Significance
Surface proteins are linked to the cell wall of Gram-positive bacterial pathogens by a mechanism requiring LPXTG motif sorting signals and sortase. Here we show that surface proteins are also released from the bacterial surface into the extracellular milieu and may fulfill functions similar to secreted polypeptides. We demonstrate that protein A of Staphylococcus aureus, a B cell superantigen, is released with peptidoglycan linked to its C terminus. Release of protein A involves murein hydrolases that remove immunostimulatory N-acetylmuramic acid and GlcNAc residues and liberate the polypeptide from the envelope. This mechanism of surface protein release may be universal for surface proteins of Gram-positive bacteria.
Keywords: surface protein, Gram-positive bacteria, sortase-anchored protein
Abstract
Staphylococcal protein A (SpA) is anchored to the cell wall envelope of Staphylococcus aureus by sortase A, which links the threonyl (T) of its C-terminal LPXTG motif to peptidoglycan cross-bridges (i.e., Gly5). SpA binds the Fcγ domains of IgG and protects staphylococci from opsonophagocytic clearance. Moreover, SpA cross-links B-cell receptors to modify host adaptive immune responses. The mechanisms whereby SpA is released from the bacterial surface to access the host’s immune system are not known. Here we demonstrate that SpA is released with murein tetrapeptide-tetraglycyl [l-Ala-d-iGln-(SpA-Gly5)l-Lys-d-Ala-Gly4] linked to its C-terminal threonyl. LytN, a cross-wall murein hydrolase, contributes to the release of SpA by removing amino sugars [i.e., N-acetylmuramic acid-N-acetylglucosamine (MurNAc-GlcNAc)] from attached peptidoglycan, whereas LytM, a pentaglycyl-endopeptidase, triggers polypeptide release from the bacterial envelope. A model is proposed whereby murein hydrolases cleave the anchor structure of released SpA to modify host immune responses.
The Gram-positive bacterium Staphylococcus aureus is a pathogen of humans (1). Cells of S. aureus are surrounded by a thick layer of highly cross-linked cell wall peptidoglycan (2). The peptidoglycan layer is formed from lipid II precursors, C55-(PO3)2-N-acetylmuramic acid (MurNAc)-(l-Ala-d-iGln-(Gly5)l-Lys-d-Ala-d-Ala)-GlcNAc (3), via the transpeptidation and transglycosylation reactions of cell wall synthesis to generate [MurNAc-(l-Ala-d-iGln-(Gly5)l-Lys-d-Ala)-GlcNAc]n polymer (4). Assembled peptidoglycan is a single large macromolecule that protects bacteria against osmotic lysis (5) and also functions as scaffold for the anchoring of wall teichoic acids (6) and proteins (7). These secondary cell wall polymers promote specific interactions between staphylococci and host tissues (8). Cell wall-anchored surface proteins are synthesized as precursors with N-terminal signal peptides and C-terminal LPXTG motif sorting signals (9). Following cleavage of the N-terminal signal peptide by signal peptidase, the C-terminal sorting signal is cleaved by sortase A between the threonyl (T) and the glycyl (G) of the LPXTG motif (10). Sortase A forms an acyl enzyme, capturing the C-terminal carboxyl group of cleaved surface proteins with its active site cysteine thiol (11). These acyl intermediates are relieved by the nucleophilic attack of the amino group of pentaglycyl within lipid II and incorporated into the cell wall via the transpeptidation and transglycosylation reactions (7, 12).
The genomes of S. aureus isolates harbor 17 to 22 genes encoding LPXTG motif surface proteins, which can be further classified as precursors with canonical or YSIRK-G/S signal peptides (13). Surface proteins with canonical signal peptides are secreted and immobilized to peptidoglycan near the cell poles of dividing staphylococci (14). In contrast, precursors with YSIRK-G/S signal peptides are secreted into the cross-wall, a membrane enclosed compartment for the de novo synthesis of peptidoglycan that separates daughter cells during division (14). When precursors with YSIRK-G/S signal peptides and LPXTG motif sorting signals have been deposited at the cross-wall and its peptidoglycan has been split, surface proteins are displayed over the staphylococcal surface (14). YSIRK-G/S precursors include proteins with important virulence functions that are synthesized in large abundance, including clumping factor A (15), fibronectin binding proteins (16, 17), iron-regulated surface protein B (18), and staphylococcal protein A (SpA) (19).
SpA binds human or animal Ig via its Ig-binding domains that capture the Fcγ domain of IgG or the Fab domain of VH3-clan IgG and IgM antibodies (20, 21). SpA binding to the Fcγ domain blocks the ability of antibodies with specific binding activities for the staphylococcal surface to promote Fc receptor-mediated opsonophagocytosis and bacterial killing (22). SpA binding to the Fab domain of VH3-clan IgM triggers B-cell receptor cross-linking and clonal expansion of B lymphocytes, which eventually undergo apoptotic collapse (23). During infection, this B-cell superantigen activity of SpA ablates host adaptive immune responses against many staphylococcal antigens (24). Although S. aureus disease predominantly manifests as localized skin or soft-tissue infection, its suppressive effects on the immune system appear to be general (25). If so, we wondered whether SpA, a key factor for staphylococcal immune evasion, is released from the bacterial surface.
Results
Protein A Release from S. aureus.
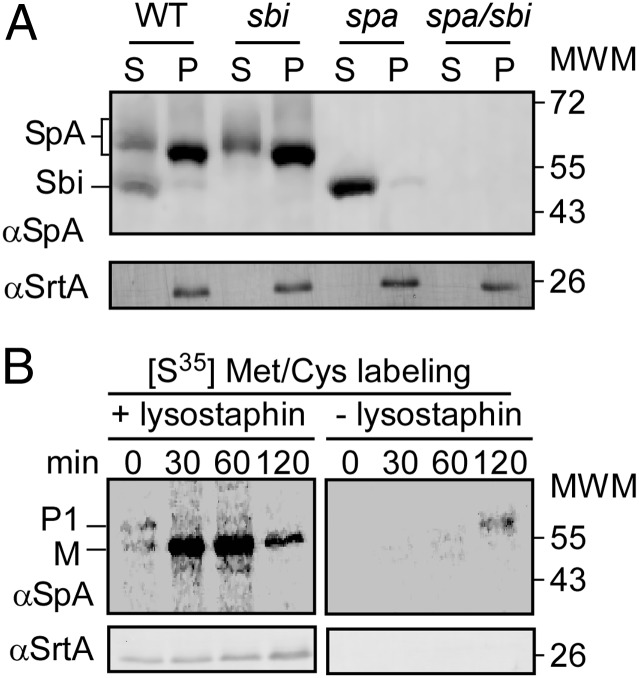
To examine the fate of protein A by immunoblotting, S. aureus Newman, as well as its isogenic spa, sbi, and spa/sbi mutants, were grown as tryptic soy broth (TSB) cultures and centrifuged, and extracellular medium was separated from the bacterial sediment. S. aureus sbi encodes staphylococcal binder of Ig, a secreted polypeptide that binds the Fcγ domain of IgG, similar to protein A (spa encoded) (26). Proteins in both fractions, extracellular medium and the bacterial sediment, were precipitated with trichloroacetic acid (TCA). Proteins tethered to the envelope were released with lysostaphin (27), and samples were analyzed by immunoblotting with SpAKKAA-mAb 3F6 (28). S. aureus Newman cultures harbored 84.90% (±1.28) of SpA in the bacterial envelope, whereas 15.10% (±1.28) were released into the extracellular medium (Fig. 1A). As controls, the spa and spa/sbi mutants did not express spa. The sbi variant was not affected for spa expression, SpA anchoring to the cell wall, or SpA release into the extracellular medium (Fig. 1A). SrtA, the membrane-anchored transpeptidase (29), was not released into the extracellular medium (Fig. 1A).
Fig. 1.
S. aureus releases protein A from the cell wall envelope. (A) S. aureus Newman WT or sbi, spa, or spa/sbi cultures were centrifuged and the extracellular medium separated with the supernatant (marked as “S”) from the bacterial pellet (marked as “P”). After treatment of the staphylococcal cell wall envelope with lysostaphin, proteins in both fractions were analyzed by immunoblotting with polyclonal antibodies against protein A (αSpA) or sortase A (αSrtA). (B) S. aureus Newman cells were washed in PBS solution, pulse-labeled with [35S]Met/Cys, and mixed with fresh culture media. At timed intervals after labeling (0, 30, 60, 120 min), two aliquots from the culture were precipitated with TCA. One sample was treated with lysostaphin to release sortase (cell wall)-anchored proteins (Left), whereas the other was treated with hot SDS to solubilize released protein A molecules (Right). Radioactive samples were immunoprecipitated with αSpA and analyzed by SDS/PAGE and PhosphorImager. Immunoblotting with αSrtA was used as fractionation control.
Following ribosomal synthesis, SpA precursors are secreted within 2 min across the membrane and processed by sortase A (9). Are SpA molecules initially anchored to the cell wall and subsequently released into the medium, or do staphylococci steadily release some portion of protein A? To address this, S. aureus Newman was pulse-labeled with [35S]methionine/cysteine. At timed intervals, culture aliquots were precipitated with TCA, suspended in buffer, and split into two samples. One sample was treated with hot SDS to solubilize proteins released into the medium; the other sample was treated with lysostaphin to solubilize anchored protein A (30). Immediately following the chase (0 min), and 30 min thereafter, all pulse-labeled protein A required lysostaphin treatment to gain solubility (Fig. 1B). Beginning at 60 min, 1.46% of protein A was found soluble without lysostaphin treatment, and 15.2% of pulse-labeled SpA was released at 120 min (Fig. 1B). Sortase A served as loading control and was not solubilized without lysostaphin treatment (Fig. 1B). Taken together, these data indicate that SpA precursors are first anchored to the cell wall envelope and subsequently released into the extracellular medium.
Structure of Released Protein A.
SpA was purified from the culture medium of S. aureus Newman lacking sbi by affinity chromatography on human IgG-Sepharose (31). Edman degradation released amino acid residues of protein A that matched the N-terminal sequence of mature SpA (32). The first Edman cycle released fourfold higher molar amounts of amino acid, i.e., alanine, the N-terminal residue of SpA and a constituent of peptidoglycan, than the next cycle (glutamine). These data suggest that phenylthiohydantoin cleaved alanine from the N terminus of SpA and from wall peptides linked to its C terminus [NH2-Ala-iGln-(Gly5)-Lys-Ala-COOH; Table S1].
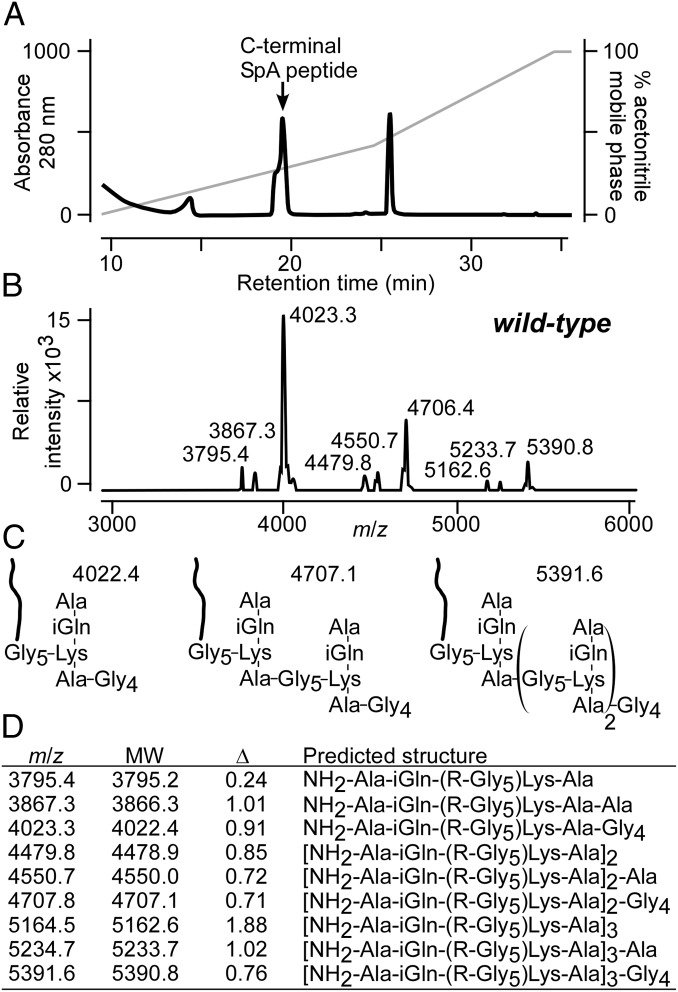
Purified protein A molecules were treated with cyanogen bromide, which cleaves polypeptide chains at methionyl residues. Peptide fragments were purified by RP-HPLC and analyzed by Edman degradation. The absorbance peak at 19 min yielded the sequence NH2-IKPGQ consistent with the C-terminal fragment of protein A (NH2-IKPGQELVVDKKQPANHADANKAQALPET; Fig. 2A). MALDI-TOF MS identified ion signal m/z as 4,023.35, 4,706.46, and 5,390.89 (Fig. 2B). These data cannot be explained on the basis of the predicted protein sequence but must be accounted for by the mass of protein A plus the cell wall component to which it is linked. We therefore built models for protein A linked to peptidoglycan fragments and interpreted m/z 4,023.35 as released SpA (R = NH2-IKPGQELVVDKKQPANHADANKAQALPET) linked to the pentaglycyl cross-bridge (Gly5) of a peptidoglycan tetrapeptide-tetraglycine [NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Gly4; Fig. 2C]. Compounds m/z 4,706.46 and 5,390.89 were interpreted as SpA tethered to cross-linked peptidoglycan with the same structure [NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Gy4]2–3; Fig. 2C. Minor ion signals were interpreted as SpA tethered to higher cross-linked wall peptides [NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Gy4]n or to peptidoglycan with cell wall pentapeptides NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Ala, respectively (Fig. 2 C and D).
Fig. 2.
Structure of released protein A. (A) Protein A was purified by affinity chromatography from the culture medium and cleaved with cyanogen bromide, and C-terminal peptides were isolated by RP-HPLC and identified by Edman degradation (Table S2). (B) C-terminal SpA peptides were subjected to MALDI-TOF MS and ion signals were recorded. (C) Model for the predominant ions signals generated by C-terminal SpA peptides. (D) Observed and predicted (MW) m/z and differentials (Δ) for SpA peptides released by WT S. aureus with their predicted structures.
Protein A Released from S. aureus atl, sle1, and lytN Mutants.
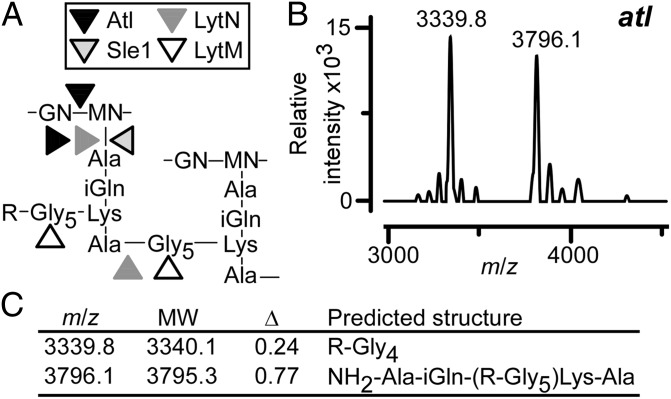
The structure of released SpA may be shaped by muralytic enzymes that split the cross-wall compartment during staphylococcal growth, i.e., Atl [N-acetylmuramoyl-l-alanine amidase (amidase) and glucosaminidase] (33, 34), Sle1 (CHAP domain amidase and d-Ala-Gly endopeptidase) (35), and LytN (CHAP domain amidase and d-Ala-Gly endopeptidase) (36, 37) (Fig. 3A). If so, S. aureus Newman mutants lacking the structural genes for these muralytic enzymes may release protein A with altered peptidoglycan structure. This was tested by purifying SpA from the medium of cultures inoculated with the S. aureus Newman atl variant that also lacks sbi (38). Following cyanogen bromide cleavage and RP-HPLC purification, protein A peptides from the atl mutant were subjected to MALDI-TOF MS, which identified ion signals with m/z 3,339.86 and 3,796.12 (Fig. 3B). Structural models for released peptide linked to tetraglycyl (R-Gly4) and peptidoglycan tetrapeptide [NH2-Ala-iGln-(R-Gly5)-Lys-Ala] fit the observed mass signals (Fig. 3 B and C).
Fig. 3.
S. aureus murein hydrolases impact the release of protein A. (A) Structure of S. aureus peptidoglycan comprised of the repeating disaccharide N-acetylglucosamine-(β1-4)-N-acetylmuramic acid (GN-MN) with linked wall peptide (Ala-iGln-Lys-Ala) and pentaglycine cross-bridge (Gly5). Arrowheads identify the cleavage sites of murein hydrolases that act at the cross-wall (Atl, Sle1, and LytN) or that function as glycyl-endopeptidases (LytM). (B) MALDI-TOF MS of C-terminal SpA peptides released from the S. aureus atl mutant. (C) Observed and predicted (MW) m/z ratios and their differentials (Δ) for SpA peptides released by the S. aureus atl mutant with their predicted structures.
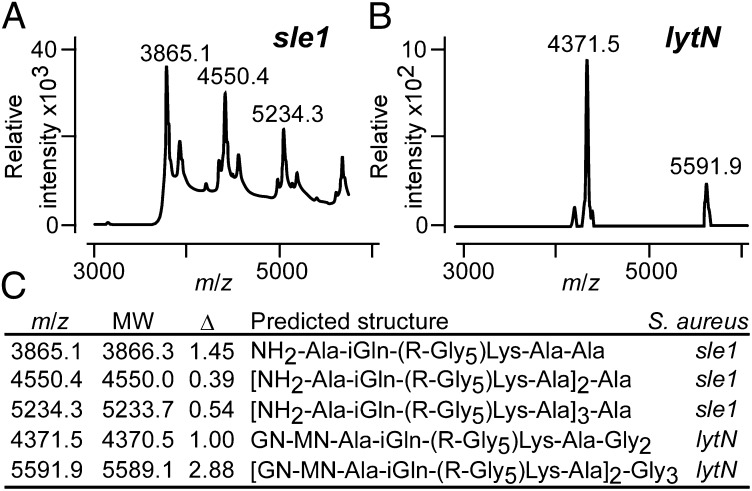
Sle1 is secreted into medium and subsequently binds to the cross-wall section of peptidoglycan via its LysM domains, connecting with glycan strands (MurNAc-GlcNAc)n that have not been decorated with wall teichoic acids (polyribitol-phosphate) at the C6-hydroxyl of MurNAc (37). LytN precursor is secreted via its YSIRK-G/S motif signal peptide into the cross-wall, and its CHAP domain cleaves peptidoglycan through its amidase and endopeptidase activities (36). Staphylococcal sle1 (35) and lytN mutants display defects in cell separation with inadequate splitting of cross-wall peptidoglycan (35, 36). Protein A was purified from the extracellular medium of S. aureus Newman sle1 and lytN mutants (also lacking sbi) and cleaved with cyanogen bromide, and C-terminal peptides were analyzed by MS. SpA released from the sle1 mutants generated the ion signals m/z 3,865.17 and 4,023.24, which were interpreted as NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Ala and NH2-Ala-iGln-(R-Gly5)-Lys-Ala-Gly4 (Fig. 4 A and C). We also detected peptidoglycan with the similar structure and two to four cross-links [NH2-Ala-iGln-(R-Gly5)-Lys-Ala)2–4]-Ala (Fig. 4 A and C). SpA peptides released by the lytN mutant generated the ion signals m/z 4,371.57 and 5,591.98, which were interpreted as MurNAc[Ala-iGln-(R-Gly5)-Lys-Ala-Gly2]-GlcNAc as well as [MurNAc(Ala-iGln-[R-Gly5]Lys-Ala)-GlcNAc]2-Gly3 (Fig. 5 B and C). These data suggest that LytN is responsible for cleaving the amide bonds between MurNAc and wall peptides, which enables the release of protein A with cross-linked peptidoglycan that lacks the amino sugars of the cell wall.
Fig. 4.
Cross-wall murein hydrolases shape the structure of released protein A. MALDI-TOF mass spectrometry of C-terminal SpA peptides released from the S. aureus sle1 (A) and lytN (B) mutants. (C) Observed (m/z) and predicted (MW) mass-to-charge ratios and their differentials (Δ) for SpA peptides released by the S. aureus sle1 and lytN mutants with their predicted structures.
Fig. 5.
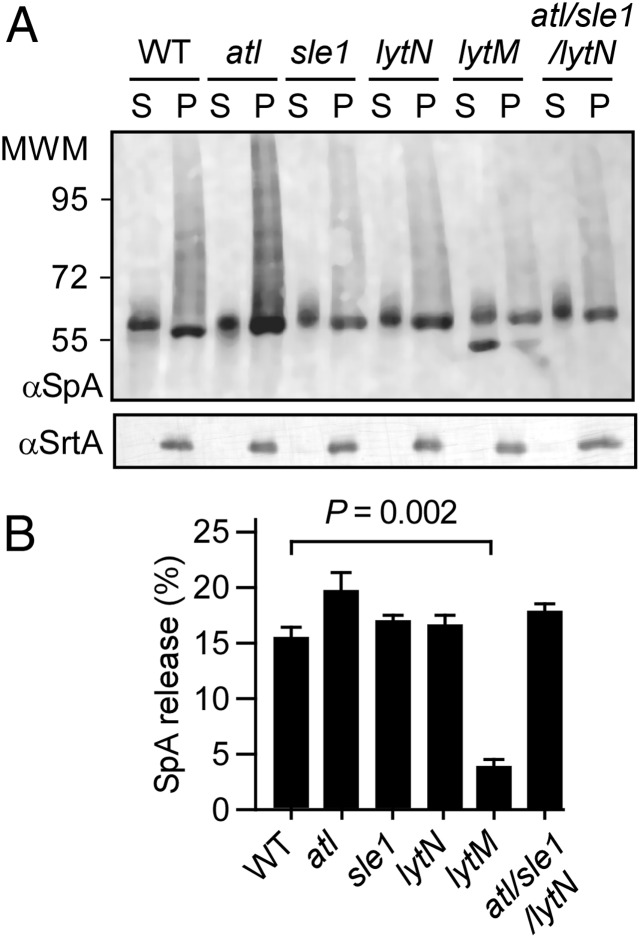
LytM endopeptidase is required for protein A release from the staphylococcal envelope. (A) S. aureus WT and atl, sle1, lytN, lytM, or atl/sle1/lytN mutant cultures were centrifuged. Proteins in the culture media were separated with the supernatant (marked “S”) from the bacterial pellet (marked “P”). After treatment of the staphylococcal cell wall envelope with lysostaphin, proteins in both fractions were analyzed by immunoblotting with polyclonal antibodies against protein A (αSpA) or sortase A (αSrtA). (B) Immune reactive signals from three different samples analyzed as shown in A were averaged, SEMs determined, and statistical significance analyzed with the two-tailed Student t test. Only the lytM mutant strain released less protein A than WT S. aureus (P = 0.002).
LytM Releases Protein A from the S. aureus Envelope.
LytM is a secreted glycyl-glycine endopeptidase of S. aureus (39), and we wondered whether this enzyme may be involved in the release of protein A. Unlike Atl, Sle1, and LytN, which are targeted to the cross-wall, the subcellular location of LytM is not yet known. SpA peptides released by the lytM mutant were not abundant. Nevertheless, these peptides generated m/z 3,795.87, 3,866.87, and 4,022.94, i.e., the same structures as WT S. aureus: Ala-iGln-(R-Gly5)-Lys-Ala, Ala-iGln-(R-Gly5)-Lys-Ala-Ala, and Ala-iGln-(R-Gly5)-Lys-Ala-Gly4 (Fig. S1).
To quantify the release of SpA, S. aureus strains were grown to similar densities and protein A release was analyzed by immunoblotting. Cultures were centrifuged, and staphylococci were treated with lysostaphin to determine the total amount of SpA associated with the envelope. The supernatant was also TCA-precipitated. Both samples were analyzed by immunoblotting in triplicate samples, and averages and SEMs were determined. Compared with S. aureus Newman (15.10% ± 1.28 of released SpA), the atl (19.78% ± 1.72; P < 0.1), lytN (16.69% ± 1.44; P < 0.5), sle1 (16.88% ± 0.29; P < 0.3), or atl/sle1/lytN (17.80% ± 0.49; P < 0.5) variants released similar amounts of protein A as WT staphylococci (Fig. 5 A and B). In contrast, the lytM mutant strain released significantly less SpA into the culture medium than WT S. aureus (3.96% ± 0.82; P < 0.01; Fig. 5 A and B). These data indicate that the cross-wall murein hydrolases shape the cell wall anchor structure of SpA to ensure that released molecules lack the amino sugars of peptidoglycan. Nevertheless, cross-wall murein hydrolases are not responsible for the ultimate release of protein A from the bacterial envelope. This function can be assigned in part to LytM, which cuts the pentaglycyl cross-bridge of the protein A cell wall anchor structure.
Protein A Release During Staphylococcal Growth.
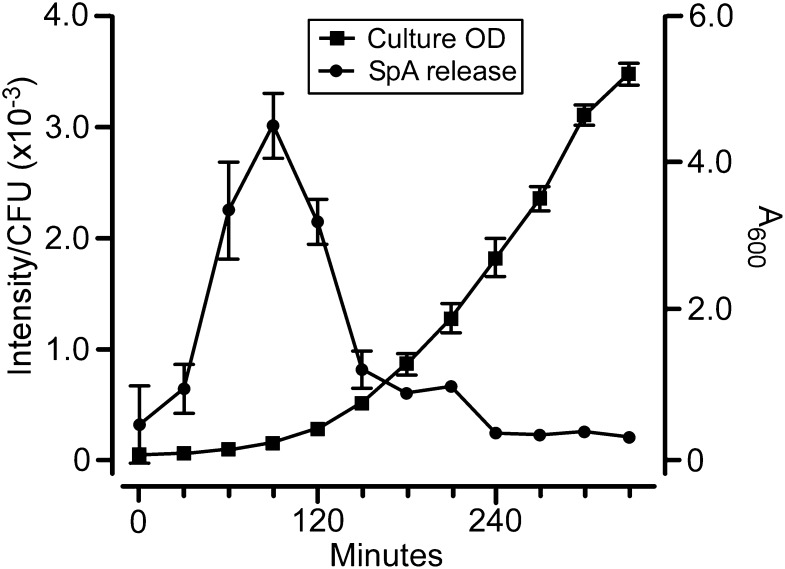
We wondered whether SpA release occurs evenly during staphylococcal growth and followed stationary-phase S. aureus cells that were washed and diluted into fresh media in 30-min time intervals through logarithmic growth. Shortly following its dilution, i.e., during early logarithmic growth, S. aureus released greater amounts of SpA per cfu than during later growth phases (Fig. 6). These results suggest that the release of SpA may be controlled by the growth phases of staphylococci, presumably with the intent of modifying host immune responses early during infection when numbers for this bacterial invader are small.
Fig. 6.
SpA release during staphylococcal growth. S. aureus Newman (sbi) cells were washed and diluted into fresh TSB medium to A600 0.05 and incubated with rotation at 37 °C. At 30-min intervals, the absorbance (600 nm) was measured and cfus were enumerated. SpA release was quantified by immunoblotting of culture supernatant samples and recorded as signal intensity divided by cfu (×10−3). Experiments were performed in triplicate to calculate average values and SEM (error bars).
Discussion
Bacterial peptidoglycan is a potent activator of the mammalian immune system (40). Muropeptides released during bacterial growth are recognized by nucleotide-binding and oligomerization domain containing proteins 1 (NOD1) and 2 (NOD2), two members of the NLR family in the cytoplasm of mammalian cells that specialize in recognizing peptidoglycan fragments from Gram-negative (i.e., NOD1) or Gram-positive bacteria (i.e., NOD2) (41, 42). Activated by muropeptides with MurNAc, NOD1 and NOD2 initiate intracellular signaling pathways that induce NF-κB and the expression of immune response genes encoding proinflammatory cytokines and antimicrobial peptides (40, 43). For example, neutrophil-derived IL-1β production, a key defense against S. aureus (44), requires staphylococcal peptidoglycan-induced activation of NOD2 (45). The importance of this pathway is illustrated by defects in NOD2 signaling, which increases the disease susceptibility of mutant mice to S. aureus infection (46).
The NOD2 signaling pathway presents a challenge to the pathogenesis of S. aureus infections, in particular the suppression of B-cell responses (24, 25). How can pathogen-released protein A activate the clonal expansion of B cells and their subsequent apoptotic collapse without simultaneously triggering a systemic activation of the immune system? We show here that protein A is released from the envelope with linked peptidoglycan fragments, i.e., l-Ala-d-iGln-(Gly5)l-Lys-d-Ala-Gly4 and l-Ala-d-iGln-(Gly5)l-Lys-d-Ala-d-Ala. Of note these peptidoglycan fragments lack the amino sugars (MurNAc-GlcNAc) of peptidoglycan, which function as potent inducers of NOD2 (42). Pulse-labeling experiments indicate that protein A is initially tethered to the bacterial envelope, which occurs at the cross-wall between dividing daughter cells (14). Mutants lacking the cross-wall murein hydrolases—Atl, Sle1, or LytN—released SpA with altered peptidoglycan structures but did not affect the overall release of protein A. Thus, cross-wall murein hydrolases shape the cell wall anchor structure of protein A without affecting its release. The purpose of these activities appears to be the synthesis of SpA anchor structures that cannot stimulate host immune responses. When these tasks are completed, protein A remains tethered to the murein sacculi via its anchoring point, the peptidoglycan pentaglycine cross-bridge, which also connects neighboring wall peptides (7). LytM eventually cleaves these cross-bridges and releases protein A into the extracellular medium.
LytM is a secreted glycyl-glycine endopeptidase that cuts the cross-bridges of staphylococcal peptidoglycan without contributing to autolysis (39). Recent work revealed that lytM is a WalKR-regulated gene (47). In WalKR mutants, in which expression of the essential two-component regulator is limiting, LytM can suppress associated growth defects presumably by relaxing the cross-linking of peptidoglycan (48). Here, we suggest an important function for LytM: the release of biologically active sortase-anchored proteins from the bacterial envelope. As lytM mutants continue to release small amounts of protein A from the envelope, we surmise the existence of additional enzymes with gycyl-glycine endopeptidase activity that may impact the release of surface proteins.
Materials and Methods
Bacterial Strains and Growth Conditions.
S. aureus strains were grown in TSB at 37 °C with agitation. S. aureus Newman WT (49) and variants with deletions in specific murein hydrolases atl, sle1, or lytN were generated by allelic replacement using the pKOR1 shuttle vector (50). Briefly, 1-kb flanking regions up- and downstream of atl, sle1, or lytN were amplified, ligated, and inserted into pKOR1 via lambda red recombination (50). Recombinant plasmids were electroporated into S. aureus Newman for integration into the chromosome and selection of deletion mutants. The spa mutant has been described earlier (51). To avoid contamination of protein A with the staphylococcal binder of Ig during affinity chromatography experiments (52), the sbi::erm allele described earlier (53) was transduced with φ85 phage into S. aureus Newman as well as its atl, sle1, and lytN variants. Transductants sbi::erm, atl/sbi::erm, sle1/sbi::erm, and lytN/sbi::erm were confirmed by PCR and DNA sequencing. The bursa aurealis transposon mutation ΦΝΞ02987 (lytM::erm) was transduced with φ85 into WT S. aureus Newman to generate the lytM mutant. Note that this mutant encodes the WT sbi gene.
Purification of SpA.
S. aureus Newman and its variants were grown overnight in TSB and used to inoculate 8 L of fresh TSB with a 1:100 dilution inoculum. Cultures were grown for 4 h at 37 °C to absorbance at 600 nm (A600) 0.2 and then centrifuged at 9,000 × g for 20 min. The supernatant was decanted and cooled on ice to 4 °C, and 200 mL 1.5 M Tris⋅HCl (pH 8.8) added to adjust to pH 7.5. SpA molecules in the culture supernatant were purified by affinity chromatography. Briefly, a gravity-feed column with 4 mL bed volume IgG Sepharose6 Fast Flow was equilibrated with 14 mL elution buffer (0.1 M glycine⋅HCl, pH 3.0) followed by 30 mL Tris–saline–Tween 20 [TST; 50 mM Tris⋅HCl (pH 7.6), 150 mM NaCl, 0.05% Tween 20]. A total of 8 L of culture supernatant was loaded onto the column at 4 °C over the course of 12 h. The column was washed with 100 mL TST and eluted with 14 mL elution buffer. The eluate was immediately neutralized with 0.35 mL 1.5 M Tris⋅HCl (pH 8.8). SpA molecules in 14 mL eluate were concentrated to a volume of 2 mL with a 10,000 molecular weight cut-off centrifugal filter unit (Millipore), dialyzed against PBS solution at 4 °C, and stored at −20 °C until further use. SpA purification was analyzed by Coomassie-stained SDS/PAGE, and protein concentration was determined with the BCA protein assay kit (Pierce).
MS.
For structural analysis, 150 µg purified SpA was dried, dissolved in 50 mL 5% (vol/vol) 2-mercaptoethanol, 0.2 M NH4HCO3, and incubated overnight at room temperature. The protein sample was again dried and dissolved in 1 mL 70% formic acid. Approximately 1 mg cyanogen bromide was added to the solution. The reaction was incubated for 18 h in the dark at room temperature, and quenched by diluting the mixture 1:5 in water. The sample was then dried, dissolved in 30% acetonitrile, and desalted by reverse-phase HPLC via an acetonitrile/water gradient on C8 Hypersil solid-phase column. Eluate peaks were dried and dissolved in 5 to 20 µL 30% acetonitrile. Protein samples were spotted on a ground platinum target plate (Bruker) and mixed 1:1 on the plate with α-cyano-4-hydroxycinnamic acid matrix (Sigma). Fractions were analyzed on a MALDI-TOF mass spectrometer (Autoflex Speed; Bruker) by using a reflectron-positive or linear-positive detection method. The detector was calibrated before each experiment with protein standards.
Measurement of SpA Release.
S. aureus strains were inoculated from frozen stocks into TSB cultures and grown overnight. The culture was diluted to A600 5.0, and 1 mL was centrifuged at 20,000 × g for 1 min. The supernatant was discarded and staphylococci were washed two times in PBS solution. The bacterial suspension was inoculated into 100 mL TSB to yield A600 0.05, and incubated at 37 °C. Before incubation and at timed intervals thereafter, 1.5 mL samples were removed from cultures and centrifuged at 20,000 × g for 1 min. One milliliter of the supernatant was removed, placed in a fresh tube, and stored on ice. The cell pellet was washed with 1 mL PBS solution and suspended in 1.5 mL TSM-L [50 mM Tris⋅HCl (pH 7.5), 0.5 M sucrose, 10 mM MgCl2, 5 µg⋅mL−1 lysostaphin (AMBI)] and placed at 37 °C for 15 min to digest the cell wall envelope with lysostaphin. A 1-mL aliquot of the staphylococcal protoplast suspension was then placed on ice.
Proteins in supernatant and protoplast samples were precipitated by adding 100% TCA/0.1% deoxycholic acid to a final concentration of 20%. Samples were vortexed and incubated on ice for 20 min. Precipitated proteins were sedimented by centrifugation at 22,000 × g for 15 min. The supernatant was discarded, and protein pellets were washed twice with ice-cold acetone, air-dried, and solubilized in 25 µL 0.5 M Tris⋅HCl (pH 8.0), 4% SDS at 4 °C overnight. An equal volume of sample buffer [125 mM Tris⋅HCl (pH 6.8), 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.01% bromophenol blue] was added, and protein samples were boiled for 10 min. Proteins were separated on 10% SDS/PAGE gel and electrotransferred to a PVDF membrane. The membrane was incubated in blocking buffer (TBS, 5% milk) for 1 h at room temperature. SpAKKAA monoclonal antibody 3F6 was used at a 1:10,000 dilution in TBS-Tween 20. After 1 h, the membrane was washed three times and incubated in a 1:10,000 solution of goat anti-mouse 680 antibody (Licor). After 1 h, the membrane was washed three times, and fluorescence was measured at 700 nm on an infrared scanner (Licor Odyssey). Signal integration was performed by Licor software. To ensure consistency between blots, two standard dilutions of purified SpA were included in every blot as a fluorescence control.
SpA Release During Staphylococcal Growth.
Overnight cultures of S. aureus Newman sbi were centrifuged, and bacterial cells were washed and diluted into fresh TSB medium to A600 0.05 and incubated with rotation at 37 °C. At 30-min intervals, the absorbance at 600 nm was measured and culture aliquots were plated to enumerate cfus. Additionally, 1.5-mL culture aliquots were removed to quantify protein A release. Following centrifugation of culture samples, 1 mL supernatant was withdrawn and proteins precipitated with TCA. The precipitate was solubilized in sample buffer and subjected to immunoblotting to quantify the abundance of SpA. The rate of SpA release was calculated by dividing the intensity of SpA-immunoreactive signals with the cfu measurements for each time interval. Experiments were performed in triplicate to calculate average values and SEM.
Supplementary Material
Acknowledgments
The authors thank members of their laboratory for critical discussion. This work was supported by US National Institute of Allergy and Infectious Diseases, Infectious Diseases Branch, Grants AI052474 and AI038897 (to O.S.). D.M. and O.S. acknowledge membership within, and support from, the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institutes of Health Award 1-U54-AI-057153).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317181111/-/DCSupplemental.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: Morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62(4):1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higashi Y, Strominger JL, Sweeley CC. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: A derivative of a C55 isoprenoid alcohol. Proc Natl Acad Sci USA. 1967;57(6):1878–1884. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strominger JL, Izaki K, Matsuhashi M, Tipper DJ. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: Penicillin-sensitive enzymatic reactions. Fed Proc. 1967;26(1):9–22. [PubMed] [Google Scholar]
- 5.Park JT, Strominger JL. Mode of action of penicillin. Science. 1957;125(3238):99–101. doi: 10.1126/science.125.3238.99. [DOI] [PubMed] [Google Scholar]
- 6.Kojima N, Araki Y, Ito E. Structure of linkage region between ribitol teichoic acid and peptidoglycan in cell walls of Staphylococcus aureus H. J Biol Chem. 1983;258(15):9043–9045. [PubMed] [Google Scholar]
- 7.Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268(5207):103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 8.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63(1):174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70(2):267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 11.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96(22):12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277(18):16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 13.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci USA. 2002;99(4):2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeDent AC, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27(20):2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11(2):237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 16.Flock JI, et al. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 1987;6(8):2351–2357. doi: 10.1002/j.1460-2075.1987.tb02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Signäs C, et al. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: Use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86(2):699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazmanian SK, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299(5608):906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 19.DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol. 2007;189(12):4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsgren A, Sjöquist J. “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol. 1966;97(6):822–827. [PubMed] [Google Scholar]
- 21.Inganäs M. Comparison of mechanisms of interaction between protein A from Staphylococcus aureus and human monoclonal IgG, IgA and IgM in relation to the classical FC gamma and the alternative F(ab’)2 epsilon protein A interactions. Scand J Immunol. 1981;13(4):343–352. doi: 10.1111/j.1365-3083.1981.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 22.Peterson PK, Verhoef J, Sabath LD, Quie PG. Effect of protein A on staphylococcal opsonization. Infect Immun. 1977;15(3):760–764. doi: 10.1128/iai.15.3.760-764.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodyear CS, Silverman GJ. Staphylococcal toxin induced preferential and prolonged in vivo deletion of innate-like B lymphocytes. Proc Natl Acad Sci USA. 2004;101(31):11392–11397. doi: 10.1073/pnas.0404382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falugi F, Kim HK, Missiakas DM, Schneewind O. The role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio. 2013;4:e00575-13. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman GJ, Goodyear CS. Confounding B-cell defences: Lessons from a staphylococcal superantigen. Nat Rev Immunol. 2006;6(6):465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. A second IgG-binding protein in Staphylococcus aureus. Microbiology. 1998;144(Pt 4):985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]
- 27.Sjöquist J, Meloun B, Hjelm H. Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur J Biochem. 1972;29(3):572–578. doi: 10.1111/j.1432-1033.1972.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim HK, et al. Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect Immun. 2012;80(10):3460–3470. doi: 10.1128/IAI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA. 2000;97(10):5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson B, Abrahmsén L, Uhlén M. Immobilization and purification of enzymes with staphylococcal protein A gene fusion vectors. EMBO J. 1985;4(4):1075–1080. doi: 10.1002/j.1460-2075.1985.tb03741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjödahl J. Repetitive sequences in protein A from Staphylococcus aureus. Arrangement of five regions within the protein, four being highly homologous and Fc-binding. Eur J Biochem. 1977;73(2):343–351. doi: 10.1111/j.1432-1033.1977.tb11324.x. [DOI] [PubMed] [Google Scholar]
- 33.Oshida T, et al. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: Cloning, sequence analysis, and characterization. Proc Natl Acad Sci USA. 1995;92(1):285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada S, et al. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178(6):1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajimura J, et al. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol. 2005;58(4):1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x. [DOI] [PubMed] [Google Scholar]
- 36.Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem. 2011;286(37):32593–32605. doi: 10.1074/jbc.M111.258863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frankel MB, Schneewind O. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem. 2012;287(13):10460–10471. doi: 10.1074/jbc.M111.336404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: Repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17(16):4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramadurai L, Jayaswal RK. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol. 1997;179(11):3625–3631. doi: 10.1128/jb.179.11.3625-3631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inohara N, Chamaillard M, McDonald C, Nuñez G. NOD-LRR proteins: Role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 41.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 42.Tanabe T, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23(7):1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi K, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416(6877):194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 44.Cho JS, et al. Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog. 2012;8(11):e1003047. doi: 10.1371/journal.ppat.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volz T, et al. Natural Staphylococcus aureus-derived peptidoglycan fragments activate NOD2 and act as potent costimulators of the innate immune system exclusively in the presence of TLR signals. FASEB J. 2010;24(10):4089–4102. doi: 10.1096/fj.09-151001. [DOI] [PubMed] [Google Scholar]
- 46.Hruz P, et al. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci USA. 2009;106(31):12873–12878. doi: 10.1073/pnas.0904958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189(22):8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delaune A, et al. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE. 2011;6(2):e17054. doi: 10.1371/journal.pone.0017054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190(1):300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55(1):58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Kim HK, Kim HY, Schneewind O, Missiakas DM. Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J. 2011;25(10):3605–3612. doi: 10.1096/fj.11-187963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Jacobsson K, Ström K, Lindberg M, Frykberg L. Staphylococcus aureus expresses a cell surface protein that binds both IgG and beta2-glycoprotein I. Microbiology. 1999;145(pt 1):177–183. doi: 10.1099/13500872-145-1-177. [DOI] [PubMed] [Google Scholar]
- 53.Bae T, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA. 2004;101(33):12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.