Abstract
Bartonella florenciae sp. nov. strain R4T is the type strain of B. florenciae sp. nov., a new species within the genus Bartonella. This strain, whose genome is described here, was isolated in France from the spleen of the shrew Crocidura russula. B. florenciae is an aerobic, rod-shaped, Gram-negative bacterium. Here we describe the features of this organism, together with the complete genome sequence and its annotation. The 2,010,844 bp-long genome contains 1,909 protein-coding and 46 RNA genes, including two rRNA operons.
Keywords: Bartonella florenciae, genome, France, shrew, Crocidura russula, taxonogenomics
Introduction
Bartonella is the monotypic genus of the family Bartonellaceae, classified among the α-Proteobacteria. To date, 29 Bartonella species have been officially validated [1,2], and many isolates have yet to be described [3,4]. Species of this genus share many general characteristics. They are small (usually less than 1μm), Gram-negative, pleomorphic coccobacilli. All members of the genus are fastidious and grow slowly in vitro. These bacteria are facultatively intracellular and use hemotrophy (infection of erythrocytes) as a parasitic strategy [5,6]. Bartonella species infect a wide range of animal species, including domestic animals such as cats, dogs, rodents, rabbits and cattle as well as a diverse group of wild animals including wildcats, coyotes, deer, elks, foxes, insectivores, bats, etc. The epidemiological cycle of bartonellae consists of a reservoir host with a chronic intravascular infection and sustained bacteremia, and a vector that transfers the bacteria from the reservoir to a susceptible host. Thus, bartonellae may be identified and isolated from a number of blood-sucking arthropods associated with the vertebrate hosts of bacteria. Proven vectors include sandflies, hippoboscids, fleas, soft and hard ticks, lice and mites. Many Bartonella species are associated with human diseases. Bartonella bacilliformis, B. quintana and B. henselae are relatively common human pathogens. Other less common pathogenic species include rodent-associated species, such as B. elizabethae, B. grahamii and B. vinsonii [7-9]. The shrew Crocidura russula is an insectivore mammal in which a Bartonella strain was once identified in Korea [10]. To date, only one officially recognized Bartonella species, B. talpae, was detected in insectivores. However, no type strain is available for this species and its genetic characterization was not achieved [1,11].
In 2003, La Scola et al. proposed a multilocus sequence analysis based on 4 genes and one intergenic spacer as a tool for the description of new Bartonella species [12]. Two of these markers, i.e., gltA and rpoB, were particularly discriminatory, with new Bartonella isolates considered as new species if they exhibit <96.0% and <95.4% sequence identity with other validated species for the 327- and 825-bp fragments of the gltA and rpoB genes, respectively. This strategy being congruent with the “gold-standard” DNA–DNA reassociation for several bacterial genera [13], these criteria have since been regularly applied for the description of new Bartonella species [2,14].
In this study, we used both the genetic criteria of La Scola et al. and the genome sequence, as well as the main phenotypic characteristics of strain R4T to present a summary classification and a set of features for B. florenciae sp. nov. strain R4T(DSM 23735 = CSUR B627). These characteristics support the circumscription of the B. florenciae sp. nov.
Classification and features
In February 2010, an adult Crocidura russula shrew was found dead without evident signs of trauma near the parking lot of the calanque d’En-Vau close to Marseille, France. The shrew was brought to the laboratory where the cardiac blood and the organs (spleen, liver and brain) were collected. The organs ground in Rinaldini solution were inoculated on Columbia agar (BioMerieux, Marcy l'Etoile, France) as previously described [15]. Strain R4 (Table 1) was obtained from the spleen following a 7-day incubation at 37°C in 5% CO2-enriched atmosphere on Columbia agar. Three other morphologically and genetically indistinguishable strains were isolated from the blood, brain and liver from the same shrew.
Table 1. Classification and general features of Bartonella florenciae strain R4T.
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Domain Bacteria | TAS [16] | ||
| Phylum Proteobacteria | TAS [17] | ||
| Class Alphaproteobacteria | TAS [18,19] | ||
| Current classification | Order Rhizobiales | TAS [19,20] | |
| Family Bartonellaceae | TAS 21-23] | ||
| Genus Bartonella | TAS [21,22,24-26] | ||
| Species Bartonella florenciae | IDA | ||
| Type strain R4T | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Not motile | IDA | |
| Sporulation | Nonsporulating | IDA | |
| Temperature range | Mesophilic | IDA | |
| Optimum temperature | 37°C | IDA | |
| MIGS-6.3 | Salinity | Growth in BHI medium + 5% NaCl | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| Carbon source | Unknown | IDA | |
| Energy source | Unknown | IDA | |
| MIGS-6 | Habitat | Crocidura russula | IDA |
| MIGS-15 | Biotic relationship | Facultative intracellular | IDA |
| Pathogenicity | Unknown | ||
| Biosafety level | 3 | ||
| MIGS-14 | Isolation | Spleen of the shrew Crocidura russula | IDA |
| MIGS-4 | Geographic location | France | IDA |
| MIGS-5 | Sample collection time | February 2010 | IDA |
| MIGS-4.1 | Latitude | 43.216667 | IDA |
| MIGS-4.2 | Longitude | 5.5 | IDA |
| MIGS-4.3 | Depth | Surface | IDA |
| MIGS-4.4 | Altitude | 40 m above sea level | IDA |
a Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample but based on a generally accepted property for the species or anecdotal evidence). Evidence codes come from the Gene Ontology project [27]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
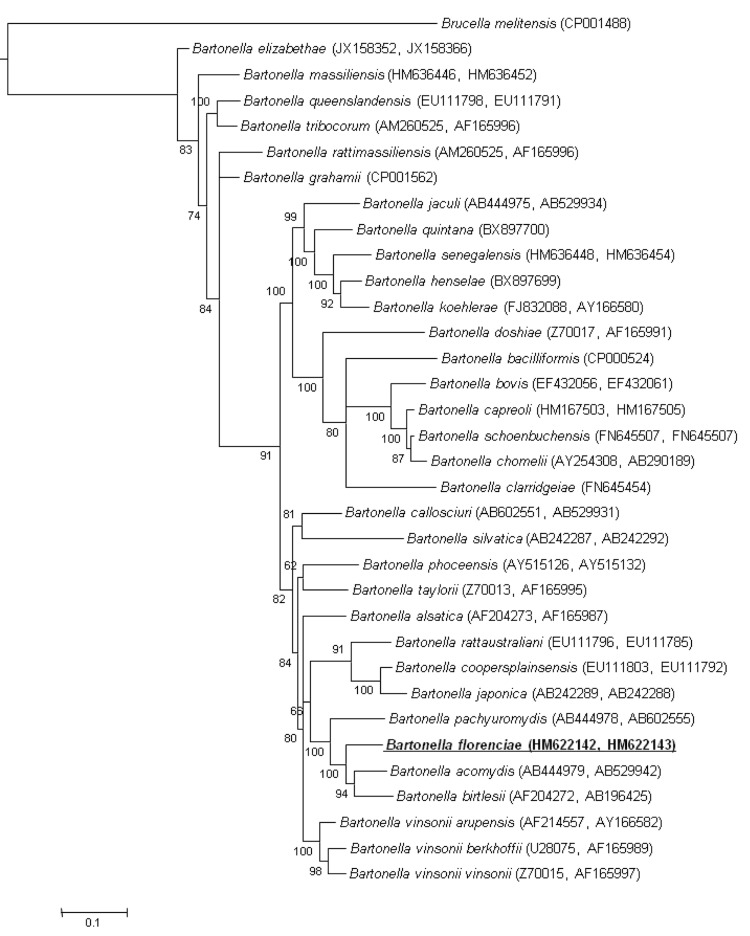
In addition to gltA and rpoB partial gene sequencing, we also sequenced the intergenic transcribed spacer (ITS) along with the 16S rRNA and ftsZ genes as previously described [10,28-31]. The ITS and 16S rRNA of strain R4T exhibited nucleotide sequence similarities of 63.8% and 99.4% with those of Bartonella tribocorum strain CIP 105476, respectively (GenBank accession number AF312505 and NR_074354, respectively); 94.4% with Bartonella birtlesii strain IBS 325 for ftsZ (AM690313), 92.6% with Bartonella acomydis strain KS2-1 for rpoB (AB529942) and 90.7% with Bartonella taylorii strain M6 for gltA (Z70013). Phylogenetically, strain R4T formed a separate branch among the rodent-associated species (Figure 1).
Figure 1.
Phylogenetic tree highlighting the position of B. florenciae strain R4T relative to other type strains within the genus Bartonella. Concatenated gltA and rpoB sequences were aligned using CLUSTALW and phylogenetic inferences obtained using Bayesian phylogenetic analysis [32] with the TOPALi 2.5 software (Biomathematics and Statistics Scotland, Edinburgh, UK) with the integrated MrBayes application [33] with the following substitution models: HKY for the first codon position, GTR+Г for the second codon position and GTR+Г+I for the third codon position. GenBank accession numbers are indicated in parentheses as (gltA, rpoB). Numbers at the nodes are bootstrap values obtained by repeating the analysis 100 times to generate a majority consensus tree. There were a total of 1,044 positions in the final dataset. The scale bar indicates a 10% nucleotide sequence divergence.
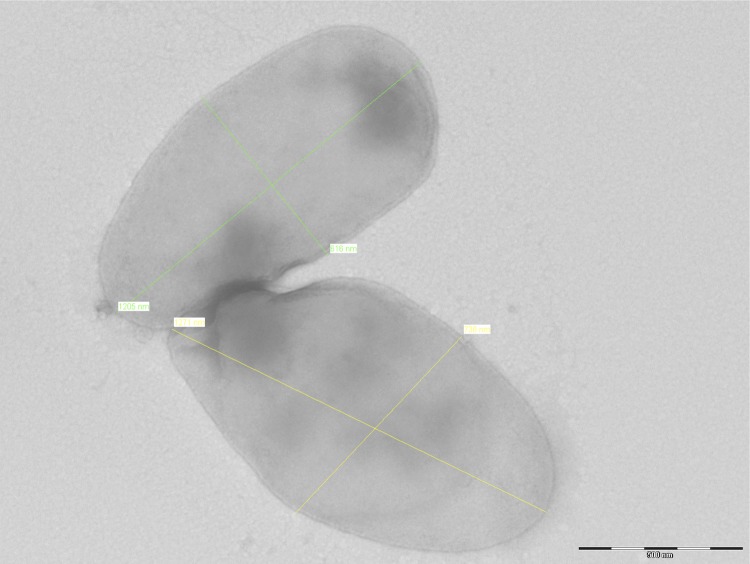
Different growth temperatures (32, 37, 42°C) were tested. Growth only occurred at 37°C in 5% CO2 atmosphere. Colonies were gray, opaque and 0.3 mm to 1 mm in diameter on blood-enriched Columbia agar. Cells grown on agar are Gram-negative and have a mean length and width of 1.39± 0.3 µm and 0.63±0.1 µm, respectively, by electron microscopy (Figure 2). No flagella or pili were observed.
Figure 2.
Transmission electron micrograph of B. florenciae strain R4T, using a Morgagni 268D (Philips) transmission electron microscope at an operating voltage of 60 kV. The scale bar represents 500 nm.
Strain R4T exhibited neither catalase nor oxidase activities. Biochemical characteristics were assessed using an Anaerobe Identification Test Panel AN MicroPlate™ (Biolog Inc., Hayward, CA, USA). None of the 95 biochemical tests available (including D-mannose, D-fructose and D-galactose) were positive. Similar profiles were previously observed for other Bartonella species [14].
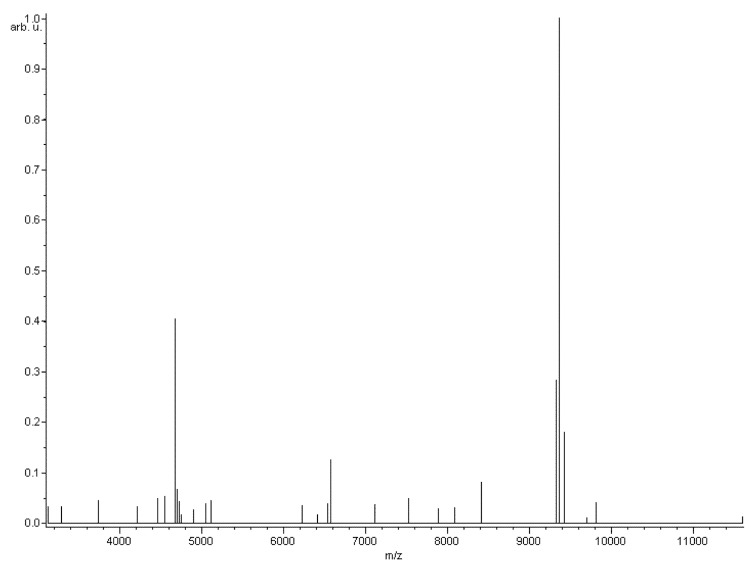
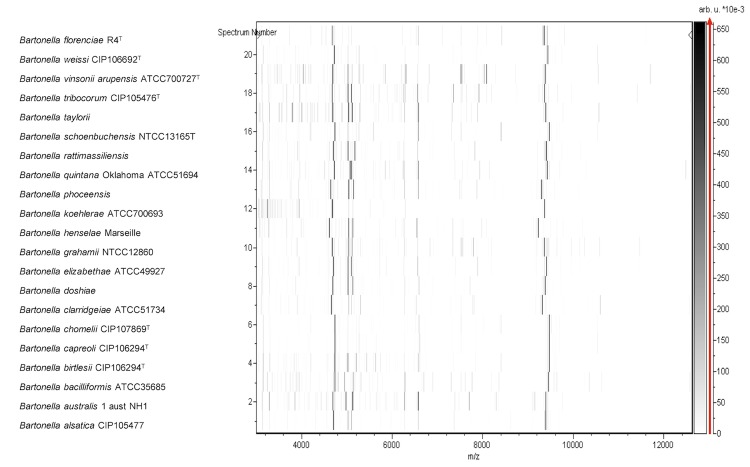
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry protein analysis was carried out as previously described using a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) [34]. Twelve individual colonies were deposited on a MTP 384 MALDI-TOF target plate (Bruker). Each smear was overlaid with 2 μL of matrix solution (a saturated solution of alpha-cyano-4-hydroxycinnamic acid) in 50% acetonitrile/2.5% trifluoroacetic acid, and allowed to dry for five minutes. The twelve R4T spectra were imported into the MALDI BioTyper software (version 2.0, Bruker) and analyzed by standard pattern matching (with default parameter settings) against the main spectra of 4,613 bacteria, including 241 spectra from 20 validly named Bartonella species, used as reference data in the BioTyper database. A score enabled the presumptive identification and discrimination of the tested species from those in a database: a score > 2 with a validated species enabled the identification at the species level; and a score < 1.7 did not enable any identification. For strain R4T, no significant score was obtained, suggesting that our isolate was not a member of any known species (Figures 3 and 4). The gel view shows the spectrum differences with other species within the Bartonella genus (Figure 4).
Figure 3.
Reference mass spectrum from B. florenciae strain R4T. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Figure 4.
Gel view comparing B. florenciae sp. nov., strain R4T with other members of the genus Bartonella. The gel view displays the raw spectra of all loaded spectrum files arranged in a pseudo-gel like fashion. The x-axis records the m/z value. The left y-axis displays the running spectrum number originating from subsequent spectra loading. The peak intensity is expressed by a Gray scale scheme code. The color bar and the right y-axis indicate the relation between the color a peak is displayed with and the peak intensity in arbitrary units.
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of the similarity of its 16S rRNA, ITS, ftsZ, gltA and rpoB to other members of the genus Bartonella. Nucleotide sequence similarity levels of these genes suggested that strain R4T represents a new species within the genus Bartonella. It was the eleventh genome of a Bartonella species and the first genome of Bartonella florenciae sp. nov. A summary of the project information is shown in Table 2. The GenBank accession number is CALU00000000 and consists of 62 contigs (14 scaffolds). Table 3 shows the project information and its association with MIGS version 2.0 compliance.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | One paired-end 3-kb library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium |
| MIGS-31.2 | Fold coverage | 36x |
| MIGS-30 | Assemblers | Newbler version 2.5.3 |
| MIGS-32 | Gene calling method | Prodigal |
| EMBL ID | CALU00000000 | |
| EMBL Date of Release | August, 17, 2012 | |
| MIGS-13 | Project relevance | Biodiversity of the Crocidura russula microbial flora |
Table 3. Nucleotide content and percentage of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 2,010,844 | 100 |
| DNA coding region (bp) | 1,624,868 | 80.8 |
| DNA G+C content (bp) | 774,294 | 38.5 |
| Total genes | 1,955 | 100 |
| RNA genes | 46 | 2.35 |
| Protein-coding genes | 1,909 | 100 |
| Protein with predicted function | 1,135 | 60 |
| Genes assigned to COG | 1,328 | 69.4 |
| Genes with peptide signal | 84 | 4.4 |
| Genes with transmembrane helices (≥3) | 193 | 10 |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Growth conditions and DNA isolation
B. florenciae sp. nov. strain R4T (DSM 23735, CSUR B627) was grown on 5% sheep blood-enriched Columbia agar at 37°C in a 5% CO2 atmosphere. Four Petri dishes were spread and resuspended in 3×100 μl of G2 buffer (EZ1 DNA Tissue kit, Qiagen). A first mechanical lysis was performed by glass powder on the Fastprep-24 device (Sample Preparation system; MP Biomedicals, USA) using 2×20-second cycles. DNA was then treated with 2.5 μg/μL lysozyme (30 minutes at 37°C) and extracted through the BioRobot EZ 1 Advanced XL (Qiagen). The DNA was then concentrated and purified on a Qiamp kit (Qiagen). The yield and concentration were measured by the Quant-it Picogreen kit (Invitrogen) on the Genios_Tecan fluorometer at 131 ng/μl.
Genome sequencing and assembly
DNA (5 μg) was mechanically fragmented on a Hydroshear device (Digilab, Holliston, MA, USA) with an enrichment size of 3-4 kb. The DNA fragmentation was visualized using the Agilent 2100 BioAnalyzer on a DNA labchip 7500 with an optimal size of 3.375 kb. The library was constructed according to the 454 GS FLX Titanium paired-end protocol. Circularization and nebulization were performed and generated a pattern with an optimal at 622 bp. After PCR amplification over 17 cycles followed by double size selection, the single-stranded paired-end library was then quantified with the BioAnalyzer on a DNA labchip RNA pico 6000 at 179 pg/μL. The library concentration equivalence was calculated as 1E+08 molecules/μL. The library was stored at -20°C until further use. The library was clonally amplified with 1.5 cpb in 3 emPCR reactions with the GS Titanium SV emPCR Kit (Lib-L) v2 (Roche). The yield of the 1.5 cpb emPCR was determined to be 8.8%, in the 5 to 20% range recommended in the Roche procedure. Approximately 790,000 beads were loaded on a ¼ region on the GS Titanium PicoTiterPlate PTP Kit 70×75 and sequenced with the GS FLX Titanium Sequencing Kit XLR70 (Roche). The run was analyzed on the cluster through the gsRunBrowser and Newbler assembler (Roche). A total of 232,038 passed filter wells were obtained and generated 72.01 Mb of DNA sequence with an average read length of 310 bp.
The passed filter sequences were assembled using Newbler with 90% identity and 40 bp as overlap. The final assembly identified 14 scaffolds and 62 large contigs (>1.5kb) which corresponds to 36× as an equivalence genome.
Genome annotation
Coding sequences (CDSs) were predicted using PRODIGAL with default parameters [35], but predicted ORFs were excluded if they spanned a sequencing gap region. The functional annotation of protein sequences was performed against the non-redundant GenBank database using BLASTP and functional categories of these proteins was searched against the Clusters of Orthologous Groups (COG) database using COGNITOR [36]. The prediction of RNAs genes, i.e., rRNAs, tRNAs and other RNAs was carried out using RNAmmer [37] and ARAGORN [38] algorithms. The transmembrane helices and signal peptides were identified using TMHMM [39] and SignalP [40] tools, respectively.
Genome properties
The genome is 2,010,844 bp long (one chromosome, one plasmid) with a 38.5% GC content (Table 3, Figure 5). Of the predicted genes, 1,909 were protein-coding genes, and 46 were RNAs including two rRNA operons. The plasmid was 25 kb-long and had a total of 28 genes. A total of 1,135 genes (60%) were assigned a putative function. The remaining genes were annotated as either hypothetical proteins or proteins of unknown functions. The distribution of genes into COGs functional categories is presented in Table 4. The properties and the statistics of the genome are summarized in Tables 3 and 4.
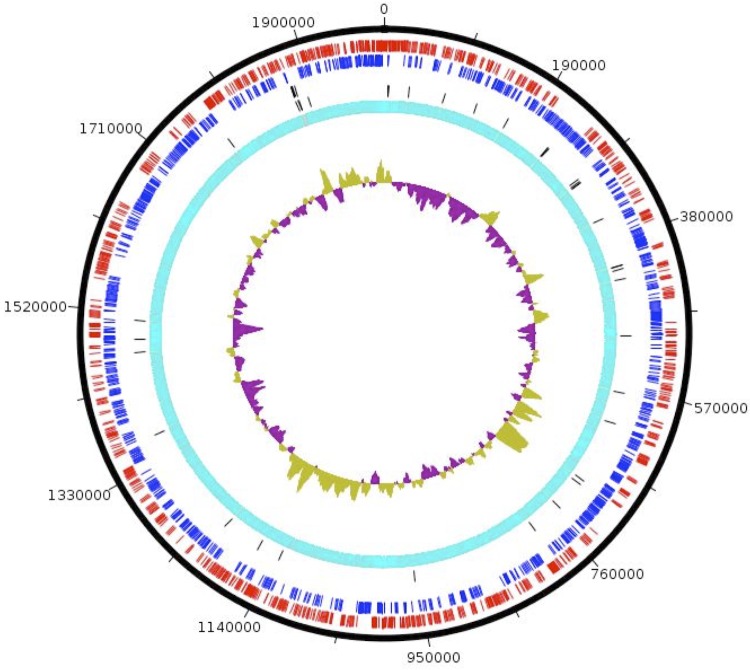
Figure 5.
Graphical circular map of the chromosome. From the outside in, the outer two circles show open reading frames oriented in the forward and reverse directions, respectively. The third circle shows rRNA genes. The inner-most circle shows GC skew, purple and olive indicating negative and positive values, respectively.
Table 4. Number of genes associated with the 25 general COG functional categories†.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 141 | 7.43 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 71 | 3.74 | Transcription |
| L | 98 | 5.17 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 25 | 1.32 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 19 | 1.00 | Defense mechanisms |
| T | 39 | 2.06 | Signal transduction mechanisms |
| M | 95 | 5.01 | Cell wall/membrane biogenesis |
| N | 9 | 0.47 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 7 | 0.37 | Extracellular structures |
| U | 85 | 4.48 | Intracellular trafficking and secretion |
| O | 74 | 3.90 | Posttranslational modification, protein turnover and chaperones |
| C | 77 | 4.06 | Energy production and conversion |
| G | 62 | 3.27 | Carbohydrate transport and metabolism |
| E | 129 | 6.80 | Amino acid transport and metabolism |
| F | 46 | 2.42 | Nucleotide transport and metabolism |
| H | 59 | 3.11 | Coenzyme transport and metabolism |
| I | 42 | 2.21 | Lipid transport and metabolism |
| P | 75 | 3.95 | Inorganic ion transport and metabolism |
| Q | 14 | 0.74 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 189 | 9.96 | General function prediction only |
| S | 134 | 7.06 | Function unknown |
| X | 581 | 30.63 | Not in COGs |
† The total is based on the total number of protein coding genes in the annotated genome.
Comparison with the Bartonella tribocorum genome
Compared to B. tribocorum strain CIP105476 (GenBank accession number NC_010161), B. florenciae strain R4T had a much smaller genome (2,010,844 and 2,619,061 bp, respectively), less genes (1,955 and 2,135 genes, respectively) and a lower G+C content (38.5% and 38.8%, respectively). Comparative genomics of the proteomes of these bacteria showed that 188 protein-coding genes present in B. florenciae were absent or present as pseudogenes in B. tricoborum. These included genes encoding the multidrug resistance efflux pump VceA protein, phage proteins, SAM-dependent methyltransferase, tolA protein, transcriptional repressor Arc, Cytosine-specific methyltransferase NlaX, Glycoside hydrolase, conjugal transfer protein TraC/D, major facilitator superfamily (MFS) proteins, NADPH-dependent FMN reductase, lytic transglycosylase, mccB proteins, transcriptional regulator proteins, membrane protein, D-isomer specific 2-hydroxyacid dehydrogenase, putative phosphoribosylglycinamide synthetase and putative type II restriction endonuclease as well as several hypothetical proteins.
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Bartonella florenciae sp. nov. that contains strain R4T. This bacterium has been isolated in France.
Description of Bartonella florenciae sp. nov.
Bartonella florenciae (flo.ren´ci.ae. N.L. gen. fem. n. florenciae of Florence, named in honor of Florence Fenollar, the prominent French microbiologist who found the Crocidura russula shrew from which the type strain was isolated).
Colonies are opaque, grey, and 0.5 to 1.0 mm in diameter on blood-enriched Columbia agar. Cells are rod-shaped without flagellae. Length and width are 1.39 ± 0.3 µm and 0.63 ± 0.1 µm, respectively. Growth is achieved at 37°C in aerobic atmosphere enriched with 5% CO2. Cells stain Gram-negative, are non-endospore-forming, and are not motile. Catalase and oxidase activities are absent. Using the Anaerobe Identification Test Panel AN MicroPlate, no biochemical activity is observed.
The genome is 2,010,844-bp long (one chromosome and one plasmid) and contains 1,909 protein-coding and 46 RNA genes, including two rRNA operons. The G+C content is 38.5%. Sequences from the ITS, 16S rRNA, ftsZ, rpoB and gltA genes, and the genome are deposited in GenBank under accession numbers HM622140, HM622139, HM622141, HM622143, HM622142 and CALU00000000, respectively. The type strain R4T (DSM 23735, CSUR B627) was isolated from a C. russula shrew found dead in calanque d’En-Vau near Marseille, France.
Acknowledgements
We are grateful to Denis Pyak, Audrey Borg, and Geetha Subramanian for their technical help. The present work was funded by the Agence Nationale de Recherche grant 2010 MALEMAF (research on emergent pathogens in Africa) and the Mediterranée-Infection Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.LPSN http://www.bacterio.cict.fr/b/bartonella.html 12-04-2013
- 2.Sato S, Kabeya H, Fujinaga Y, Inoue K, Une Y, Yoshikawa Y, Maruyama S. Bartonella jaculi sp. nov., Bartonella callosciuri sp. nov., Bartonella pachyuromydis sp. nov., and Bartonella acomydis sp. nov. isolated from wild Rodentia. Int J Syst Evol Microbiol 2013; 63:1734-1740 10.1099/ijs.0.041939-0 [DOI] [PubMed] [Google Scholar]
- 3.Billeter SA, Hayman DT, Peel AJ, Baker K, Wood JL, Cunningham A, Suu-Ire R, Dittmar K, Kosoy MY. Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology 2012; 139:324-329 10.1017/S0031182011002113 [DOI] [PubMed] [Google Scholar]
- 4.Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, Agwanda B, Breiman RF, Rupprecht CE. Bartonella spp. in bats, Kenya. Emerg Infect Dis 2010; 16:1875-1881 10.3201/eid1612.100601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minnick MF, Anderson BE. The genus Bartonella In: Dworkin M (editor), The Procaryotes, Springer, New York, 2006, p. 467-493. [Google Scholar]
- 6.Birtles RJ. Bartonellae as elegant hemotropic parasites. Ann N Y Acad Sci 2005; 1063:270-279 10.1196/annals.1355.044 [DOI] [PubMed] [Google Scholar]
- 7.Angelakis E, Billeter SA, Breitschwerdt EB, Chomel BB, Raoult D. Potential for tick-borne bartonelloses. Emerg Infect Dis 2010; 16:385-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O'Connor SP. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol 1993; 31:872-881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerkhoff FT, Bergmans AM. van der ZA, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol 1999; 37:4034-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim CM, Kim JY, Yi YH, Lee MJ, Cho MR, Shah DH, Klein TA, Kim HC, Song JW, Chong ST, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci 2005; 6:327-334 [PubMed] [Google Scholar]
- 11.Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol 1995; 45:1-8 10.1099/00207713-45-1-1 [DOI] [PubMed] [Google Scholar]
- 12.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol 2003; 11:318-321 10.1016/S0966-842X(03)00143-4 [DOI] [PubMed] [Google Scholar]
- 13.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, Maiden MC, Nesme X, Rosselló-Mora R, Swings J, Trüper HG, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 2002; 52:1043-1047 10.1099/ijs.0.02360-0 [DOI] [PubMed] [Google Scholar]
- 14.Inoue K, Kabeya H, Shiratori H, Ueda K, Kosoy MY, Chomel BB, Boulouis HJ, Maruyama S. Bartonella japonica sp. nov. and Bartonella silvatica sp. nov., isolated from Apodemus mice. Int J Syst Evol Microbiol 2010; 60:759-763 10.1099/ijs.0.011528-0 [DOI] [PubMed] [Google Scholar]
- 15.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piémont Y. Bartonella tribocorum sp.nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol 1998; 48:1333-1339 10.1099/00207713-48-4-1333 [DOI] [PubMed] [Google Scholar]
- 16.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergeys's Manual of systematic Bacteriology, second edition, vol.2 (The Proteobacteria), part B (the Gammaproteobacteria). New York: Springer; 2005. p.1. [Google Scholar]
- 18.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergeys's Manual of systematic Bacteriology, second edition, vol.2 (The Proteobacteria), part C (the Alpha-, Beta-, Delta-, and Epsilonproteobacteria).New York: Springer; 2005. p. 1-324. [Google Scholar]
- 19.Validation List No. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 20.Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 324. [Google Scholar]
- 21.Brenner DJ, O'Connor SP, Winkler HH, Steigerwalt AG. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb. nov., Bartonella henselae comb. nov., and Bartonella elizabethae comb. nov., and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol 1993; 43:777-786 10.1099/00207713-43-4-777 [DOI] [PubMed] [Google Scholar]
- 22.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 23.Gieszczykiewicz M. Zagadniene systematihki w bakteriologii - Zur Frage der Bakterien-Systematic. Bull Acad Pol Sci Biol 1939; 1:9-27 [Google Scholar]
- 24.Strong RP, Tyzzer EE, Sellards AW. Oroya fever, second report. J Am Med Assoc 1915; 64:806-808 10.1001/jama.1915.02570360022007 [DOI] [Google Scholar]
- 25.Weinman D. Genus I. Bartonella Strong, Tyzzer and Sellards 1915, 808. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 904-905. [Google Scholar]
- 26.Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol 1995; 45:1-8 10.1099/00207713-45-1-1 [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol 1995; 45:1-8 10.1099/00207713-45-1-1 [DOI] [PubMed] [Google Scholar]
- 29.Birtles RJ, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bact 1996; 46:891-897 10.1099/00207713-46-4-891 [DOI] [PubMed] [Google Scholar]
- 30.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene aalysis for detection and identification of Bartonella species. J Clin Microbiol 2001; 39:430-437 10.1128/JCM.39.2.430-437.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J Clin Microbiol 2002; 40:3641-3647 10.1128/JCM.40.10.3641-3647.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572-1574 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 33.http://mrbayes.csit.fsu.edu/
- 34.Seng P, Drancourt M, Gouriet F, La SB, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49:543-551 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 35.Prokaryotic Dynamic Programming Genefinding Algorithm (PRODIGAL) http://prodigal.ornl.gov/
- 36.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 2000; 28:33-36 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 2004; 32:11-16 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.TMHMM http://www.cbs.dtu.dk/services/TMHMM/
- 40.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; 8:785-786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]