution data sets, will be a critical resource for future investigative efforts to understand its pathogenesis mechanisms, inform tumor biology and ultimately develop effective therapies against this deadly cancer.
Toward those ends, TCGA has expanded the scope and depth of molecular data on GBM, including adoption of next-generation sequencing technology (TCGA, 2011, 2012a). Here, we report the efforts of the TCGA GBM Analysis Working Group (AWG) to further our understanding of GBM pathobiology by constructing a detailed somatic landscape of GBM through a series of comprehensive genomic, epigenomic, transcriptomic and proteomic analysis.
RESULTS
Samples and Clinical Data
As summarized in Table 1, the dataset contains molecular and clinical data for a total of 543 patients. Note that different subsets of patients were assayed on each technology platform. The most significant additions to the GBM dataset include sequencing of GBM whole genomes, coding exomes and transcriptomes, expanded DNA methylomes as well as profiling of a targeted proteome. In particular, 291 pairs of germline-tumor native DNAs (e.g. without whole-genome amplification) were characterized by hybrid-capture whole-exome sequencing (WES) and of these, 42 pairs underwent deep coverage whole-genome sequencing (WGS). The transcriptomes of 164 RNA samples were profiled by RNA-sequencing (RNA-seq). Protein expression profiles were generated from 214 patient samples using reverse phase protein arrays (RPPA). The data package associated with this report was frozen on 7/15/2013 and is available at the Data Portal: https://tcga-data.nci.nih.gov/docs/publications/gbm_2013/.
Table 1.
Characterization platforms and data availability
| Data Type | Platforms | Cases in 2008 | Cases in 2013 |
|---|---|---|---|
| DNA sequence of exome | Illumina on native DNA | 0 | 291 |
| Sanger on native DNA | 91 | 148 | |
| Illumina on whole genome amplified DNA | 0 | 163 | |
| DNA sequence of whole genome | Illumina on native DNA | 0 | 42 |
| DNA copy number/genotype | Affymetrix SNP6 | 206 | 578 |
| Agilent 224K/415K | 206 | 413 | |
| mRNA expression profiling | Affymetrix U133A | 206 | 544 |
| Affymetrix Exon | 201 | 417 | |
| mRNA sequencing | Illlumina on native cDNA | 0 | 164 |
| CpG DNA Methylation | Illumina GoldenGate | 242 | 242 |
| Illumina 27K | 0 | 285 | |
| Illumina 450K | 0 | 113 | |
| miRNA expression profiling | Agilent | 205 | 491 |
| Protein expression profiling | Reverse phase protein arrays | 0 | 214 |
| Clinical characteristics | Tier 1/Tier 2 | 206 | 543 |
TCGA sample collection spanned 17 contributing sites (SI Table S1). Tier 1 clinical data elements (including age, pathology and survival) are available on 539 of 543 patients (99.6%) and Tier 2 data including treatment information on 525 patients (96.7%) (Figure S1, see Data Portal). Clinical characteristics of this patient cohort are similar to our previous report in 2008 (TCGA, 2008) with a median age of 59.6 years and a male to female ratio of 1.6 (333:209). Median overall survival was 13.9 months with 2-year survival of 22.5% and 5-year survival of 5.3%. Due to TCGA selection of primary GBM, IDH1 mutation is infrequent in the TCGA cohort compared to other published series. Of the 423 patients with adequate sequencing coverage (by either whole exome next-generation sequencing or previously reported Sanger-based sequencing), 28 (6%) had the IDH1-R132H mutation, while one individual had an R132G and one had an R132C mutation. No IDH2 mutations were found. The associated G-CIMP methylation pattern was present in all cases of IDH1 mutation (R132H/G/C) while seven G-CIMP cases lacked IDH1 mutations. Overall, G-CIMP pattern was present in 42 out of 532 cases (7.9%). Clinically-relevant MGMT DNA methylation status was estimated from CpG islands as previously described (Bady et al., 2012). Conventional positive prognostic factors were confirmed by univariate analysis: age < 50 (OS 21.9 vs. 12.3 months, p=2.4e-11), MGMT DNA methylation (16.9 vs. 12.7, p=0.0018), IDH1 mutation (35.4 vs. 13.3, p=1.55e-5) and G-CIMP DNA methylation (38.3 vs. 12.7, p=8.3e-9). Age, MGMT and IDH1/G-CIMP status were independently significant in multivariate analysis (SI Table S1).
Patients in this TCGA cohort were diagnosed between 1989 and 2011, with 414 patients (76%) receiving their diagnosis in or after 2002 when the use of concurrent temozolomide (TMZ) with adjuvant radiation became widely adopted. Combined TMZ chemotherapy and radiation treatment is documented for 40% of all patients (217/543), and for 50.2% of the 414 patients diagnosed in or after 2002. Summaries of treatment classification classes are provided in SI..
Whole-exome sequencing identifies significantly mutated genes in glioblastomas
Solution-phase hybrid capture and whole-exome sequencing were performed on paired tumor and normal native genomic DNA obtained from 291 patients. Overall, 138-fold mean target coverage was achieved, with 92% of bases covered at least 14-fold in the tumor and 8-fold in the normal – the threshold which offers 80% power to detect mutations with an allelic fraction of 0.3 (Carter et al., 2012) (see Extended Experimental Procedures). Overall, of the 291 tumor exomes sequenced, 21,540 somatic mutations were identified, with a median rate of 2.2 coding mutations per megabase (lower-upper quartile range: 1.8 – 2.3). Among the somatic mutations were 20,448 single nucleotide variants (SNVs), 39 dinucleotide mutations and 1,153 small insertions and deletions (indels). The SNVs mutations included 5,379 silent, 3,901 missense, 831 nonsense, 360 splice-site and 760 mutations resulting in a frame shift.
Mutations were evaluated across samples to distinguish genes which appear targeted by driver rather than passenger mutations using both MutSig (TCGA, 2008, 2011, 2012a) and InVEx algorithms (Hodis et al., 2012). MutSig assesses mutation significance as a function of gene size, trinucleotide context, gene structure and background mutation rates. InVEx compares the ratio of non-silent exonic mutations to synonymous and intronic/UTR nucleotide variants, an algorithm that is particularly effective for genomes with elevated mutation rates such as melanoma and lung adenocarcinoma. When both InVEx and MutSig algorithms were run on the same dataset, a total of 71 genes were identified as significantly mutated genes (SMG). To validate mutation calls, all 757 SNVs and indels detected by exome sequencing in these 71 SMGs were subject to orthogonal validation by targeted re-sequencing in 259 tumor/normal pairs. At sites with adequate coverage to detect the mutant alleles, 98% of SNVs, 84% of insertions, and 82% of deletions were validated (see Extended Experimental Procedures).
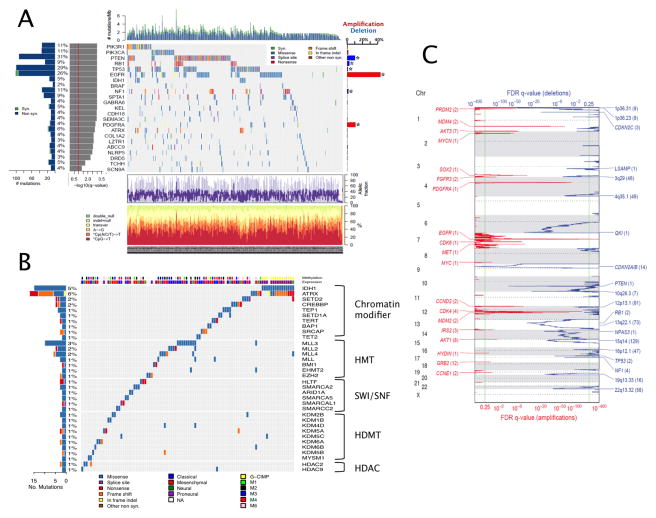
As summarized in Figure 1A, both InVEx and MutSig algorithms identified previously reported genes as significantly mutated in GBM, namely PTEN, TP53, EGFR, PIK3CA, PIK3R1, NF1, RB1, IDH1 and PDGFRA (Figure 1A). In addition, both algorithms identified the leucine-zipper-like transcriptional regulator 1 (LZTR1), mutated in ten samples, as a novel significantly mutated gene in GBM (SI Table S2, SI Figure S2). LZTR1, a putative transcriptional regulator associated with the DiGeorge congenital developmental syndrome (Kurahashi et al., 1995), has not previously been implicated in cancer. It is located at chromosome 22q, and in five of six samples with available copy number data it was simultaneously targeted by hemizygous deletion.
Figure 1. Somatic genomic alterations in glioblastoma.
(A) Summary of significantly mutated genes from 291 exomes. Specific mutations for LZTR1, SPTA1, KEL, and TCHH are shown in SI Figure S2.
Upper histogram: Number of mutations per sample (substitutions and indels). Left histogram, rate of mutations per gene and percentage of samples affected.
Central heat map: Distribution of significant mutations across sequenced samples, color coded by mutation type.
Left histograms: Overall count and significance level of mutations as determined by log(10) transformation of the MutSig q-value. Red line indicates a q-value of 0.05.
Right histogram: Summary of focal amplifications (red) and deletion (blue) determined from DNA copy number platforms (asterisk denotes inclusion in statistically significant recurrent CNA by GISTIC).
Lower chart: Average fraction of tumor reads versus total number of reads per sample. Bottom chart: top, rates of non-silent mutations within categories indicated by legend; bottom, mutation spectrum of somatic substitutions of samples in each column.
(B) Mutations in 38 genes related to specific epigenetic function categories (out of 161 genes linked to chromatin modification) across 98 GBMs (out of 292 GBM). IDH1 mutation status is included to illustrate its co-occurrence with ATRX mutation. An additional 37 GBMs harbored mutations in one of the remaining 129 CMGs.
(C) Recurrent sites of DNA copy number aberration determined from 543 samples by the GISTIC algorithm. Statistically significant, focally amplified (red) and deleted (blue) regions are plotted along the genome. Significant regions (FDR<0.25) are annotated with the number of genes spanned by the peak in parentheses. For peaks that contain a putative oncogene or tumor suppressor, the gene is noted.
MutSig additionally identified 61 additional genes (71 overall) with mutation frequency above background with a q-value of < 0.1 (SI Table S2). These included spectrin alpha 1 (SPTA1, mutated in 9%), which encodes a cell motility protein that interacts with the ABL oncogene and is related to various hereditary red blood cell disorders; ATRX (6%), a member of the SWI/SNF family of chromatin remodelers recently implicated in pediatric and adult high-grade gliomas (Kannan et al., 2012; Liu et al., 2012; Schwartzentruber et al., 2012); GABRA6 (4%), an inhibitory neurotransmitter in the mammalian brain; and KEL (5%) which codes for a transmembrane polymorphic antigen glycoprotein (SI Figure S2). Albeit at low frequency, several hotspot mutations were found to be significant in this cohort of GBM, most notably the IDH1 R132H mutation. The BRAF V600E sequence variant, which confers sensitivity to vemurafenib in melanoma (Chapman et al., 2011), was detected in five of 291 GBMs (1.7%). Mutation of H3.3 histones, reported in pediatric gliomas (Schwartzentruber et al., 2012), were not observed in this cohort of primary GBM.
To facilitate exploration of mutation data by non-computational biologists, we developed a patient-centric table (PCT) that categorizes each gene in each sample by the type of mutation (silent, missense, InDel, etc.) observed, and describes the confidence of each call based on the coverage in normal and tumor samples (see Data Portal, Extended Experimental Procedures). To illustrate one potential use of this table, we interrogated the mutation pattern of 161 genes functionally linked to chromatin organization (hereafter referred to as CMG or “chromatin modification genes”, see Extended Experimental Procedures) using this PCT. In total, 135 samples or 46% of the sample cohort harbored at least one non-synonymous mutation in this CMG gene set (Figure 1B). Importantly, CMG mutations were found to be mutually exclusive overall by MEMo analysis (p=0.0008) (Ciriello et al., 2012), suggesting potential biological relevance of chromatin modification in GBM.
Genomic gains and losses in GBM
We expanded our previously reported DNA copy number analysis from 206 GBMs (TCGA, 2008) to 543 samples. The larger data set, coupled with improvement of the analytical algorithm GISTIC (Mermel et al., 2011), resulted in a significant refinement of previously-defined amplification and deletion peaks, thus allowing improved nomination of candidate gene targets for several recurrent somatic copy number aberrations (SCNA) (Figure 1C). The most common amplification events on chromosome 7 (EGFR/MET/CDK6), chromosome 12 (CDK4 and MDM2) and chromosome 4 (PDGFRA) were found at higher frequencies than previously reported (SI Table S3), and often contained only a single gene in the common overlapping region. Additionally, frequent gains of genes such as SOX2, MYCN, CCND1 and CCNE2 were precisely established. Except for the highly recurrent homozygous deletions in CDKN2A/B, all statistically significant DNA losses were hemizygous. Losses were more frequent than amplifications, as has been reported as a general pattern in cancer (Beroukhim et al., 2010). We were able to pinpoint single genes as deletion targets in some cases, most notably in recurrent deletion of 6q26. While the 6q26 deletion has previously been associated with other candidates such as PARK2, our analysis unequivocally defined QKI as the sole gene within the minimal common region and the target of homozygous deletion in 9 cases. The QKI gene was also mutated in 5 cases without evidence of deletion (two frame-shift, two missense and one splice-site mutation). This is consistent with a recent publication demonstrating that QKI functions as a tumor suppressor in GBM by acting as a p53-responsive regulator of mature miR-20a stability to regulate TGFβR2 expression and TGFβ network signaling (Chen et al., 2012). Other single gene deletion targets include LRP1B, NPAS3, LSAMP and SMYD3. Similar to the mutation data, we have also algorithmically generated a Patient-Centric Table summarizing DNA copy number aberration and DNA methylation status for each gene and miRNA for each of the cases in the cohort (see Data Portal).
Recurrent structural rearrangements defined by genomic and transcriptomic sequencing
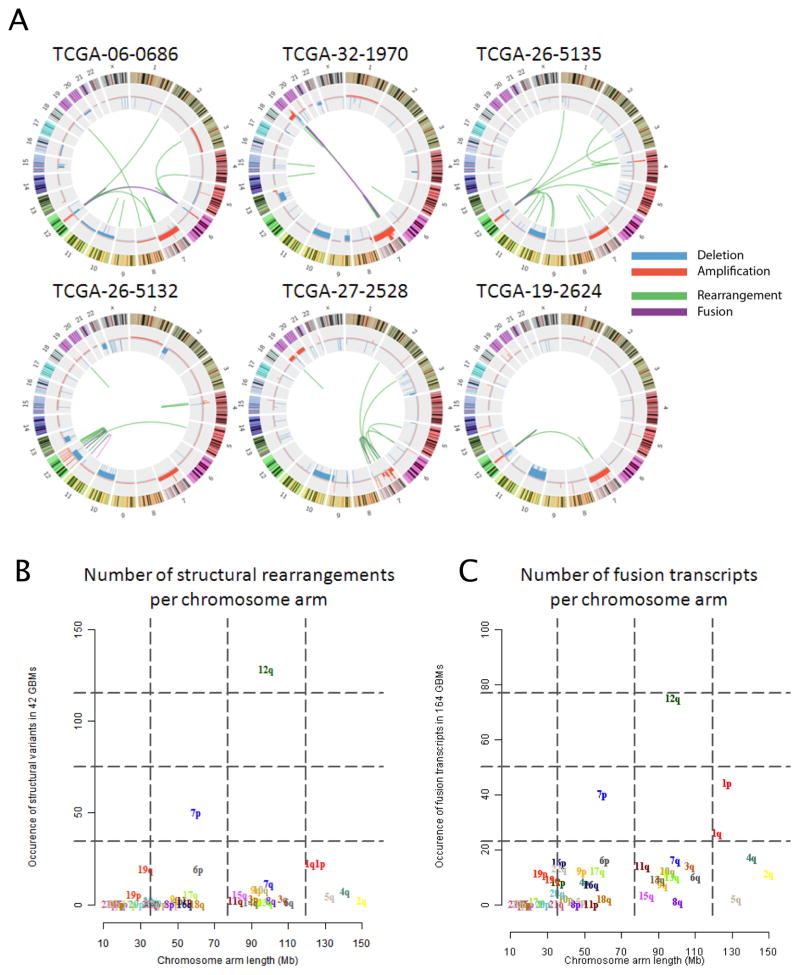
To explore genomic and transcriptomal structural rearrangements, we performed whole-genome paired-end sequencing with deep coverage on 42 pairs of tumor and matched germline DNA samples as well as RNA sequencing (RNA-seq) of 164 GBM transcriptomes (SI Table S4). We detected genomic rearrangements using BreakDancer and BamBam (Sanborn et al., 2013) (see Extended Experimental Procedures), in addition to expressed RNA fusions using PRADA (http://sourceforge.net/projects/prada/). In total, we identified 238 high confidence candidate somatic rearrangements, including 49 interchromosomal, 125 intrachromosomal and 64 intragenic structural variants (Figure 2A and B; SI Table S4). The number of events per sample ranged from 0 to 32 (median: 2), with one sample containing a distinctively high number of rearrangements in the context of local chromothripsis involving a 7.5 Mb region on chromosome 1. No rearrangements were detected in eight samples. Overall, the number of rearrangements generally appeared lower than what has been previously reported for prostate cancer (Sanborn et al., 2013), lung adenocarcinoma (Imielinski et al., 2012) and melanoma (Berger et al., 2012). Recurrent intragenic events were detected in seven genes: EGFR (n=12), CPM (n=3), PRIM2 (n=3), FAM65B (n=2), PPM1H (n=2), RBM25 (n=2), and HOMER2 (n=2). Since unbalanced structural rearrangements in DNA can be detected as breakpoints in DNA copy number profiles, we investigated whether CNA breakpoints could indicate potential sites of recurrent structural rearrangement using all 492 samples with aCGH data (n=492). Of note, 41 of 129 high-confidence rearrangement events from whole-genome sequencing (WGS) involved genes identified as significant targets of recurrent intragenic copy number breakpoints (iCNA) in the larger cohort of GBM based on DNA copy number profiles (SI Table S4, Data Portal).
Figure 2. Structural rearrangements and transcript variants in GBM.
(A) Circos plots of structural DNA and RNA rearrangements in six GBMs, selected from 28 cases with available whole genome and RNA sequencing based on their rearrangement frequency. Outer ring indicates chromosomes. Copy number levels are displayed along the chromosome map in red (copy number gain) and blue (copy number loss). Each line in the center maps a single structural variant to the site of origin for both genes (see SI Figure S3 for additional analysis of fusion transcripts derived from RNA sequencing).
(B) The chromosome arm of origin of both ends of each rearrangement detected in whole genome sequencing data from 42 GBM were counted and compared to chromosome arm length.
(C) The chromosome arm of both partners in fusion transcripts detected from RNA sequencing data from 164 GBM were counted and compared to chromosome arm length.
RNA seq analysis identified 48 interchromosomal and 180 intrachromosomal mRNA fusion transcripts in 106 of 164 samples (Figure 2C; SI Table S4). Approximately 37% of these were in-frame transcripts, 35% were out-of-frame and the remaining 29% were involved a 3′ or 5′ untranslated region (SI Figure S3A). A substantial portion (44%) of the intrachromosomal events resulted from recombination of genomic loci located less than 1Mb apart. A notable example is the recently reported oncogenic FGFR3-TACC3 inversion (Singh et al., 2012), which was detected in two cases. Interestingly, the FGFR3/TACC3 locus was focally amplified in both samples, suggesting that CNA could serve as a marker of FGFR3-TACC3 rearrangement. Overall, focal amplifications involving FGFR3 or TACC3 were detected in 14 of 537 GBM copy number profiles (2.6%).
Ten of the 42 GBMs with WGS analyses demonstrated rearrangements between EGFR and adjacent genes such as BRIP (n=2) and VOPP1 (n=2), or structural variants of genes surrounding the EGFR locus, such as LANCL2 and PLEXHA (n=2) (SI Table S4). Both types of 7p11 rearrangements were detected in six samples. This pattern was confirmed in the RNA-seq data where eighteen samples of 164 samples showed evidence of transcribed fusion transcripts, such as EGFR-SEPT14 (n=6), SEC61G-EGFR (n=4), LANCL2-SEPT14 (n=1) and COBL-SEPT14 (n=1) (SI Table S4). These fusions tended to be part of a focal gain, suggesting a complex rearrangement (SI Figure S3B).
Genomic rearrangements pertaining to chromosome arm 12q were identified in 11 of 42 whole genomes and 12q-associated fusion transcripts were found expressed in 25 of 164 transcriptomes. A variety of different genomic and transcriptomic variants were found on 12q though none were recurrent (SI Table S4). The majority of 12q lesions occurred in tandem, i.e. as adjacent events in the same GBM. As an illustration, a single sample showed a pattern in which 15 non-adjacent segments (14 from chromosome 12 and one fragment from chromosome 7) were highly amplified (>40 copies) with eight 12q rearrangement events, including the MDM2, CDK4 and EGFR oncogenes (SI Figure S3C). WGS analysis reconstructed two independent circular paths that accounted for all of the amplified segments (SI Figure S3C). Each circle contained at least one oncogene, with one circle (0152-DM-A) containing one copy of CDK4 and two copies of MDM2 and the other circle (0152-DM-B) containing one copy of EGFR. These reconstructed circles are most consistent with extrachromosomal double minute chromosomes (Kuttler and Mai, 2007). Recently, the same data set was used to identify enrichment of genomic breakpoints relating to chromosome 12q14–15, a locus harboring the MDM2 and CDK4 oncogenes, which pertained to less favorable outcome (Zheng et al., 2013), and the reconstruction of double minutes confirmed using orthogonal methods (Sanborn et al., 2013).
EGFR is frequently targeted by multiple alterations of DNA and RNA
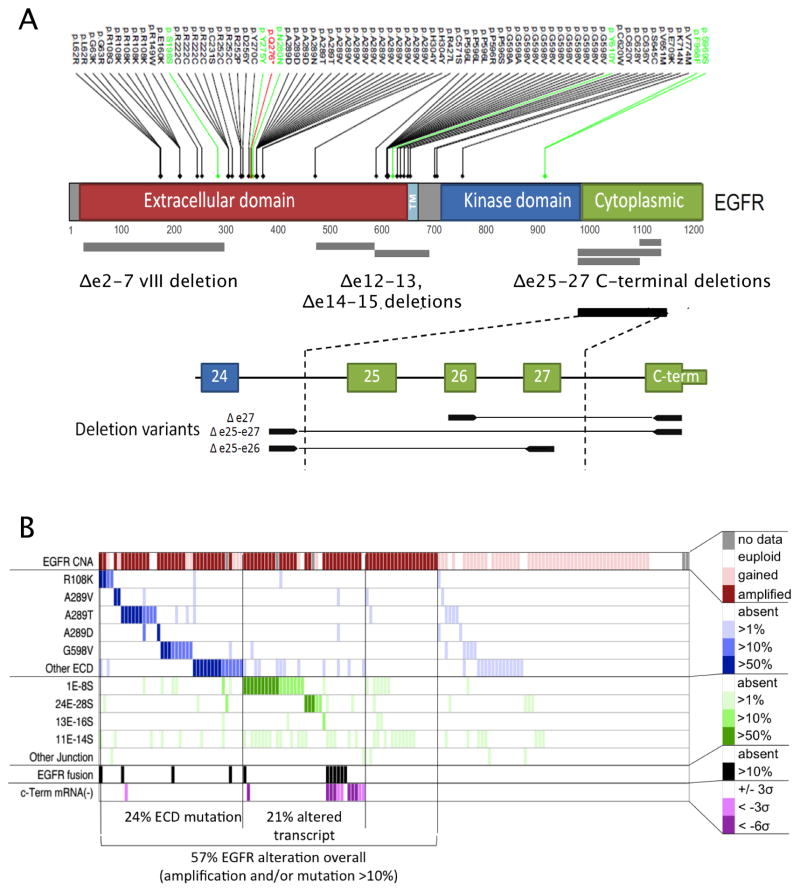
As anticipated, EGFR was among the most frequently mutated genes and RNA-seq detected a diversity of altered transcripts (Figure 3A). EGFR mutations were accompanied by regional DNA amplification in the majority of cases, leading to a wide range of mutation allelic frequencies. Comparing the allelic frequencies of point mutations in DNA- and RNA-seq data revealed a high degree of concordance between the type and prevalence of mutations at the DNA level and the composition of expressed mRNA transcripts (SI Figure S4A).
Figure 3. Somatic alterations of the EGFR locus.
(A) EGFR protein domain structure with somatic mutations summarized from 291 GBMs with exome sequencing and transcript alterations identified across 164 GBMs with RNA sequencing.
(B) EGFR alterations are summarized by transcript prevalence in 164 GBMs with RNA sequencing. Red, top: focal amplification or regional gain inferred from DNA copy number. Blue: Prevalence of sequencing reads with EGFR point mutation. Green: prevalence of reads with aberrant exon-exon junctions (e.g., 1E–8S is a junction spanning from the end of exon 1 to the start of exon 8, consistent with EGFRvIII mutation). Black: EGFR fusion transcript detected (see rearrangements). See related SI Figure S4 for comparison of EGFR mutations in DNA and RNA and for a summary of EGFR rearrangements.
RNA-seq also provided a complete picture of aberrant exon junctions and a semi-quantitative assessment of their expression levels. Transcript allelic fraction (TAF) was calculated as the ratio of each aberrant exon junction to the sum of aberrant and wild-type junctions at the 3′ junction end, corrected for read depth (80% confidence, binomial confidence interval). TAFs for recurrent point mutations and junctions are summarized in SI Table S5. In 11% of tumors, the aberrant exon 1–8 junction characteristic of EGFRvIII was highly expressed (≥10% TAF), while 19% showed at least a low level expression (≥1%). The results were concordant with an independent assessment of EGFRvIII by digital mRNA assay using barcoded probes (nCounter, Nanostring Technologies and by real-time PCR (see Data Portal). While the biological or clinical relevance of low-level EGFRvIII expression remains to be demonstrated, EGFRvIII expression in a minor population of GBM cells has been shown to confer a more aggressive tumor phenotype through paracrine mechanisms (Inda et al., 2010).
A variety of other recurrent non-canonical EGFR transcript forms were detected in the RNA-seq data (Figure 3A, SI Figure S4B). Three different C-terminal rearrangements targeting the cytoplasmic domain of the EGFR were detected at ≥10% TAF in 3.7% of cases and at ≥1% TAF in another 9%. Comparison with WGS data confirmed the presence of C-terminal deletions in 9 cases where sequence data was available. C-terminal deletion variants have previously been associated with gliomagenesis in experimental rodent systems in vivo (Cho et al., 2012). The prevalence of EGFR C-terminal deletion reported here is likely an underestimate since complete loss of the C-terminus may yield aberrant terminal junctions not mappable by transcriptome sequencing. Relative under-expression of C-terminus exons 27–29 (< 3 standard deviations) was readily apparent in another 7.3% of cases without detectable aberrant junctions (Figure 3B).
We identified two relatively uncharacterized recurrent EGFR variants, namely deletions of exons 12–13 (Δ12–13) in 28.7% and exons 14–15 (Δ14–15) in 3%. EGFR Δ12–13 has been previously identified by RT-PCR analysis of glioma (Callaghan et al., 1993). Both Δ12–13 and Δ14–15 appear to be expressed in minor allelic fractions (<10%), raising the question of whether they result from splicing aberration or genomic deletion. Among tumors expressing Δ12–13mRNA, analysis of aberrant junctions in WGS data (BamBam) failed to identify concordant DNA deletion in 14/15 cases where data was available. One case showed a concordant breakpoint as a minor component of a highly rearranged locus. By comparison, EGFRvIII-expressing tumors had concordant deletion spanning exons 2–7 in all 7 cases where WGS data was available (SI Table S5).
In total, 38.4% of cases harbored an EGFR genomic rearrangement or a point mutation expressed in at least 10% of transcripts (Figure 3B; SI Table S5). Overall, 57% of GBM showed evidence of mutation, rearrangement, altered splicing and/or focal amplification of EGFR. While PDGFRA showed no recurrent gene fusions, intragenic deletion of exons 8 and 9 (PDGFRA Δ8,9) was highly expressed (≥10% TAF) in 1 of the 164 samples with RNA sequencing data. Low-level expression of PDGFRA Δ8,9 was far more prevalent in the RNA-seq data (n=29 of 163) and could represent a splice variant. This result is concordant with previously reported estimates of Δ8,9 expression (Ozawa et al., 2010). A novel PDGFRA variant with deletion of exons 2–7 was found highly expressed in a single case (TCGA-28-5216).
The landscape of somatic alterations in glioblastoma
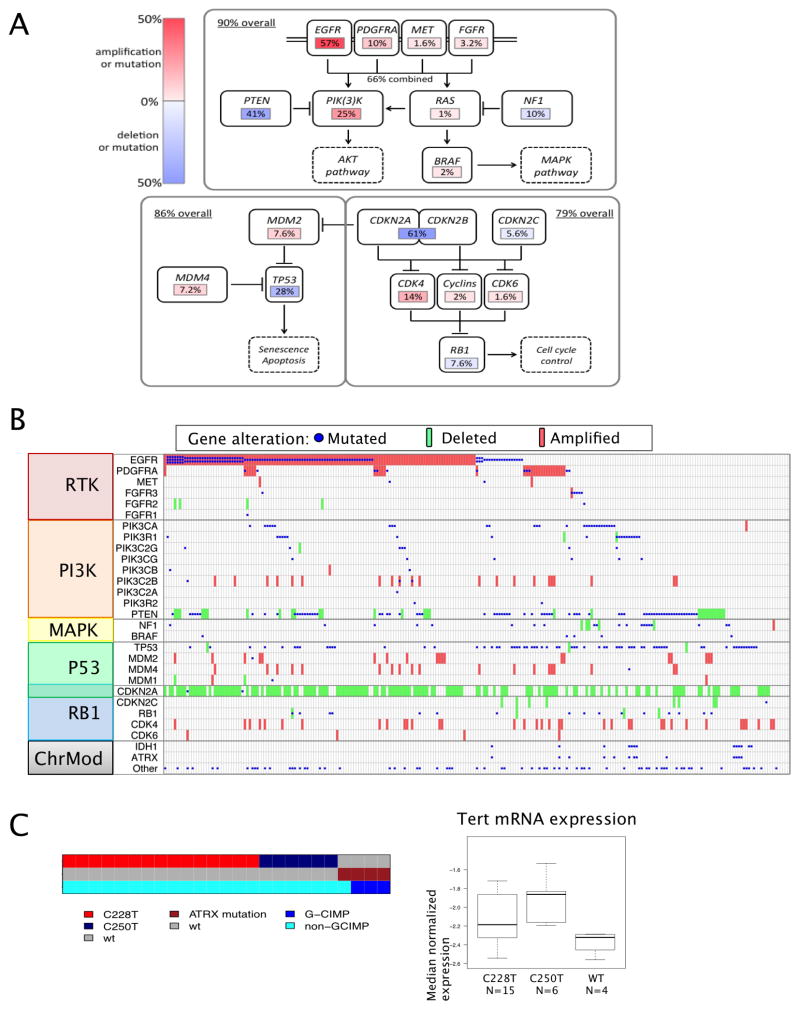
The addition of whole exome and transcriptomal sequencing data has extended the palette of somatic alterations affecting major cancer pathways in GBM. Figure 4 presents a landscape view of the canonical signal transduction and tumor suppressor pathways in GBM based on whole exome sequencing data of 291 patients. Unsupervised analysis of 251 GBMs with both copy number and WES mutation data identified genes sets (modules) in which somatic alterations were significantly mutually exclusive (MEMo, (Ciriello et al., 2012)). This analysis confirmed mutual exclusivity among alterations affecting the p53 pathway (MDM2, MDM4 and TP53), the Rb pathway (CDK4, CDK6, CCND2, CDKN2A/B and RB1), and various components influencing the PI3K pathway (PIK3CA, PIK3R1, PTEN, EGFR, PDGFRA, NF1) (SI Table S6).
Figure 4. Landscape of Pathway Alterations in GBM.
Alterations affecting canonical signal transduction and tumor suppressor pathways are summarized for 251 GBM with both exome sequencing and DNA copy number data. Rearrangements are underestimated in this summary since RNA-seq data were available for only a subset of cases with exome sequencing data (153/291, 61%).
(A) Overall alteration rate is summarized for canonical PI3K/MAPK, p53 and Rb regulatory pathways.
(B) Per-sample expansion of alterations summarized in 5A. Mutations (blue), focal amplifications (red) and homozygous deletions are selected from the patient-centric tables and organized by function. All missense, nonsense and frame-shift mutations are included. EGFRvIII is inferred from RNA data and included as a mutation if >=10% transcribed allelic frequency. Deletions are defined by log2 ratios < −1 or <−0.5 and focally targeting the gene (see Extended Experimental Procedures). Amplifications are defined by log2 ratio>2 or >1 and focal.
(C) Left: For a cohort of 25 GBMs for which whole genome sequencing allowed genotyping, TERT promoter C228T and C250T mutations occurred in a mutually exclusive fashion. All four TERT promoter wildtype GBM harbored ATRX mutation, and were enriched in G-CIMP group.
Right: TERT promoter mutations are associated with elevated expression.
As shown, at least one RTK was found altered in 67.3% of GBM overall: EGFR (57.4%), PDGFRA (13.1%), MET (1.6%) and FGFR2/3 (3.2%). Half of the tumors with focal amplification and/or mutation of PDGFRA harbored concurrent EGFR alterations (42.4%, 14/33), as did the majority of MET-altered tumors (3/4), reflecting a pattern of intratumoral heterogeneity that has been previously documented by in situ hybridization (Snuderl et al., 2011; Szerlip et al., 2012).
PI3-kinase mutations were found in 25.1% of GBM (63/251), with 18.3% affecting p110alpha and/or p85alpha subunits and 6.8% in other PI3K family genes. PI3K mutations were mutually exclusive of PTEN mutations/deletions (p=0.0047, Fisher’s Exact), with 59.4% of GBM showing one or the other (149/251). Considering the RTK genes, PI3-kinase genes and PTEN, 89.6% of GBM had at least one alteration in the PI3K pathway and 39% had two or more. The NF1 gene was deleted or mutated in 10% of cases, and never co-occurred with BRAF mutations (2%).
Concordant with the previous TCGA GBM report, the p53 pathway was found to be dysregulated in 85.3% of tumors (214/251), through mutation/deletion of TP53 (27.9%), amplification of MDM1/2/4 (15.1%) and/or deletion of CDKN2A (57.8%). As expected, TP53 alterations were mutually exclusive with amplification of MDM family genes (p=0.0003) and CDKN2A (p=1.99e–7). Concurrently, 78.9% of tumors had one or more alteration affecting Rb function: 7.6% by direct RB1 mutation/deletion, 15.5% by amplification of CDK4/6, and the remainder via CDKN2A deletion.
As reported for lower grade gliomas (Ichimura et al., 2009), 12 of the 13 GBMs with IDH1 hotspot mutations harbored concurrent TP53 mutations. Consistent with recent reports, mutations in SWI/SNF complex gene ATRX often co-occurred in these cases (Figure 4B). Mutations in IDH1 and ATRX appear to be more prevalent in GBM samples without RTK alteration (p=7.2e-5 and 7.3e-4, respectively), tumors genotypically more consistent with secondary GBM (Ohgaki and Kleihues, 2007).
Telomerase reverse transcriptase (TERT) promoter mutations were recently reported in glioma, mapping to positions 124 (C228T) and 146bp (C250T) upstream of the TERT ATG start site (Killela et al., 2013). Of the 42 cases with deep coverage WGS data, 25 samples had adequate coverage (read count >10) of the TERT promoter for mutational analysis. We detected the C228T mutation in 15 of the 25 cases, while the C250T variant was found in another 6 cases (Figure 4C). TERT promoter mutations at these two hot spots were correlated with up-regulated TERT expression at the RNA level (Figure 4C). Interestingly, the four GBMs with non-mutated TERT promoters all harbored ATRX mutations and these were concurrent with IDH1 and TP53 mutations as recently described (Liu et al., 2012). Finally, in line with the role of ATRX in alternative lengthening of telomeres (ALT) (Lovejoy et al., 2012), ATRX-mutant GBM tumors do not exhibit elevated TERT RNA expression compared to tumors with TERT promoter mutations (Figure 4C). Taken together, these data suggest that maintenance of the telomere either through reactivation of telomerase by TERT promoter mutation-induced increased TERT expression or ALT as a result of ATRX mutation is a requisite step in GBM pathogenesis.
While reported median survival for patients with GBM ranges from 12–18 months, a subset of individuals will survive for more than three years (Dolecek et al., 2012; Dunn et al., 2012). We cross-referenced our data set to identify any factor(s) associated with long-term survival (n=39 or 7.7% of the cohort). Although no specific genomic alteration was significantly over-represented in this subset, amplifications of CDK4 and EGFR and deletion of CDKN2A were observed at decreased frequencies in these long survivors (see Data Portal). Age at diagnosis was found to be a major determinant, with 79% of long-term survivors being diagnosed at younger than 50 years of age. Despite their relatively favorable prognosis, only one third of patients with G-CIMP+ GBM survived beyond three years, suggesting that other factors yet to be identified are contributing to overall long-term survival of GBM patients.
Molecular subclasses defined by global mRNA expression and DNA Methylation
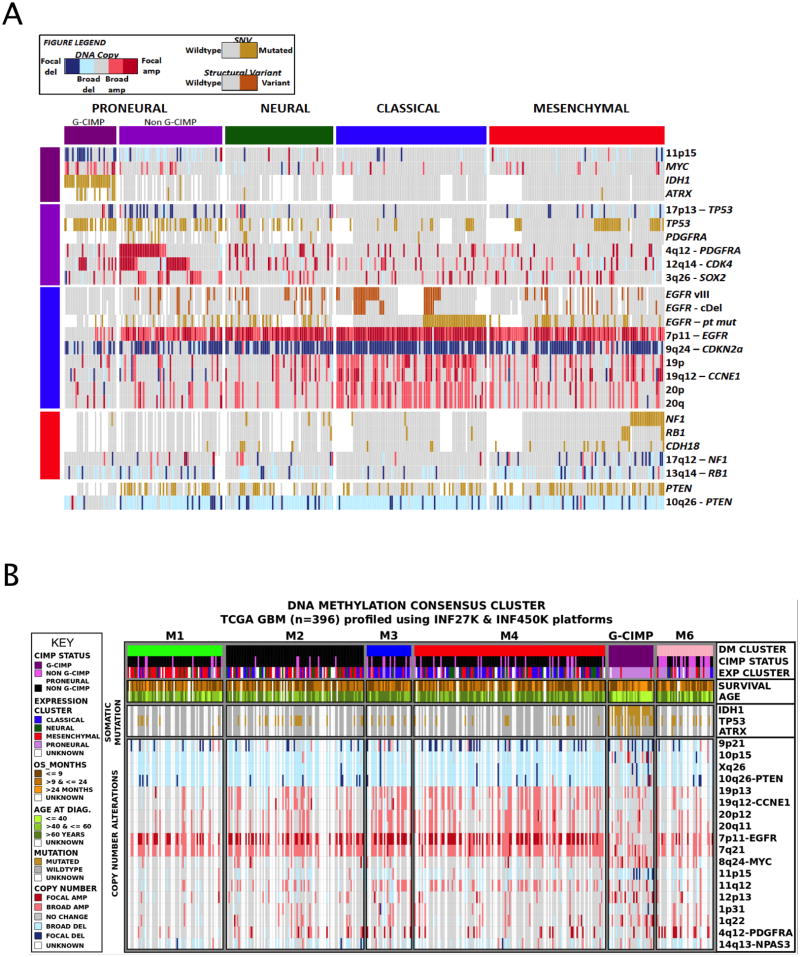
Widespread differences in gene expression have previously been reported in GBM, grouping TCGA tumors into proneural, neural, classical and mesenchymal transcriptomic subtypes (Phillips et al., 2006; Verhaak et al., 2010). Samples not included in previously published analysis (n=342) were classified into one of classes using single sample gene set enrichment analysis (Figure 5A, SI Table S7) Similarly, we sought to assign each case in the TCGA cohort to one of the DNA methylation subclasses. The promoter DNA methylation array platforms used by TCGA have evolved with increasing resolution from the Illumina GoldenGate (n=238), Infinium HumanMethylation27 (HM27, n=283) and Infinium HumanMethylation450 (HM450, n=76) platforms (SI Figure S5A). We re-analyzed a total of 396 GBM samples, comprised of 305 new GBM samples profiled on the HM27 (n=192) and HM450 (n=113) platforms in addition to 91 cases profiled on HM27 that were included previously (Noushmehr et al., 2010). Hierarchical consensus clustering of the DNA methylation profiles stratified these 396 GBM cases into six classes, including G-CIMP (Figure 5B, SI Figures S5B and S5C, and SI Table S7). Cluster M1 (35/58, 60%) is enriched for mesenchymal GBMs while cluster M3 (18/31, 58%) is enriched for classical subtype (Figure 5B, red and blue, respectively). As expected, the G-CIMP cluster is enriched for proneural subtype tumors.
Figure 5. Molecular subclasses of GBM and their genomic molecular correlates.
(A) Genomic alterations and survival associated with five molecular subtypes of GBM. Expression and DNA methylation profiles were used to classify 332 GBMs with available (native DNA and whole genome amplified DNA) exome sequencing and DNA copy number levels. The most significant genomic associations were identified through Chi-square tests, with p-values corrected for multiple testing using the Benjamini-Hochberg method.
(B) Genomic alterations and sample features associated with six GBM methylation clusters. Epigenomic consensus clustering was performed on 396 GBM samples profiled across two different platforms (Infinium HM27 and Infinium HM450). Six DNA methylation clusters were identified (see related SI Figure S5), represented as M1 to M6, where M5 is G-CIMP. These DNA methylation signatures are correlated with 27 selected features composed of clinical, somatic and copy number alterations; DM cluster, G-CIMP status, four TCGA GBM gene expression subclasses, two clinical features (Age at diagnosis/overall survival in months), somatic mutations (IDH1, TP53, ATRX) and 18 selected copy number alterations.
To be able to perform more robust exploration of the relationship of G-CIMP phenotype to other genomic alterations, we incorporated the previously reported G-CIMP status (Noushmehr et al., 2010) on 175 additional GBM cases profiled on the GoldenGate platform. A total of 534 GBM cases, were used in the following integrative analyses. The age of GBM diagnosis was statistically different (41yrs vs. 56yrs; p-value = 0.008) between proneural G-CIMP (n=28) and proneural non-G-CIMP (n=22) subtypes, reinforcing the notion that the epigenomics of these transcriptomically similar patients mark distinct etiologies and/or disease characteristics. We observed seven G-CIMP(+) cases lacking IDH1 mutation. These were similar to G-CIMP cases harboring IDH1 mutations with respect to their median age at diagnosis (40yrs vs. 37yrs, p-value = 0.58) and overall survival (mean 913 days vs. 1248 days, p-value = 0.45). IDH2 mutation was not detected in these seven G-CIMP+/IDH1 wildtype GBM, suggesting that alternative pathway(s) responsible for the hypermethylator phenotype.
Next, to identify genomic alterations enriched in each of the transcriptomic or epigenomic subtypes, we referenced the Patient-Centric Tables to count DNA mutation and copy number aberration events per subtype. This analysis confirmed previous reports, demonstrating significant associations between PDGFRA amplification and the non-G-CIMP+ proneural subgroup, as well as NF1 inactivation and the mesenchymal subtype (Figure 5A). Additionally, the enhanced power of the larger data set identified an enrichment of ATRX mutations and MYC amplifications in the G-CIMP+ subtype, CDK4 and SOX2 amplifications in proneural subtype, and broad amplifications of chromosomes 19 and 20 in the classical subtype (Figure 5A). In contrast to G-CIMP, cluster M6 was relatively hypomethylated, with a predominance of non-mutated IDH1 cases belonging to the proneural subtype (22/37, 59%) with concurrent PDGFRA amplification (Figure 5B).
To explore a plausible connection between chromatin deregulation and DNA methylation, we counted mutations in the 161 CMGs (Figure 1B) per each methylation subclass,. In addition to the association of IDH1 and ATRX mutations and G-CIMP, mutations of other CMGs were enriched across the M2, M4 and M6 subclasses (38% of cases in these three subclasses harbor at least one CMG mutation vs. 18% among the other classes, p=0.0015). Furthermore, cases with missense mutation or deletion of MLL genes (n=18) or HDAC family genes (n=4) clustered in the M2 DNA methylation subtype (10/21). These patterns of co-occurrence suggest a functional relationship between chromatin modification and DNA methylation that remains to be elucidated. Recently, Sturm et al. reported that adult and pediatric GBM with alterations of IDH1, H3F3A and receptor tyrosine kinases (RTK) were associated with epigenetic subtypes (Sturm et al., 2012). We compared the Sturm et al methylation-based classification with ours using the 74 TCGA cases that were also classified by by those authors. We found that four tumors classified as “IDH” subtype in Sturm et al. were assigned to G-CIMP subtype in our classification scheme (SI Figure S5D). The “Mesenchymal” tumors were assigned to M1 and M2 (21/25), “RTK II ‘classic’” tumors were assigned to M3 and M4 (30/34) and the “RTK I ‘PDGFRA’” tumors were assigned to M6. No TCGA samples were clustered in the Sturm et al’s “G34” or “K27” classes and we found the corresponding histone mutations to be absent across the TCGA sample set.
Lastly, we explored the relationship of molecular subclasses with clinical parameters such as treatment response or survival. In the current larger TCGA cohort, the survival advantage of proneural subtype GBM (Phillips et al., 2006) was definitively shown to be conferred by G-CIMP status, with non-G-CIMP proneural GBMs and not mesenchymal GBM tending to show less favorable outcomes in the first twelve months following initial diagnosis compared to other subtypes (p-value 0.07; SI Figure S6A). While most of the samples clustered in the M6 group were classified as proneural, this methylation subclass was not associated with adverse survival overall (SI Figure S6B) (Noushmehr et al., 2010). This observation reinforces the notion that target genes affected by the G-CIMP phenotype likely contribute to the improved prognosis for this subset of proneural GBM.
DNA methylation of the MGMT gene promoter is a known marker for treatment response (Hegi et al., 2005). We found that the MGMT locus was methylated in 48.5% of patients in our cohort (174 of 359 assessed), and that G-CIMP cases showed an increased likelihood of having MGMT DNA methylation (79% of G-CIMP vs. 46% for non-G-CIMP; SI Figure S6C). When correlated with outcome, MGMT status distinguished responders from non-responders amongst samples classified as classical (n = 96; p = 0.01) but not among samples classified as proneural (n = 66; p = 0.57), mesenchymal (n = 104; p = 0.62) and neural (n = 55; p = 0.12) (SI Figures S6D and E). In summary, our data provides evidence for MGMT DNA methylation as a predictive biomarker in the GBM Classical subtype of GBM, but not other subtypes.
Regulatory networks of miRNA and mRNA in gliomagenesis
MicroRNAs (miRs) have been found to promote or suppress oncogenesis through modulation of gene expression via mRNA degradation or inhibition of translation (Bartel, 2004; Krol et al., 2010). Recent studies have proposed additional mechanisms of miR-mRNA regulation, including modulation of competing endogenous RNA (ceRNA), which are mRNA with competitive miR binding sites (Sumazin et al., 2011; Tay et al., 2011). Leveraging the existence of matched mRNA and miR profiling data on a large number of samples, we sought to define the salient interactions between specific pairs of miRs and mRNAs through both of these mechanisms.
We employed a relevance network based approach to infer miR:mRNA associations in GBMs with matched miR and mRNA profiles (n=482). Putative regulatory targets of individual miRs were defined as those genes having strong negative correlation with the miR (< −0.3) and prediction support in three commonly used databases (Miranda, Pictar, TargetScan). 133 miR:mRNA associations defined the final putative miR regulatory network (see Data Portal). The most prevalent associations related to molecular subtypes. For instance, hsa-mir-29a (part of the miR29 family, thought to play a role in the TP53 pathway (Park et al., 2009) was predicted to regulate 23 genes. 17 of these 23 genes were expressed at distinctively high levels in the non-G-CIMP+ proneural tumors only, and not in the G-CIMP+ tumors. Interestingly, three (BCL11A, PCFG3, SS18L1) of the 23 genes in this subnetwork are predicted to act as PDGFRA ceRNAs (see below).
Competitive endogenous mRNAs (ceRNAs) are mRNAs co-regulated in trans by a common miR (Sumazin et al., 2011; Tay et al., 2011). Here, we used a correlation- and NLS-based approach, integrating miRNA and mRNA expression and copy number profiles to predict ceRNAs for four GBM signature genes: PDGFRA, EGFR, NF1, and PTEN. Interestingly, predicted PDGFRA ceRNAs significantly overlapped with proneural GBM signature genes (p-value <1e-15), while EGFR ceRNAs significantly overlapped with classical GBM signature genes (p-value=1.2e-14) (see Data Portal). Predicted ceRNAs of NF1 overlapped with proneural signatures (P<1e-15) and PTEN-associated ceRNAs were correlated with the mesenchymal signature. This provocative finding raises the possibility that ceRNA regulation by miR may contribute to the transcriptomic signature that defines the molecular subtypes in GBM, although this hypothesis remains to be tested.
Signaling pathway activation in different molecular subtypes of GBM
To assess whether enrichment of genomic alterations in molecular subtypes translates into downstream pathway activation, we performed targeted proteomic profiling by reverse-phase protein arrays (RPPA). 214 sample lysates were probed with 171 antibodies targeting phospho- and/or total-protein levels among signaling pathways as previously described (TCGA, 2012c). After normalization, co-clusters of correlated signaling molecules within specific signaling pathways were observed (see Extended Experimental Procedures, Data portal) and were utilized as readout of pathway activity status for correlative analyses.
Unsupervised clustering of RPPA data failed to produce a consistent partitioning of the sample cohort into clearly-defined subtypes. However, 127 out of the 171 antibodies were found to correlate significantly with transcriptomal subtype (Kruskal-Wallis, p<0.05; Extended Experimental Procedures). As anticipated, EGFR amplification/mutation was associated with significant elevations in total EGFR expression (p=3.74E-15) and phosphorylation (p=1.44E-12, SI Figure S7A), both prominent in classical subtype tumors (SI Figure S7B). Classical GBMs also showed relative downregulation of pro-apoptotic proteins (including cleaved caspase 7, cleaved caspase 9, Bid and Bak) as well as MAP kinase signaling, including its downstream target p90RSK. Notch1 and Notch3 expression were moderately increased in classical tumors, consistent with previous reports linking EGFR and Notch activation in GBM (Brennan et al., 2009).
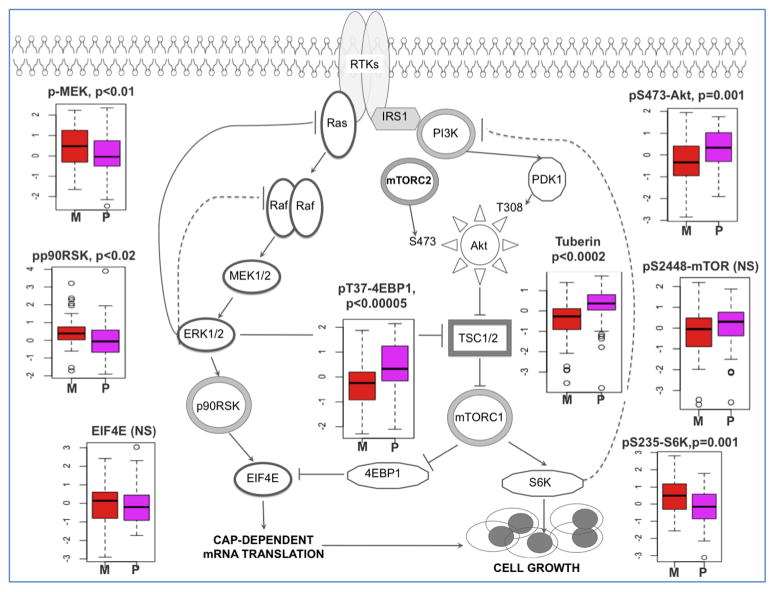
Mesenchymal subtype tumors exhibited elevated levels of endothelial markers, such as CD31 and VEGFR-2, consistent with previous findings (Phillips et al., 2006), as well as markers of inflammation (e.g., Fibronectin and its downstream target COX-2). Mesenchymal tumors showed moderately increased activation of the MAPK pathway, as evidenced by higher levels of phospho-Raf, phospho-MEK and phospho-ERK (Figure 6). These tumors also exhibited decreased levels of the mTOR regulatory protein, tuberin (TSC2 gene product), which is inhibited by ERK phosphorylation.
Figure 6.
Canonical PI3K and MAPK pathway activation determined by reverse phase protein arrays and compared between GBM subclasses: Proneural (P, purple, n=55) and Mesenchymal (M, red, n=45). Activation/expression levels are plotted for principal signaling nodes of the MAPK (phospho-MEK and phospho-p90RSK), PI3 kinase (pS473-Akt) and mTOR (TSC1/2, phospho-mTOR, p235/236 S6, phospho-4EBP1 and EIF4E) pathways (p-values, two-tailed T-test). Mesenchymal tumors showed increased activation of the MAPK pathway (evidenced by higher levels of phospho-MEK and downstream phospho-p90RSK) and decreased levels of phospho-ERK inhibitory target TSC2. In contrast, proneural tumors showed relatively elevated expression and activation of members of the PI(3) kinase pathway including Akt PDK1 target site threonine 308 (p=0.01, not shown) and Akt mTORC2 target site (serine 473). Phospho-ERK levels were not significantly different between these two subtypes.
In contrast to the mesenchymal subtype, proneural GBMs showed relatively elevated expression and activation of the PI3K pathway including the Akt-regulated mTorc1 activation site (Figure 6). Proneural tumors showed greater inhibition of the 4EBP1 translation repressor, whereas mesenchymal tumors display elevated S6 kinase activation (indicative of mTOR effector pathway activation). Therefore, both subtypes achieve mTOR pathway activation although the specific patterns of steady-state protein activation differ.
G-CIMP+ tumors shared characteristics with their proneural superfamily, but also showed decreased expression of several proteins, including Cox-2, IGFBP2 and Annexin 1. Among the 171 antibodies tested in the TCGA dataset, these three proteins were the most negatively prognostic (Cox proportional hazard test, p<0.0004–0.0013). IGFBP2 and Cox-2 have been independently reported as poor prognostic markers in diffuse gliomas (Holmes et al., 2012; Shono et al., 2001), and low IGFBP2 expression has been associated with global DNA hypermethylation in glioma (Zheng et al., 2011). Members of the annexin family have been associated with glioma growth and migration, and annexin-1 is known to be under-expressed in secondary but not primary GBM (Schittenhelm et al., 2009). Together, the correlations of these proteins with G-CIMP status suggest that their prognostic significance is not independent. Analysis of DNA methylation for IGFBP2, COX2 and ANXA1 found no evidence of hyper-methylation in G-CIMP tumors.
Interestingly, samples with RTK amplification had lower levels of canonical RTK-target pathway activities as measured by phospho-AKT, phospho-S6 kinase and phospho-MAPK co-cluster levels (SI Figure S7C). While PTEN loss and deletion were each associated with incremental increases in AKT pathway activity, PI3K-mutant samples had lower AKT activity than samples lacking PI3K mutations, concordant with findings in breast cancer (TCGA, 2012c). Samples harboring NF1 mutation/deletion showed elevated MAP kinase activity (p-ERK and p-MEK, p-value<0.001), and trended towards decreased PKC pathway activity. These examples of non-linear relationship between protein signaling and underlying genetic mutations speak to complex and likely dynamic signaling in cancers.
DISCUSSION
In this study, we provided a comprehensive catalogue of somatic alterations associated with glioblastoma, constructed through whole genome, exome and RNA sequencing as well as copy number, transcriptomic, epigenomic and targeted proteomic profiling. With the availability of detailed clinical information including treatment and survival outcome for nearly the entire cohort, this rich data set offers new opportunity to discover genomics-based biomarkers, validate disease-related mechanisms and generate novel hypotheses.
In addition to alterations in signature oncogenes of GBM, such as EGFR and PI3K, we found that over 40% of tumors harbor at least one non-synonymous mutation among the chromatin-modifier genes. A role for chromatin organization in GBM pathology, which has been described for cancer types such as ovarian carcinoma (Wiegand et al., 2010) and renal carcinoma (Varela et al., 2011), is suggested. We also detected mutations in genes for which targeted therapies have been developed, such as BRAF (Chapman et al., 2011), and FGFR1/FGFR2/FGFR3 (Singh et al., 2012), demonstrating the potential clinical impact of this TCGA dataset.
Structural rearrangements that contributed to the overall complexity of the genome and transcriptome were detected in the majority of GBM. A high frequency of structural variants on the q arm of chromosome 12, involving the MDM2 and CDK4 genes, was observed and associated with the presence of double minute, extrachromosomal DNA fragments, which may be functionally relevant (Zheng et al., 2013). The identification of complex EGFR fusion and deletion variants in nearly half of GBM confirm relevance of this category of somatic alterations to the disease. While the development of a therapeutic strategy targeting mutated EGFR could have a major impact on survival and continues to be a topic of great interest (Vivanco et al., 2012), strategies will need to address the possibility that different EGFR alterations might exist concurrently in a tumor and yield differential biological activities and/or responses to any given targeted inhibitor.
Another level of biological complexity is revealed by targeted proteomic profile, which showed that the impact of specific genomic alterations on downstream pathway signaling is not linear. The discordance between genomic features and proteomic activation status speak to a complex, and likely dynamic, relationship between signaling and molecular alterations. This observation has provocative clinical implication as it directly challenges the notion that therapeutic inhibition of downstream signaling components along a pathway would yield similar efficacy of targeting the mutated gene itself. Additionally, this observation highlights the limitation of TCGA data, namely its inherent static nature given a single time point analysis, and its inability to map specific genetic or protein changes to the individual cells or cell population given its approach to whole-tumor tissue analysis.
In summary, this report reaffirms the power and value of TCGA’s comprehensive multidimensional and clinically annotated GBM reference dataset in enabling hypothesis generation based on unanticipated observations and relationships emerged from unbiased data-driven analyses. We believe that this public resource will serve to facilitate discovery of new insights that can advance our molecular understanding of this disease.
EXPERIMENTAL PROCEDURES
Patient and Sample Characteristics
Specimens were obtained from patients, with appropriate consent from institutional review boards. Details of sample preparation are described in the Extended Experimental Procedures.
Data generation
In total, 599 patients were assayed on at least one molecular profiling platform, which platforms included: (1) exome sequencing, (2) DNA copy number and single nucleotide polymorphism arrays, (3) whole genome sequencing (4) gene expression arrays, (5) RNA sequencing, (6) DNA methylation arrays, (7) reverse phase protein arrays and (8) miRNA arrays. Details of data generation are described in the Extended Experimental Procedures.
Whole Genome and Exome Sequencing Data Analysis
Massively Parallel Sequencing Exome capture was performed by using Agilent SureSelect Human All Exon 50 Mb according the manufacturer’s instructions. All exome and whole genome sequencing was performed on the Illumina GA2000 and HiSeq platforms. Basic alignment and sequence quality control were done by using the Picard and Firehose pipelines at the Broad Institute. Mapped genomes were processed by the Broad Firehose pipeline to perform additional quality control, variant calling, and mutational significance analysis.
RNA Sequencing Data Analysis
Libraries were generated from total RNA and constructed using the manufacturers protocols. Sequencing was done on the Illumina HiSeq platform. Read mapping and downstream data analysis (expression profiles, fusion transcripts, structural transcript variants) were performed using the PRADA pipeline.
Array Data Preprocessing and Analysis
To ensure across-platform comparability, features from all array platforms were compared to a reference genome as previously described (TCGA, 2008). Both single platform analyses and integrated cross-platform analyses were performed, as described in detail in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Exome DNA sequencing in 291 glioblastomas, 42 with whole genome sequencing
RNA sequencing of 164 glioblastomas identifies recurrent gene rearrangements
Copy number, DNA methylation, protein, mRNA and miRNA expression profiles of 543 GBMs
Integrated analysis of somatic alterations, molecular subtypes and affected pathways
Acknowledgments
The TCGA research network contributed collectively to this study. Biospecimens were provided by the Tissue Source Sites and processed by the Biospecimen Core Resource. Data generation and analyses were performed by the Genome Sequencing Centers, Cancer Genome Characterization Centers, and Genome Data Analysis Centers. All data were released through the Data Coordinating Center. Project activities were coordinated by NCI and NHGRI Project Teams.
This work was supported by the following grants from the USA National Institutes of Health: U24CA143883, U24CA143858, U24CA143840, U24CA143799, U24CA143835, U24CA143845, U24CA143882, U24CA143867, U24CA143866, U24CA143848, U24CA144025, U24CA143843, U54HG003067, U54HG003079, U54HG003273, U24CA126543, U24CA126544, U24CA126546, U24CA126551, U24CA126554, U24CA126561, U24CA126563, U24CA143731, U24CA143843.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, Dietrich PY, Weller M, Mariani L, Heppner FL, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta neuropathologica. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan T, Antczak M, Flickinger T, Raines M, Myers M, Kung HJ. A complete description of the EGF-receptor exon structure: implication in oncogenic activation and domain evolution. Oncogene. 1993;8:2939–2948. [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012 doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Pastorino S, Zeng Q, Xu X, Johnson W, Vandenberg S, Verhaak R, Cherniack AD, Watanabe H, Dutt A, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2012;71:7587–7596. doi: 10.1158/0008-5472.CAN-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome research. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KM, Annala M, Chua CY, Dunlap SM, Liu Y, Hugen N, Moore LM, Cogdell D, Hu L, Nykter M, et al. Insulin-like growth factor-binding protein 2-driven glioma progression is prevented by blocking a clinically significant integrin, integrin-linked kinase, and NF-kappaB network. Proc Natl Acad Sci U S A. 2012;109:3475–3480. doi: 10.1073/pnas.1120375109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, Heguy A, Petrini JH, Chan TA, Huse JT. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Akagi K, Inazawa J, Ohta T, Niikawa N, Kayatani F, Sano T, Okada S, Nishisho I. Isolation and characterization of a novel gene deleted in DiGeorge syndrome. Hum Mol Genet. 1995;4:541–549. doi: 10.1093/hmg/4.4.541. [DOI] [PubMed] [Google Scholar]
- Kuttler F, Mai S. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin Cancer Biol. 2007;17:56–64. doi: 10.1016/j.semcancer.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta neuropathologica. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS genetics. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. The American journal of pathology. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Sanborn JZ, Salama SR, Grifford M, Brennan CW, Mikkelsen T, Jhanwar S, Katzman S, Chin L, Haussler D. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm J, Trautmann K, Tabatabai G, Hermann C, Meyermann R, Beschorner R. Comparative analysis of annexin-1 in neuroepithelial tumors shows altered expression with the grade of malignancy but is not associated with survival. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:1600–1611. doi: 10.1038/modpathol.2009.132. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shono T, Tofilon PJ, Bruner JM, Owolabi O, Lang FF. Cyclooxygenase-2 expression in human gliomas: prognostic significance and molecular correlations. Cancer Res. 2001;61:4375–4381. [PubMed] [Google Scholar]
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012a;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012b;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012c;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, Kubek S, Oldrini B, Chheda MG, Yannuzzi N, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Fu J, Vegesna R, Mao Y, Heathcock LE, Torres-Garcia W, Ezhilarasan R, Wang S, McKenna A, Chin L, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27:1462–1472. doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Houseman EA, Morrison Z, Wrensch MR, Patoka JS, Ramos C, Haas-Kogan DA, McBride S, Marsit CJ, Christensen BC, et al. DNA hypermethylation profiles associated with glioma subtypes and EZH2 and IGFBP2 mRNA expression. Neuro Oncol. 2011;13:280–289. doi: 10.1093/neuonc/noq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.