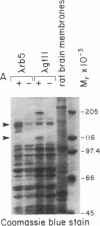
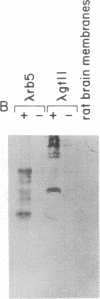
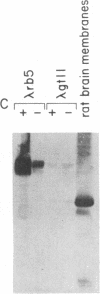
Abstract
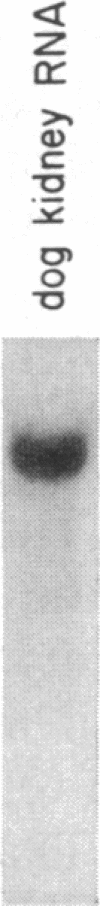
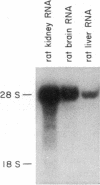
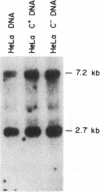
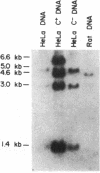
We have isolated a cDNA clone for the rat brain Na,K-ATPase alpha subunit. A lambda gt11 cDNA expression library constructed from mRNA of 1- and 2-week-old rat brains was screened with an antibody reactive with rat brain Na,K-ATPase. A positive phage clone, lambda rb5, containing a 1200-base-pair cDNA insert expressed a beta-galactosidase-cDNA fusion protein that was reactive by immunoblotting with the Na,K-ATPase antibody. This fusion protein was also reactive in ELISA with a monoclonal antibody directed against the alpha subunit of the Na,K-ATPase. A 27S mRNA species exhibiting sequence hybridization to the cDNA insert of lambda rb5 was identified in rat brain, kidney, and liver, as well as in dog kidney. This 27S mRNA exhibited a tissue-specific pattern of abundance consistent with the relative abundance of Na,K-ATPase polypeptides in vivo: kidney greater than brain greater than liver. In a ouabain-resistant HeLa cell line, C+, which contains minute chromosomes and at least a 10-fold greater number of sodium pumps than parental HeLa cells, DNA sequences complementary to lambda rb5 cDNA were amplified approximately 40-fold. Analysis of the lambda rb5 cDNA sequence demonstrated a perfect nucleotide sequence match between a portion of the cDNA and the amino acid sequence of the Na,K-ATPase alpha-subunit fluorescein isothiocyanate binding site. Taken together, the data presented here demonstrate that the lambda rb5 cDNA clone is a portion of the gene coding for the rat brain Na,K-ATPase alpha subunit. The ATPase gene appears to be present in one or very few copies in the rat and human genomes and to be transcriptionally regulated in different rat tissues. In a ouabain-resistant human cell line, on the other hand, ouabain resistance appears to involve an increase in the number of gene copies coding for the Na,K-ATPase.
Full text
PDF
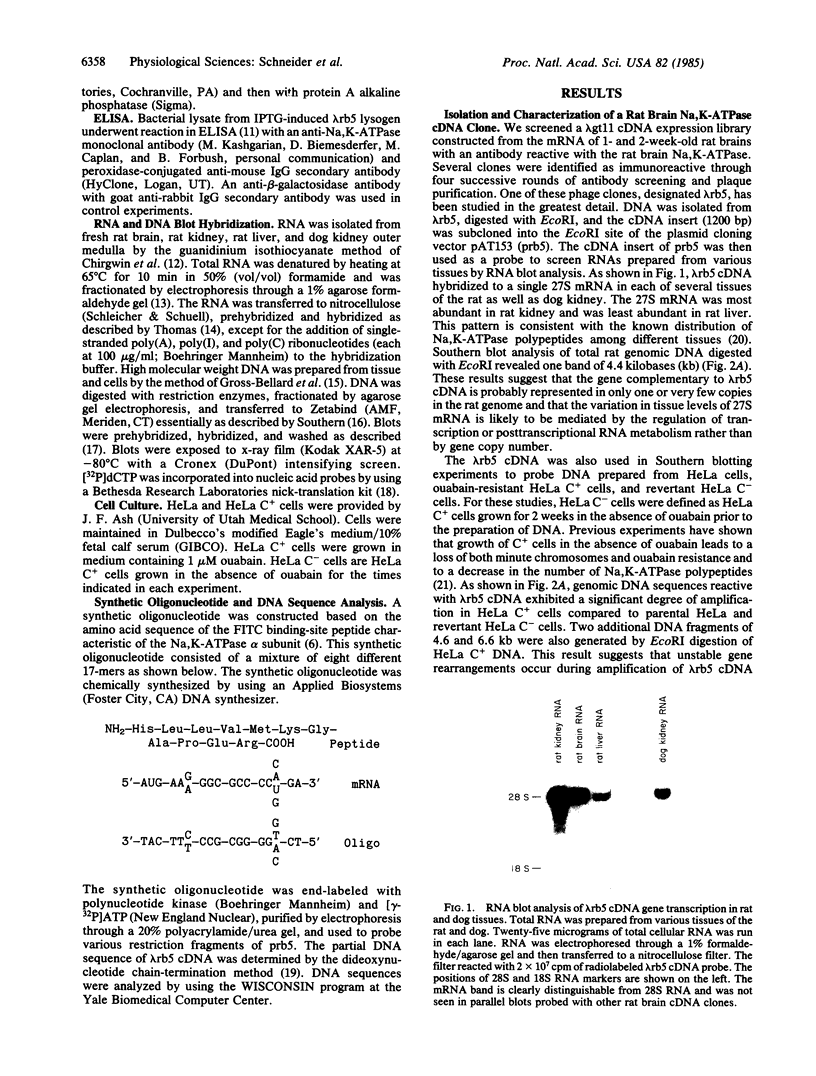


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ash J. F., Fineman R. M., Kalka T., Morgan M., Wire B. Amplification of sodium- and potassium-activated adenosinetriphosphatase in HeLa cells by ouabain step selection. J Cell Biol. 1984 Sep;99(3):971–983. doi: 10.1083/jcb.99.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- English L. H., Epstein J., Cantley L., Housman D., Levenson R. Expression of an ouabain resistance gene in transfected cells. Ouabain treatment induces a K+-transport system. J Biol Chem. 1985 Jan 25;260(2):1114–1119. [PubMed] [Google Scholar]
- Engvall E., Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971 Sep;8(9):871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- Farley R. A., Tran C. M., Carilli C. T., Hawke D., Shively J. E. The amino acid sequence of a fluorescein-labeled peptide from the active site of (Na,K)-ATPase. J Biol Chem. 1984 Aug 10;259(15):9532–9535. [PubMed] [Google Scholar]
- Goldberg D. A. Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5794–5798. doi: 10.1073/pnas.77.10.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Levenson R., Racaniello V., Albritton L., Housman D. Molecular cloning of the mouse ouabain-resistance gene. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1489–1493. doi: 10.1073/pnas.81.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchinson C., Wilderspin A. F., Trinnaman B. J., Green N. M. Identification of a labelled peptide after stoicheiometric reaction of fluorescein isothiocyanate with the Ca2+ -dependent adenosine triphosphatase of sarcoplasmic reticulum. FEBS Lett. 1982 Sep 6;146(1):87–92. doi: 10.1016/0014-5793(82)80710-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosoff P. M., Cantley L. C. Increasing the intracellular Na+ concentration induces differentiation in a pre-B lymphocyte cell line. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7547–7550. doi: 10.1073/pnas.80.24.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg P., Glaser L., Schlesinger P., Cassel D. Epidermal growth factor stimulates amiloride-sensitive 22Na+ uptake in A431 cells. Evidence for Na+/H+ exchange. J Biol Chem. 1983 Apr 25;258(8):4883–4889. [PubMed] [Google Scholar]
- Rozengurt E., Heppel L. A. Serum rapidly stimulates ouabain-sensitive 86-RB+ influx in quiescent 3T3 cells. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4492–4495. doi: 10.1073/pnas.72.11.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Macara I. G., Levenson R., Housman D., Cantley L. Evidence that a Na+/Ca2+ antiport system regulates murine erythroleukemia cell differentiation. J Biol Chem. 1982 Jan 25;257(2):773–780. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Specht S. C., Sweadner K. J. Two different Na,K-ATPases in the optic nerve: cells of origin and axonal transport. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1234–1238. doi: 10.1073/pnas.81.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweadner K. J., Goldin S. M. Active transport of sodium and potassium ions: mechanism, function, and regulation. N Engl J Med. 1980 Apr 3;302(14):777–783. doi: 10.1056/NEJM198004033021404. [DOI] [PubMed] [Google Scholar]
- Sweadner K. J. Two molecular forms of (Na+ + K+)-stimulated ATPase in brain. Separation, and difference in affinity for strophanthidin. J Biol Chem. 1979 Jul 10;254(13):6060–6067. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]