Abstract
The CD4+/CD8+ T cell ratio is altered when HIV-1 infects the human immune system. However, the exact mechanisms of how CD4+ and CD8+ T cells participate in HIV infection are still unknown. This study used bioinformatics methods to compare the transcriptional profiles between CD4+ and CD8+ T cells in HIV-1-infected patients in order to explore the potential molecular mechanisms of CD4+ and CD8+ T cells in HIV-1 infection. We found that expression patterns of differentially expressed genes (DEG) in CD4+ T cells were dramatically different from those in CD8+ T cells. We also constructed protein–protein interaction (PPI) networks to extract functional modules at each stage, and found that some of the important genes such as BRCA1 were central hubs of the modules. Finally, we applied functional annotation to the modules and found that CD4+/CD8+ T cells played critical roles in regulating the cell cycle and other cellular pathways. Thus, this study would greatly further our understanding of the roles of T cells in HIV infection, and provide potential clues for developing AIDS vaccines in the future.
Introduction
Human immunodeficiency virus (HIV) is a lentivirus that causes acquired immunodeficiency syndrome (AIDS). HIV-1 infection is characterized by changes in T cell function and homeostasis as well as extreme heterogeneity between infected and untreated individuals. The majority of HIV-infected patients develop AIDS in an average of 10 to 20 years.1,2 The differences in the clinical course of HIV-1 infection may correspond to genetic variances in HIV-1 strains, host genetic variances, or differences in the virus-specific immune responses.3–6
HIV infection can be divided into three stages: nonprogressive, chronic, and acute infection. It was found that long-term nonprogressive patients carried undetectable viral loads, had normal CD4+ T cell levels, but had virus-specific cellular responses in peripheral blood and mucosal compartments.7,8 This is in contrast to the chronically HIV-1-infected patients who had high viral loads and CD4+ T cell depletion.8 As more extensive studies were focused on the chronic infections, a new model explaining CD4+ T cell depletion during chronic HIV-1 infection suggested that activated CD4+ T cells from untreated HIV-positive individuals were in a hyperproliferative state modulated by type I interferons that were clearly different from those of HIV-negative individuals.9
CD4+ T cells mainly act as a T cell type to facilitate other T cells in resisting virus infection, while CD8 is widely spread on the surface of suppressor T lymphocytes and cytotoxic T lymphocytes, with different dynamics during HIV-1 infection than CD4.10 It was then concluded that HIV-1 infection resulted in profound disorders in the immune system with a loss of CD4+ T cells and a reduction of the CD4+/CD8+ T cell ratio.11 However, the pathogenic mechanism responsible for these consequences remains elusive.
Therefore, it is important to investigate the difference in gene expression between CD4+ and CD8+ T cells at different stages during HIV-1 infection and to explore the potential molecular mechanism. In the present study, we obtained the transcriptional profiles of CD4+ and CD8+ T cells from HIV-1-infected patients and compared them to normal controls through bioinformatics methods. Our purpose is to better understand the characteristic differences in hierarchical clustering and functions between CD4+ and CD8+ T cells. Our results may help to better classify the mechanisms in the cell immune system in response to HIV infection.
Materials and Methods
Affymetrix microarray data
The gene expression profiles were characterized from the Gene Expression Omnibus (GEO) database (ID: GSE6740), which were deposited by Hyrcza and colleagues.12 A total of 40 genechips were available, including genechips from uninfected samples, nonprogressive HIV infection samples, chronic infection HIV samples, and acute HIV infection samples (five genechips from CD4+ cells and five genechips from CD8+ cells, respectively). The annotations of chips were downloaded from the GPL96 platform (Affymetrix Human Genome U133A Array).
Data preprocessing
The downloaded original data were converted into expression measures and missing data were imputed.13 Then robust multiarray average (RMA) was used to normalize the data.14
Differentially expressed genes (DEGs) analysis
The samples in GSE6740 were divided into three groups to perform pairwise comparisons: uninfected vs. acute HIV infection, uninfected vs. chronic HIV infection, and uninfected vs. nonprogressive HIV infection. For each group, the Linear Models for Microarray Data (LIMMA) package in R language was used to identify DEGs.15 The values of p and | log (FC) | were obtained directly during the analysis and only those genes with a p-value<0.05 and | log (FC) |>1 were selected as the DEGs.
Functional classification
The database of Clusters of Orthologous Groups of proteins (COGs) (www. ncbi.nlm.nih.gov/COG)16 has been incepted as a phylogenetic classification of proteins from complete genomes. All the DEGs were aligned to the COG database through the Basic Local Alignment Search Tool (BLASTx)12,17 for functional annotation with the threshold of E-value<1e-05. The functions of DEGs in CD4+ and CD8+ T cells were observed directly at any stage.
Hierarchical clustering
Hierarchical clustering was performed to compare the gene expression profiles between CD4+ T cells and CD8+ T cells.18 First, all genes were classified by hierarchical clustering to observe the overall expression patterns. Second, the overlapping DEGs of CD4+ and CD8+ T cells among all three HIV-infected stages were extracted. Then the expression pattern of those DEGs was characterized through hierarchical clustering analysis.
Construction of the protein–protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING) database provides both experimental and predicted interaction information.19 The overlapping DEGs of CD4+ and CD8+ T cells among all three HIV-infected stages were mapped to the STRING database to analyze the interactions between two DEGs according to their confidence score. Then, the PPI networks were constructed using Cytoscape.20
Functional modules in the PPI network
Molecular Complex Detection (MCODE) can effectively detect densely connected regions in a molecular interaction network, many of which correspond to known molecular complexes based solely on connectivity data.21 MCODE detects protein complexes that are of the highest quality in terms of the function and localization similarity of proteins within predicted complexes. The functional modules from above PPI networks were identified by the method of MCODE with the threshold of degree ≥2 (degree of each node in one module must be no less than 2) and K-core ≥2 (number of neighbors of each node in one module must be no less than 2). The degree and K-core of each node could be calculated and obtained directly in MCODE.
To analyze the biological function of the modules, the Biological Networks Gene Ontology Tool (Bingo) was used to annotate the module based on Gene Ontology (GO).22 A false discovery rate (FDR) <0.01, which denotes the significant level of corresponding node enrichment, was selected as the cut-off criterion. The value of FDR could be derived directly in Bingo analysis.
Results
Differentially expressed genes analysis
The original data downloaded from the GEO database were normalized by the RMA method (Fig. 1). By the criterion of p-value<0.05 and |log (FC)|>1, 398 (acute HIV infection samples vs. uninfected samples), 280 (chronic HIV infection samples vs. uninfected samples), and 208 (nonprogressor samples vs. uninfected samples) DEGs were identified in CD4+ T cells, respectively. Similarly, 507, 314, and 270 DEGs were identified in CD8+ T cells, respectively. The numbers of DEGs detected in CD8+ T cells were larger than those in CD4+ T cells in all three HIV-infected stages.
FIG. 1.
Expression box plot after data normalization.
Functional classification of DEGs
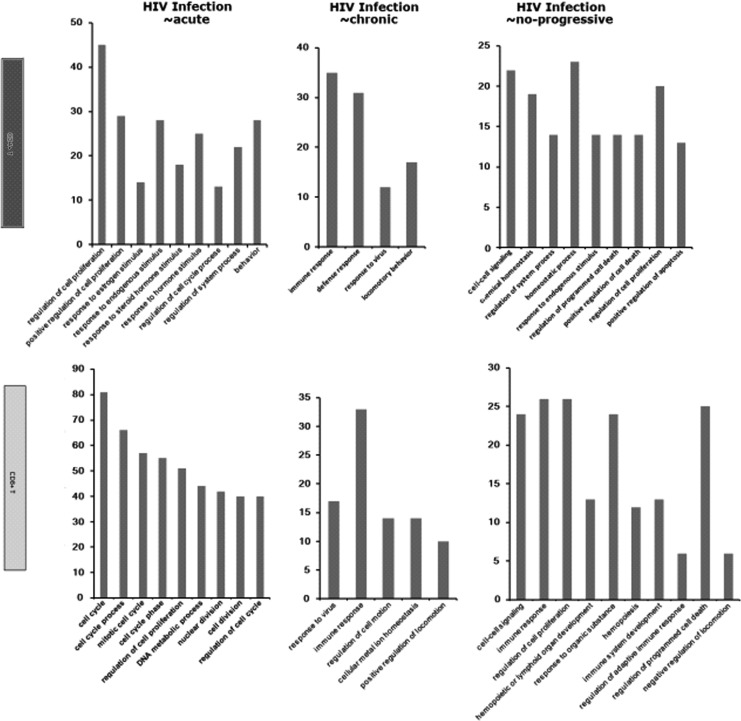
The biological function of DEGs in CD4+ and CD8+ T cells were analyzed by COG with a threshold of E-value<1e-05. According to the similarity of the gene sequence between DEGs and the gene sequence of each node recorded in GO, the DEGs in different stages were classified into different clusters (Fig. 2). The details of every cluster can be seen in Table 1. It was clear that the significant GO categories in CD4+ T cells from nonprogressive HIV infection samples were associated with a homeostatic process and cell signaling, whereas those in CD8+ T cells were associated with an immune response and regulation of cell proliferation. Interestingly, the significant GO categories in CD4+ T cells and CD8+ T cells from chronic HIV infection samples were similar in that both were related to an immune response and locomotor behavior. Also, the DEGs in CD4+ T cells and CD8+ T cells from acute HIV infection samples were classified into the same functional group, which was the function of cell proliferation regulation.
FIG. 2.
Function classification diagrams of differentially expressed genes (DEGs) in two T cell types at three different HIV infection stages. The names of the functional categories in each plot are displayed on the horizontal axis and the numbers of genes on the vertical axis. Upper panel: CD4+ T cells (black); lower panel: CD8+ T cells (gray).
Table 1.
Function Clusters (Top Three) According to the Differentially Expressed Genes in CD4+ and CD8+ T Cells
| GO-term | Term | Count | Genes (part) | |
|---|---|---|---|---|
| CD4+ | ||||
| Acute | 0042127 | Regulation of cell proliferation | 45 | BRCA1, ITGB1, GF1R, |
| 0008284 | Positive regulation of cell proliferation | 29 | ITBG1, IGF1R, ENDN3 | |
| 0009719 | Response to endogenous stimulus | 28 | BRCA1, IGF1R, CAV1 | |
| 0006955 | Immune response | 35 | IF144, IFI44L, TNFSF11 | |
| Chronic | 0006952 | Defense response | 31 | ITGB1, IFIH1, KYNU |
| 0007626 | Locomotor behavior | 17 | SERPIND, ANKH, CCRL2 | |
| 0042592 | Homeostatic process | 23 | ITGB1, IL1A, TNNI3 | |
| Nonprogressor | 0007267 | Cell–cell signaling | 22 | GAL, GLRB, SYT1 |
| 0042127 | Regulation of cell proliferation | 20 | GAL, IL1A, FLT1 | |
| CD8+ | ||||
| Acute | 0007049 | Cell cycle | 81 | BRCA1, ITBG1, NCAPG |
| 0022402 | Cell cycle process | 66 | BRCA1, ITBG1, NCAPG | |
| 0000278 | Mitotic cell cycle | 57 | BRCA1, ITBG1, NCAPG | |
| 0009615 | Response to virus | 17 | IF144,IFIH1, BST2 | |
| Chronic | 0006955 | Immune response | 33 | IF144L, IF144, IFIH1 |
| 0051270 | Regulation of cell motion | 14 | IGF1R, ACTN1, | |
| 0006955 | Immune response | 26 | CSF3, IGF1R, LST1 | |
| Nonprogressor | 0042127 | Regulation of cell proliferation | 26 | CSF3, IGF1R, LST1 |
| 0043067 | Regulation of programmed cell death | 25 | TOP2A, IGF1R, OP2 | |
Hierarchical clustering
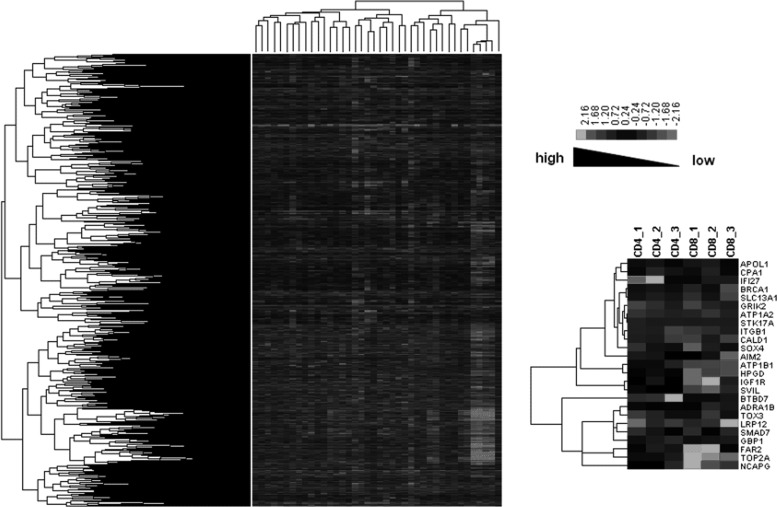
At first, we performed hierarchical clustering for all genes (Fig. 3, left panel). The result did not show any obvious difference in gene expression patterns between CD4+ T cells and CD8+ T cells. Then, we adjusted the hierarchical clustering based on the overlapping DEGs in CD4+ and CD8+ T cells at all three stages (Fig. 3, right panel). We then observed that the gene expression patterns in CD4+ T cells were dramatically different from those in CD8+ T cells. Among them, CPA1 (carboxypeptidase A1) and BRCA1 (breast cancer-associated protein-1) were both significantly up-regulated in CD4+ T cells but down-regulated in CD8+ T cells.
FIG. 3.
Hierarchical clustering diagrams. Left panel represents the whole function classification diagram of genes across all the samples. The hierarchical clustering result of overlapping DEGs in CD4+ and CD8+ T cells is shown in the right panel. The relative levels of gene expression are depicted with a color scale where gray represents a low expression level and white represents a high expression level.
PPI network construction and modules identification
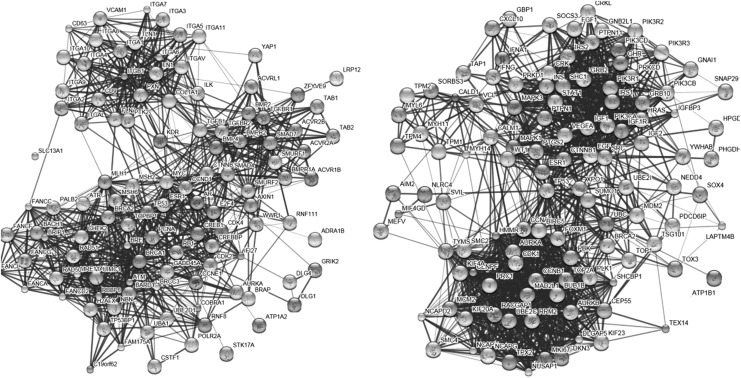
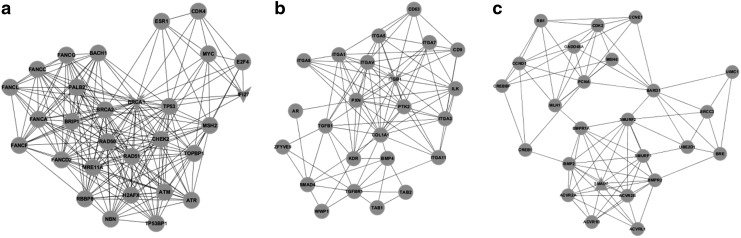
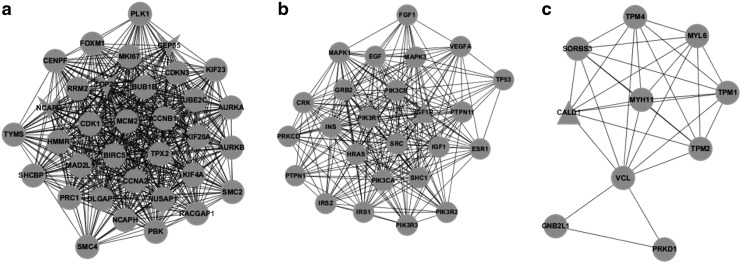
To understand the DEGs from a systemic perspective, we mapped the overlapping DEGs in CD4+ T cells and those in CD8+ T cells at all stages into STRING, and constructed two PPI networks, respectively (Fig. 4). Since the overlapping DEGs in each cell type were highly abundant, the two PPI networks were too complicated to yield any useful information. Therefore, we used MCODE to identify the functional modules in these two networks. With a threshold of degree ≥2 and k-score ≥2, three functional modules were extracted from each network (Figs. 5 and 6). In the modules derived from the PPI network of CD4+ T cells, BRCA1, IFI27 (interferon α-inducible protein 27), ITGB1 (integrin beta 1), and SMAD7 (SMAD family member 7) appeared to be hub nodes. In contrast, NCAPG (non-SMC condensin I complex subunit G), OP2 (osteogenic protein 2), GEP55, IGF1R (insulin-like growth factor 1 receptor), and CALD1 (caldesmon 1) appeared to be hub nodes in modules derived from the PPI network of CD8+ T cells.
FIG. 4.
Protein–protein interaction (PPI) network constructed in CD4+ T cells (left) and CD8+ T cells (right).
FIG. 5.
(a–c) Functional modules derived from the PPI network in CD4+ T cells. The circular node represents the predicted interactive gene (protein). The triangular node represents DEGs, among which the inverted triangular node represents the down-regulated genes and the regular triangular node represents the up-regulated genes.
FIG. 6.
(a–c) Function modules derived from the PPI network in CD8+ T cells. The circular node represents the predicted interactive gene (protein). The triangular node represents DEGs, among which the inverted triangular node represents the down-regulated genes and the regular triangular node represents the up-regulated genes.
BINGO was used to annotate the modules and as a consequence, the enriched function within each module was obtained (Tables 2 and 3). For modules of CD4+ T cells, the functions were enriched in DNA repair, response to DNA damage stimulus (top two categories of module a), cell surface receptor-linked signaling pathway, system development (top two categories of module b), positive regulation of cellular process, and signaling pathway (top two categories of module c). Correspondingly, the GO categories of nuclear division, mitosis (top two categories of module a), signal transduction and transmission (top two categories of module b), and muscle contraction and system process (top two categories of module c) were significantly enriched in modules of CD8+ T cells.
Table 2.
Function Modules (Top Five) from Interaction Network According to the Differentially Expressed Genes of CD4+ T Cells
| GO-ID | FDR | Description | |
|---|---|---|---|
| Module a | 6281 | 1.51E-29 | DNA repair |
| 6974 | 2.74E-29 | Response to DNA damage stimulus | |
| 33554 | 6.07E-25 | Cellular response to stress | |
| 6259 | 7.42E-25 | DNA metabolic process | |
| 51716 | 1.68E-20 | Cellular response to stimulus | |
| Module b | 7166 | 1.79E-13 | Cell surface receptor-linked signaling pathway |
| 48731 | 4.53E-10 | System development | |
| 23033 | 5.58E-10 | Signaling pathway | |
| 48856 | 1.91E-09 | Anatomical structure development | |
| 23052 | 2.02E-09 | Signaling | |
| 48513 | 7.60E-09 | Organ development | |
| Module c | 48522 | 3.11E-14 | Positive regulation of cellular process |
| 23033 | 7.42E-14 | Signaling pathway | |
| 48518 | 1.97E-13 | Positive regulation of biological process | |
| 31323 | 2.41E-13 | Regulation of cellular metabolic process | |
| 19222 | 7.26E-13 | Regulation of metabolic process |
FDR, false discovery rate.
Table 3.
Function Modules (Top Five) from Interaction Network According to the Differentially Expressed Genes of CD8+ T Cells
| GO-ID | FDR | Description | |
|---|---|---|---|
| Module a | 280 | 2.28E-28 | Nuclear division |
| 7067 | 2.28E-28 | Mitosis | |
| 279 | 2.28E-28 | M phase | |
| 87 | 2.61E-28 | M phase of mitotic cell cycle | |
| 48285 | 2.61E-28 | Organelle fission | |
| Module b | 7165 | 1.24E-15 | Signal transduction |
| 23060 | 8.61E-15 | Signal transmission | |
| 23046 | 8.61E-15 | Signaling process | |
| 23052 | 8.61E-15 | Signaling | |
| 23033 | 9.03E-14 | Signaling pathway | |
| Module c | 6936 | 3.36E-13 | Muscle contraction |
| 3012 | 3.42E-13 | Muscle system process | |
| 3008 | 1.01E-05 | System process | |
| 30029 | 4.53E-04 | Actin filament-based process | |
| 6928 | 1.83E-03 | Cellular component movement |
Discussion
It is well known that HIV-1 is the pathogen that can cause AIDS.23 We hypothesized that finding the difference in mutual DEGs between CD4+ and CD8+ T cells at different stages of infection would provide further insight into the mechanisms and explain why CD4+ but not CD8+ T cells undergo progressive depletion after HIV-1 infection.
There have been few studies focusing on analyzing the systemic characteristics of CD4+ T cells at different stages of HIV infection, or the difference between CD4+ and CD8+ T cells at the same stage. In this study, transcriptional profiles of CD4+ and CD8+ T cells from HIV-1-infected patients were compared with uninfected patients by DNA microarrays, in an attempt to determine the functional relationship between CD4+ and CD8+ T cells in HIV-1-infected patients.
BRCA1 was significantly enriched in several GO categories, such as cell proliferation and the cell cycle, in CD4+ and CD8+ T cells from acute HIV infection samples (Table 1) as well as the hub node of the PPI network. These results suggested that BRCA1 may play an important role in the progression of HIV-1 infection. Indeed, a previous study concerning the role of BRCA1 in HIV infection demonstrated that HIV-1 viral protein R (Vpr)-induced apoptosis was mediated via ART (Rad3-related protein) phosphorylation of BRCA1 and consequent up-regulation of GADD45α (growth arrest and DNA damage-45 protein α).24 In a recent study, Buckley et al. proposed that BRCA1 can regulate interferon-gamma (IFN-γ) signaling through a mechanism involving the type I IFNs.24 Meanwhile, evidence showed that type I IFNs were produced in response to microbial infections as part of the innate immune response.25 Thus we considered that the involvement of BRCA1 in HIV-1 infection may be partly by regulating the IFN-γ signaling pathway.
Previous studies had confirmed that virus reproduction (over 99%) occurred mainly in CD4+ T cells inside the peripheral blood and lymphoid tissue.26 And infection with HIV-1 can inhibit CD4+ T cell proliferation. Our results were consistent with the fact that the functions of DEGs in CD4+ T cells from acute HIV infection were correlated with cell proliferation.
Note that there were more down-regulated genes in CD8+ T cells than in CD4+ T cells, such as NCAPG, CEP55, OP2, and GF1R. We proposed that there was an apparent difference in DEGs between CD4+ and CD8+ T cells at various stages after HIV infection.
The functional modules in CD4+ T cells and CD8+ T cells (Figs. 5 and 6) suggested that the gene regulation patterns between CD4+ and CD8+ T cells were evidently different. In addition, the functional difference between the modules in CD4+ and CD8+ T cells that we observed in the current study was consistent with the studies indicating that specific CD8+ T cells play a leading role in directly fighting HIV-1 infection while CD4+ T cells mainly function through supporting CD8+ T cells and B cells.27
In conclusion, the immune responses of CD4+ and CD8+ T cells at different stages after HIV-1 infection were different. Even at the same stage, the DEGs of CD4+ T cells were significantly different from those of CD8+ T cells. The specific DEGs in CD4+ and CD8+ T cells would likely provide a good clue to further elucidate the functional roles of different T cells in HIV infection, and potentially lead to the development of a promising method for AIDS vaccine design.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Mourich DV, Lee S, Reyes-Teran G, Mackewicz CE, and Levy JA: Lack of differences in nef alleles among HIV-infected asymptomatic long-term survivors and those who progressed to disease. AIDS Res Hum Retroviruses 1999;15(17):1573–1575 [DOI] [PubMed] [Google Scholar]
- 2.Altfeld M, Addo MA, Rosenberg ES, et al. : Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 2003;17(18):2581–2591 [DOI] [PubMed] [Google Scholar]
- 3.Ogg GS, Jin X, Bonhoeffer S, et al. : Quantitation of HIV-1 specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 1998;279(5359):2103–2106 [DOI] [PubMed] [Google Scholar]
- 4.Rhame FS: When to start antiretroviral therapy. Curr Infect Dis Rep 2011;13(1):60–67 [DOI] [PubMed] [Google Scholar]
- 5.Gilliam BL, Patel D, Talwani R, and Temesgen Z: HIV in Africa: Challenges and directions for the next decade. Curr Infect Dis Rep 2012;14(1):91–101 [DOI] [PubMed] [Google Scholar]
- 6.Pentier JM, Sewell AK, and Miles JJ: Advances in T-cell epitope engineering. Frontiers Immunol 2013;4:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sankaran S, Guadalupe M, Reay E, et al. : Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc Natl Acad Sci USA 2005;102(28):9860–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith MZ, Bastidas S, Karrer U, and Oxenius A: Impact of antigen specificity on CD4+ T cell activation in chronic HIV-1 infection. BMC Infect Dis 2013;13:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedaghat AR, German J, Teslovich TM, et al. : Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: Type I interferon-mediated disruption of T-cell dynamics. J Virol 2008;82(4):1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogg GS, Jin X, Bonhoeffer S, et al. : Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 1998;279(5359):2103–2106 [DOI] [PubMed] [Google Scholar]
- 11.Plymale DR, Tang DSN, Comardelle AM, Fermin CD, Lewis DE, and Garry RF: Both necrosis and apoptosis contribute to HIV-1-induced killing of CD4 cells. AIDS 1999;13(14):1827–1839 [DOI] [PubMed] [Google Scholar]
- 12.Hyrcza MD, Kovacs C, Loutfy M, et al. : Distinct transcriptional profiles in ex vivo CD4(+) and CD8(+) T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8(+) T cells. J Virol 2007;81(7):3477–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Troyanskaya O, Cantor M, Sherlock G, et al. : Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17(6):520–525 [DOI] [PubMed] [Google Scholar]
- 14.Fujita A, Sato J, Rodrigues L, Ferreira C, and Sogayar M: Evaluating different methods of microarray data normalization. BMC Bioinform 2006;7(1):469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK: Limma: Linear models for microarray data. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor R (Gentleman VC, Dudoit S, Irizarry R, and Huber W, Eds.). Springer, New York, 2005, pp. 397–420 [Google Scholar]
- 16.Tatusov RL, Natale DA, Garkavtsev IV, et al. : The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 2001;29(1):22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ: Basic local alignment search tool. J Mol Biol 1990;215(3):403–410 [DOI] [PubMed] [Google Scholar]
- 18.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, and Church GM: Systematic determination of genetic network architecture. Nat Genet 1999;22(3):281–285 [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Franceschini A, Kuhn M, et al. : The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39(Database issue):D561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, et al. : Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bader GD. and Hogue CW: An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 2003;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maere S, Heymans K, and Kuiper M: BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21(16):3448–3449 [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz RJ: Reservoirs of human immunodeficiency virus type 1: The main obstacles to viral eradication. Clin Infect Dis 2002;34(1):91–97 [DOI] [PubMed] [Google Scholar]
- 24.Buckley NE, Hosey AM, Gorski JJ, et al. : BRCA1 regulates IFN-gamma signaling through a mechanism involving the type I IFNs. Mol Cancer Res 2007;5(3):261–270 [DOI] [PubMed] [Google Scholar]
- 25.Pestka S, Langer JA, Zoon KC, and Samuel CE: Interferons and their actions. Annu Rev Biochem 1987;56:727–777 [DOI] [PubMed] [Google Scholar]
- 26.Gunthard HF, Havlir DV, Fiscus S, et al. : Residual human immunodeficiency virus (HIV) type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J Infect Dis 2001;183(9):1318–1327 [DOI] [PubMed] [Google Scholar]
- 27.Migueles SA, Laborico AC, Shupert WL, et al. : HIV-specific CD8(+) T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 2002;3(11):1061–1068 [DOI] [PubMed] [Google Scholar]