Significance
Malaria infection causes a severe disease with diverse symptoms. The molecular mechanisms underlying the differences of malaria pathology remain unknown or controversial. Here we infected mice with two closely related strains of rodent malaria parasite Plasmodium yoelii and characterized host genome-wide responses to the infections. We found that in mice infected with parasite N67, type I interferon was produced to a high level, leading to suppression of parasitemia. We further characterized the molecular mechanisms and identified host receptors in recognizing parasite ligands. In contrast, mice infected with N67C parasite mounted a strong inflammatory response, leading to severe pathology and host death. This study reveals previously unrecognized mechanisms associated with strain-specific malaria infection and provides important information for studying human malaria pathogenesis.
Keywords: rodent, interferon, MDA5, MAVS, phagocytosis
Abstract
Malaria infection triggers vigorous host immune responses; however, the parasite ligands, host receptors, and the signaling pathways responsible for these reactions remain unknown or controversial. Malaria parasites primarily reside within RBCs, thereby hiding themselves from direct contact and recognition by host immune cells. Host responses to malaria infection are very different from those elicited by bacterial and viral infections and the host receptors recognizing parasite ligands have been elusive. Here we investigated mouse genome-wide transcriptional responses to infections with two strains of Plasmodium yoelii (N67 and N67C) and discovered differences in innate response pathways corresponding to strain-specific disease phenotypes. Using in vitro RNAi-based gene knockdown and KO mice, we demonstrated that a strong type I IFN (IFN-I) response triggered by RNA polymerase III and melanoma differentiation-associated protein 5, not Toll-like receptors (TLRs), binding of parasite DNA/RNA contributed to a decline of parasitemia in N67-infected mice. We showed that conventional dendritic cells were the major sources of early IFN-I, and that surface expression of phosphatidylserine on infected RBCs might promote their phagocytic uptake, leading to the release of parasite ligands and the IFN-I response in N67 infection. In contrast, an elevated inflammatory response mediated by CD14/TLR and p38 signaling played a role in disease severity and early host death in N67C-infected mice. In addition to identifying cytosolic DNA/RNA sensors and signaling pathways previously unrecognized in malaria infection, our study demonstrates the importance of parasite genetic backgrounds in malaria pathology and provides important information for studying human malaria pathogenesis.
Malaria is a deadly disease that infected ∼219 million people and killed ∼660,000 in 2010 (1). Lack of effective vaccines and poorly understood molecular mechanisms governing the host response to malaria infection have been some of the main obstacles in controlling the disease. Innate immunity plays a key role in protective response to malaria infection (2–7). Production of proinflammatory cytokines/chemokines (PICCs) such as TNF-α, IL-1β, IFN-γ, and IL-6 is necessary for controlling parasite infection, but overproduction of these PICCs can also lead to severe pathology (6). In contrast to viral and bacterial infections, from which many pattern recognition receptors (PRRs) and signaling pathways were discovered and elucidated, our understanding of how the host recognizes malaria-associated molecular patterns (MAMPs) and how the signals of pattern recognition are translated to the production of various cytokines/chemokines and to disease pathogenesis is still limited (3). Toll-like receptors (TLRs) and scavenger receptors have been implicated in the response to malaria infection and for triggering proinflammatory responses (4, 8–15); however, other studies indicated that TLR-dependent signaling is dispensable for controlling pathogenesis (16, 17). As a result, the role of TLRs in malaria infection and pathogenesis, therefore, is still controversial (3). Similarly, the NOD-like receptor containing pyrin domain 3 (NLRP3) inflammasome was implicated in the production of IL-1β, which might play a role in fever and other inflammatory responses; synthetic hemozoin could induce IL-1β and neutrophil recruitment in a NALP3-dependent manner (18, 19). However, Plasmodium berghei ANKA (PbANKA)-infected caspase1 and IL-1β KO mice displayed disease phenotypes similar to that of wild-type mice, suggesting minimal roles for these two molecules in the disease (20, 21). Plasmodium falciparum DNA containing an ATTTTAC motif was shown to induce type I IFN (IFN-I) response involving stimulator of IFN genes (STING), TANK-binding kinase 1 (TBK1), IFN regulatory factors 3 and 7 (IRF3/IRF7), and an unknown DNA recognition receptor, but not TLR9 and RIG-I (22). Recently, cGMP-AMP (cGAMP) and cGAMP synthase (cGAS) were identified as the second messenger and receptor for the STING/IRF3 signaling pathway (23, 24); however, whether cGAS can also recognize the malaria AT-rich motif remains to be determined. Additionally, CD36 was shown to be involved in the sequestration of the parasite and in phagocytosis of P. falciparum-infected red blood cells (iRBCs) by human monocytes/macrophages without inducing a proinflammatory cytokine response (25, 26). More studies are necessary to clearly identify host PRRs, the MAMPs that trigger host responses, and the signaling pathways in response to malaria infection.
Many parasite-derived factors have been reported to stimulate or modulate the host response, including glycolipid (glycosylphosphatidylinositol, GPI), hemozoin pigment, nucleic acids, and a DNA–protein complex (10, 13, 22, 27–29). Additionally, various parasite proteins can bind to host receptors directly and modulate host responses; for example, circumsporozoite protein (CSP) was reported to interfere with the nuclear translocation of NF-κB (30), and proteins with Duffy binding-like (DBL) domains such as the P. falciparum erythrocyte membrane protein 1 (PfEMP1) or erythrocyte binding ligand (EBL) are able to bind to host cells expressing CD36, ICAM1, and/or Duffy antigen receptor for chemokine (DARC) and to trigger (or suppress) the host inflammatory response (31, 32). Variations in these molecules or others in the parasite genome can influence the host response and disease outcomes. Host genetic backgrounds can also affect disease severity of malaria infection (33). Variations in genetic backgrounds of malaria parasites and their hosts may partly explain the dramatic differences in disease outcome, conflicting observations in studies of host response to malaria infection, and the difficulties in developing an effective vaccine to prevent malaria infection.
The rodent malaria parasite Plasmodium yoelii is an excellent model for studying the effect of parasite–host interaction on disease severity because parasite strains producing different disease phenotypes are available, and the influences of host genetic background on disease outcome can be minimized through the use of inbred mice. Here we investigated disease phenotypes and host signaling pathways leading to suppression of parasitemia after infection with two isogenic P. yoelii strains (P. y. nigeriensis N67 and P. y. nigeriensis N67C, designated N67 and N67C, respectively). We first measured the genome-wide transcriptional variations in host response to infection of N67 and N67C parasites using a mouse expressional microarray and showed differential host responses to the two parasites. We found that infection with N67 stimulated a strong IFN-I response mediated through RNA polymerase III (RNA Pol III) and melanoma differentiation-associated protein 5 (MDA5), leading to suppression of early parasitemia; in contrast, infection of N67C stimulated strong proinflammatory responses and early host death. Additionally, infection with N67 resulted in higher levels of surface phosphatidylserine (PS) on iRBCs, which appears to trigger stronger macrophages (MΦs) and/or dendritic cell (DC) phagocytosis and IFN-I response. Our results reveal molecular mechanisms of host innate response previously unrecognized in infection by malaria parasites and provide new avenues to analyze human immune responses to malaria infection.
Results
Parasitemia and Pathology in Mice Infected with N67 and N67C Parasites.
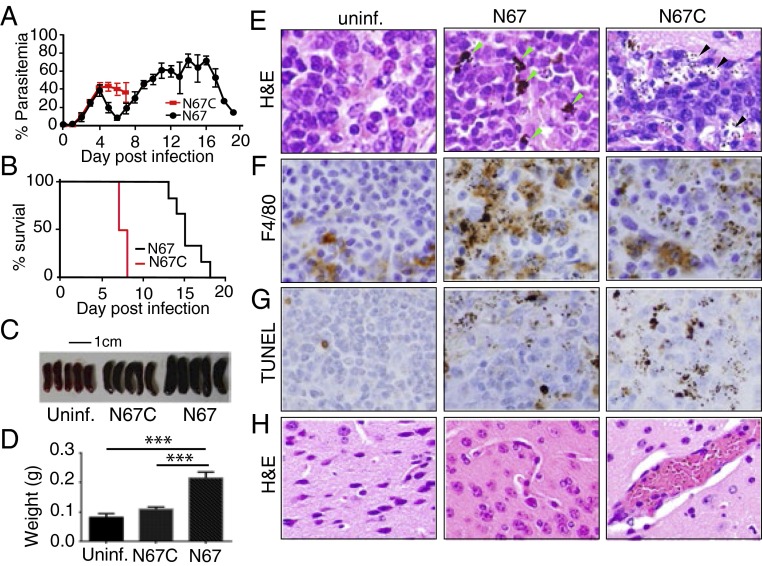
To determine strain-specific innate immune responses, we first compared disease phenotypes and host genome-wide transcriptional responses to infections with N67 and N67C. Although N67 and N67C are isogenic and have very similar genome sequences (34), they produced very different parasitemia and mortality profiles (Fig. 1 A and B). Parasitemias in mice infected with N67 rose rapidly for several days but were reduced to low levels before day 8, before climbing again; parasitemias in mice given N67C parasites also increased rapidly but the animals could not control these infections and died by day 7 postinfection. Histological examination of the infected mice showed dramatic differences in pathology in the spleen and brain (Fig. 1, Fig. S1, and Dataset S1). Mice infected with N67 had larger spleens (Fig. 1 C and D) and more active phagocytosis by MΦs with larger hemozoin pigments than those infected with N67C or uninfected controls (Fig. 1 E and F). In N67C-infected mice, most of the pigments were small and in the subcapsular sinusoids with signs of tissue necrosis (Fig. 1E and Fig. S1C). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining showed signs of apoptosis in the spleens, particularly in those infected with N67C (Fig. 1G). In the brain, sequestration of iRBCs was observed in the capillaries of mice infected with N67C, but not those infected with N67 (Fig. 1H and Fig. S1 E and F). We also counted red pulp cells positive by F4/80 or TUNEL staining and found that N67-infected spleens had significantly (P < 0.001, t test) more F4/80 positive cells (average 64.8 ± 5.9) than those infected with N67C (average 38.2 ± 10.8). Conversely, N67C-infected mice had significantly (P < 0.001, t test) more TUNEL positive cells (average 63.3 ± 3.9) than those infected with N67 (average 22.8 ± 4.2).
Fig. 1.
Disease phenotypes of mice infected with Plasmodium yoelii parasite strains N67 and N67C. (A) Parasitemia of C57BL/6 mice infected with N67 or N67C parasites (106). SDs were determined from eight mice in each group. (B) Mortality rates of mice infected with N67 or N67C. (C) Images of spleens from uninfected (uninf.), N67-, or N67C-infected mice of the same age. (D) Average weights (in grams) of the spleens in C; significant at ***P < 0.001, t test. (E) Hematoxylin and eosin (H&E) staining of splenic tissues of uninfected mice and those infected with N67 or N67C; green arrowheads point to large pigments within the spleen of N67-infected mice, and black arrowheads indicate small dots of pigments in infected red blood cells or in subcapsular sinusoid spaces. (F) Monoclonal antibody F4/80 staining showing large numbers of macrophages (brown) in the spleen of N67-infected mice. Note large numbers of dark pigments in the stained cells. (G) TUNEL staining of spleen tissues showing apoptosis in infected mice, particularly those infected with N67C. (H) H&E staining of brain tissues showing infected red blood cells in the blood vessels of N67C-infected mice but not in mice infected with N67. All of the data in C–H were from day 4 mice postinfection. For the H&E staining, seven (N67C infected), five (N67 infected), and three uninfected mice were examined; for the TUNEL and F4/80 staining, tissues from six mice for each group were processed.
Differential Gene Expression in Response to the P. yoelii Strains.
To investigate the role of gene expression differences in the dynamics of parasitemia and pathogenesis, we hybridized RNA samples from the spleens of groups of six mice injected with 1 × 106 N67, N67C, or PBS (control) 4 d postinfection (a time when N67 parasitemia begins to decline) to the Illumina MouseRef-8 v2.0 whole-genome expression BeadChip. Signals from each of the array hybridizations were normalized and processed as described in Materials and Methods. Analysis of gene descriptors using Parametric Analysis of Gene Set Enrichment (PAGE) (35) showed several major Gene Ontology (GO) terms associated with differential expression patterns in response to parasite infections (Dataset S2). Groups of genes related to chemotaxis, inflammation, and IFN responses were up-regulated in the mice infected with the two parasites. Among these, pathways of positive regulation of neutrophil chemotaxis and CCR1/CCR2/CCR5 chemokine receptor binding were expressed at higher levels in mice infected with N67C than those infected with N67, whereas pathways associated with IFN-I response and with protozoan or virus infection were expressed at higher levels in N67-infected mice. Interestingly, pathways involved in erythrocyte differentiation, heme/porphyrin synthesis, and water and CO2 transport were up-regulated in the N67-infected mice, but were dramatically down-regulated in the mice infected with N67C. Finally, both N67- and N67C-infected mice had down-regulated pathways related to host immune response such as genes in antigen processing and presentation, DC cell chemotaxis, positive regulation of monocyte chemotaxis and alpha-beta T-cell activation, and negative regulation of leukocyte apoptosis, suggesting that P. yoelii infection suppresses or regulates many arms of the host immune response. Ingenuity Pathway Analysis (IPA) identified the IL-10 signaling and inflammatory responses mediated through CD14 and IL-1R pathways as they were the most markedly up-regulated in the N67C-infected mice (Fig. S2), whereas components of the IFN-I pathways were activated to higher levels in N67-infected mice (Fig. S3). Although these response patterns such as up-regulation of proinflammatory pathways largely agreed with those of previous microarray expressional profiling studies (8, 36, 37), our results clearly showed strain-specific responses and complex disease phenotypes/pathology that were greatly influenced by parasite genetic background. We therefore focused our investigation on the role of the IFN-I and putative CD14 signaling responses in the control of parasitemia and pathogenesis.
Up-Regulation of IFN-I Linked to Suppression of Parasitemia in N67-Infected Mice.
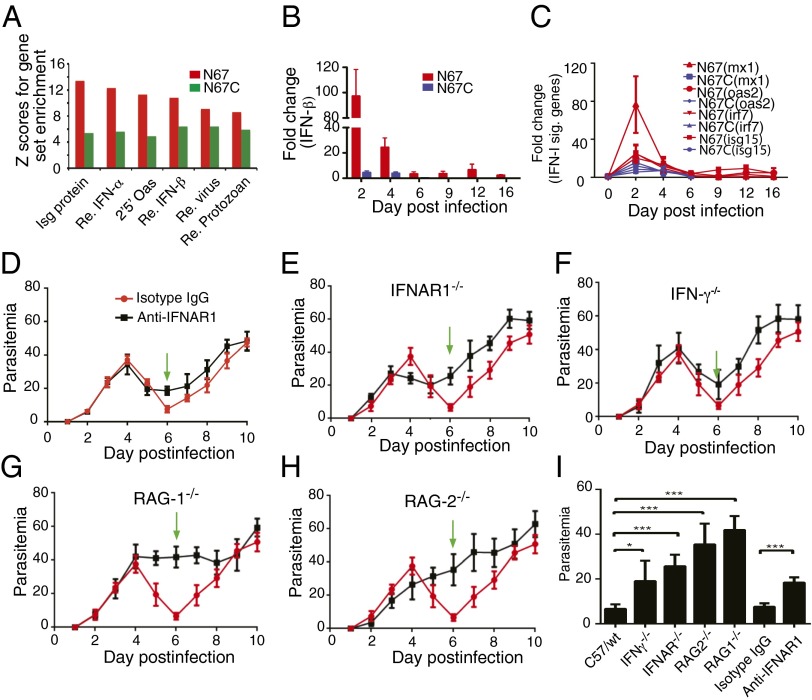
One of the most interesting observations was the decline of parasitemia after day 4 that appeared to be linked to the elevated IFN-I response in the N67 infection (Figs. 1A and 2A). To further verify that IFN-I and IFN-I “signature” genes were expressed at higher levels in the N67-infected mice, we used quantitative PCR (qPCR) to measure splenic mRNA for IFN-β, MX-1, OAS2, IRF7, and ISG15 from mice infected with N67 and N67C on day 2 to day 16 postinfection and showed that the transcripts for these genes were indeed higher in the splenocytes of N67-infected mice at days 2–4 postinfection than those infected with N67C or uninfected, particularly the increase of IFN- β and MX-1 mRNAs (Fig. 2 B and C). We next used anti-IFNAR1 antibody to block the receptor on day 2 postinfection of the mice with N67 and showed reduced ability in controlling day 6 parasitemia in the treated mice (Fig. 2D). The involvement of IFN-I and IFN-II in N67 parasitemia control (N67C-infected mice died on day 7) was further confirmed after infection of IFNAR−/− and IFN-γ−/− mice with N67; the declines of parasitemia in both KO mice were not as great as those of WT mice on day 6 after infection (Fig. 2 E and F). No obvious decline of parasitemia in RAG1−/− and RAG2−/− mice was observed (Fig. 2 G and H), suggesting that responses mediated by T and B cells were also required for the control of early parasitemia. The differences in suppression of parasitemia, as calculated by comparing the lowest (or “valley”) parasitemia between the KO and WT mice or between anti–IFNAR1- and isotype IgG-treated mice, were all statistically significant (t test, P < 0.05 for the IFN-γ−/− group and P < 0.001 for the rest) (Fig. 2I). The elevated IFN-I response and the suppression of parasitemia early in infection may also contribute to longer survival time of the N67-infected mice.
Fig. 2.
Variations in host type I IFN (IFN-I) response to infections with N67 and N67C parasites and suppression of N67 parasitemia by strong IFN-I response. (A) Higher up-regulation of type I IFN-related genes in response to N67 infection compared with those infected with N67C parasite, detected using microarray hybridization. The y axis is gene set enrichment score (Z) using the PAGE method. (B) Levels of IFN-β transcripts in N67- and N67C-infected mice detected using qPCR at various days postinfection. Fold changes were relative to uninfected mice. (C) Transcript levels of IFN-I signature genes (mx1, oas2, irf7, and isg15) as in B. (D) Daily N67 parasitemia of mice treated with anti-IFNAR1 or isotype IgG control antibodies. Green arrows point to the valley of parasitemia (or the lowest parasitemia after declining) in WT mice. (E–H) Daily N67 parasitemia in knockout (black lines) and WT mice (red lines). (E) IFNAR−/−; (F) IFN-γ−/−; (G) RAG1−/−; and (H) RAG2−/−. (I) Average valley of N67 parasitemia from WT and KO mice or mice treated with antibodies. Significance at *P < 0.05; ***P < 0.001 using t test. SDs were estimated from three to five mice. Mice were injected with 1 × 106 infected red blood cells. Blood smears were made daily, and parasites were counted under a microscope after Giemsa staining.
MDA5 and MAVS, but Not TLRs, Play a Role in the Decline of Parasitemia.
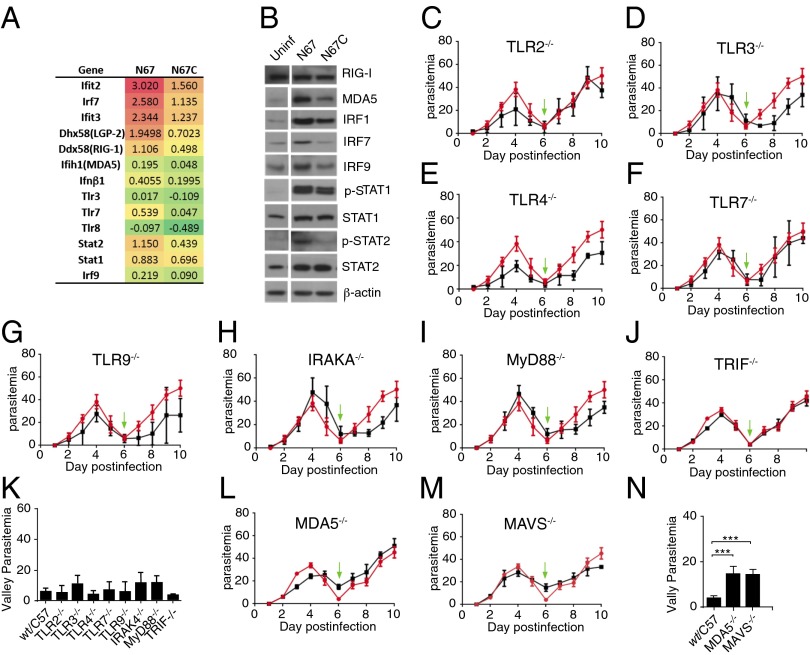
To identify the signaling pathways leading to the strong IFN-I response in the N67-infected mice, we determined the levels of protein expression and/or phosphorylation of some key molecules in the IFN-I signaling networks that also had up-regulated mRNAs (Fig. 3A), particularly in the N67-infected mice. We showed similar higher protein levels for MDA5, IRF1, IRF7, and IRF9 in the N67-infected mice than the N67C-infected or uninfected mice (Fig. 3B). STAT1 and STAT2 were also phosphorylated to higher levels in the N67-infected mice, consistent with the observation from microarray analysis. In addition to the RIG-I pathway, TLR and other signaling pathways may also contribute to production of IFN-I. We therefore investigated the potential role of TLR pathways in the IFN-I response using various KO mice, even though the mRNA transcript levels of TLRs were not up-regulated (only TLR7 was slightly up-regulated in the N67-infected mice) (Fig. 3A). Consistent with gene expression results, infections of TLR and TLR adaptor KO mice, including TLR2−/−, TLR3−/−, TLR4−/−, TLR7−/−, TLR9−/−, IRAK4−/−, MyD88−/−, and TRIF−/− (Fig. 3 C–J), did not show significant differences (P > 0.05, t tests) in the lowest valleys of parasitemia (Fig. 3K), although the peak parasitemia were slightly lower in the TLR2, TLR4, and TLR9 KO mice compared with those of WT mice (Fig. 3 C, E, and G). These results suggest that TLR pathways do not play a major role in the decline of early N67 parasitemia. In contrast, the lowest parasitemia levels of MDA5−/− and MAVS−/− KO mice were significantly different from those of WT mice (Fig. 3 L–N), indicating involvement of MDA5 in parasite ligand recognition and parasitemia control.
Fig. 3.
MDA5 signaling, but not TLRs, and higher level of type I IFN response play an important role in suppression of parasitemia in the N67-infected mice. (A) Log ratio mRNA expression differences from six mice infected with the N67 or N67C parasites. Red color indicates up-regulation and green (negative values) down-regulation compared with uninfected mice. (B) Protein expression and/or phosphorylation (p-STAT1 and p-STAT2) of key molecules that may play a role in IFN signaling pathways, most of which were expressed or phosphorylated at higher levels in mice infected with N67 than those infected N67C parasite. Uninf, spleen homogenate from uninfected mice 4 d postinfection. β-Actin was used as protein loading control. Note that the original gels contained samples from parasites that are not described here and are removed, creating spaces between the uninfected and N67-infected lanes in B. The uninfected and infected samples were run on the same gels. (C–J, L and M) Daily N67 parasitemia of WT (red lines) and knockout mice (black lines) infected with N67: TLR2−/− (C), TLR3−/−(D), TLR4−/−(E), TLR7−/−(F), TLR9−/− (G), IRAK4−/−(H), MyD88−/−(I), TRIF−/− (J), MDA5−/− (L), and MAVS−/− (M). The green arrows point to valley parasitemia of WT infection. (K and N) Average valley parasitemia between WT and KO mice of TRL pathways (K) and KO mice of the MDA5/MAVS pathway (N). Significance at ***P < 0.001 of t test. Each group consists of five to six mice.
MDA5, RIG-I, and RNA Polymerase III All Contributed to Activation of the IFN-I Pathway.
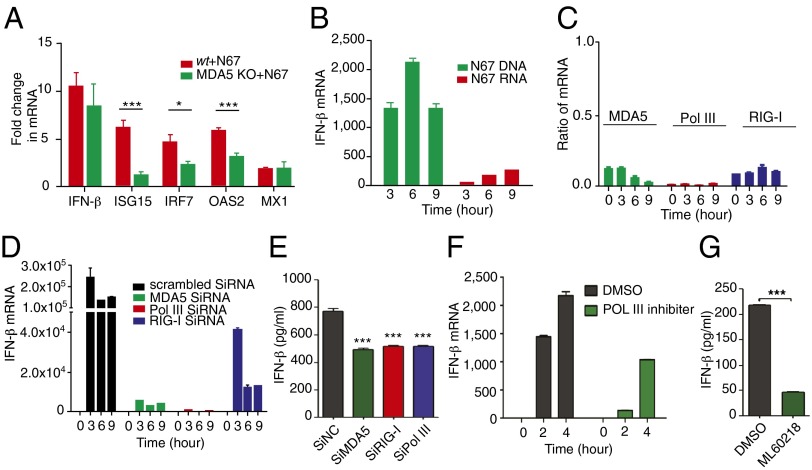
To investigate the relationship of MDA5 and IFN-I response, we infected MDA5-deficient mice and showed significantly lower levels of mRNAs for several IFN-I signature genes (isg15, irf7, and oas2) after N67 infection, although the reductions in IFN-β and MX1 mRNAs were not significant (Fig. 4A). Activation of the MDA5-mediated IFN-I pathway requires recognition of parasite DNA or RNA. To investigate the nature of parasite ligands and host innate immune receptor(s), we transfected a murine macrophage-like cell line (RAW264.7) with purified N67 genomic DNA (gDNA) or total RNA in vitro and found increased IFN-β transcript in the transfected cells compared with untransfected controls (Fig. 4B). Furthermore, parasite DNA was more potent than RNA in stimulating IFN-β expression. We next used RNAi to knockdown the expression of genes encoding DNA/RNA sensors in the RIG-I pathway, MDA5, RIG-I, and RNA polymerase III (RNA Pol III), which is known to convert cytosolic poly(dA-dT) DNA into 5′-triphosphate (5′-ppp) RNA (38), and achieved >80% reduction in gene expression in most cases (Fig. 4C). We then stimulated the cells with N67 DNA and showed that IFN-β expression in both mRNA and protein could be significantly suppressed by RNAi knockdown of the genes encoding MDA5, RIG-I, or RNA pol III (Fig. 4 D and E), but not scrambled siRNA, suggesting conversion of parasite DNA into 5′-ppp RNA by RNA Pol III before binding to MDA5. Reduction in IFN-β expression, both in mRNA (Fig. 4F) and protein (Fig. 4G) levels after treatment of the RAW264.7 cells with RNA Pol III inhibitor ML-60218 (EMD Millipore) further confirmed its role in DNA recognition and IFN-I response. Based on these results, we conclude that the MDA5/MAVS-mediated IFN-I pathway (Fig. S4) plays an important role in controlling parasitemia during N67 infection.
Fig. 4.
Reduced expression of IFN-β or signature genes after gene knockout (MDA5), inhibition, or RNAi knockdown of the key genes in the RIG-I signaling pathway. (A) Expression of INF-β and signature genes in splenocytes of MDA5 KO or WT mice. Isg15, IFN-induced 17-kDa protein; irf7, IFN regulatory factor 7; oas2, 2'–5′ oligoadenylate synthetase 2; mx1, IFN-induced GTP-binding protein. (B) IFN-β mRNA levels from Raw264.7 cells after in vitro stimulation with N67 parasite DNA or RNA. One microgram (1 μg) of DNA or total RNA was transfected into the cells using Lipofectamine. (C) RNAi knockdown of MDA5, RNA Pol III, and RIG-I gene expression, showing great reduction in transcript levels relative to uninfected cells (ratios). (D) IFN-β mRNA levels from gene knockdown cells at different time points after stimulation of Raw264.7 cells with N67 parasite DNA. (E) IFN-β protein levels in RNAi knockdown cells measured using ELISA 6 h after DNA stimulation; significant at ***P < 0.001 of t test. (F and G) Reduction in IFN-β mRNA (F) and protein (G) levels after treatment of Raw264.7 cells with RNA Pol III inhibitor ML60218 (25 μM) in dimethyl sulfoxide (DMSO) for 24 h and N67 DNA stimulation; ***P < 0.001 of t test.
Conventional DCs Are the Main Source of Early IFN-I.
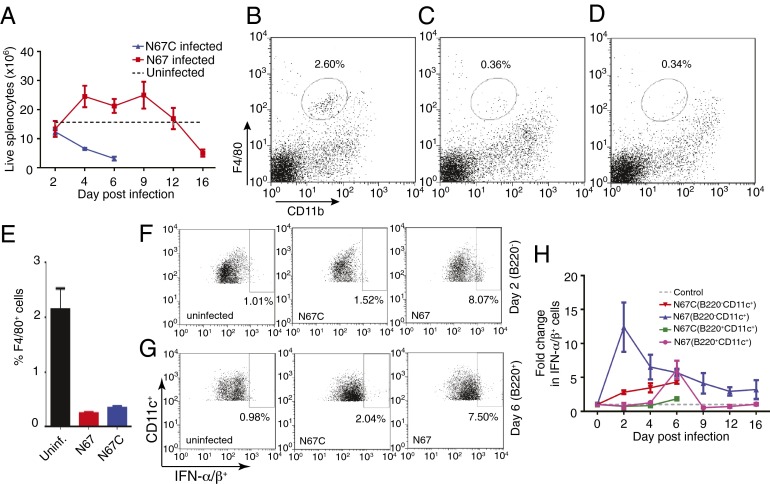
We next investigated the cellular sources of the IFN-I by counting live splenocytes in the N67- and N67C-infected mice and by staining specific cell populations for production of IFN-I. Our histochemical staining showed enlarged spleens and accumulation of a large number of activated MΦs in the spleen of N67-infected mice (Fig. 1E), but whether the cells were still alive or contributed to the higher IFN-I level remained to be determined. Trypan blue staining for live cells showed that the total splenocytes in the N67-infected mice increased from day 4 to day 9 and declined to below those of uninfected mice after day 12 (Fig. 5A). The total live cells in the N67C-infected mice, however, declined quickly from day 2 postinfection. On day 4, the live MΦs decreased from ∼2.1% of total cells in uninfected mice to ∼0.35% in N67- or N67C-infected mice (Fig. 5 B–E). The dramatic reduction in MΦ numbers of both N67- and N67C-infected mice suggested that these cells were unlikely to be the source of the elevated IFN-I observed in N67-infected mice. In fact, we had difficulty in obtaining sufficient MΦ populations for IFN-I straining.
Fig. 5.
Live splenocytes and type I IFN producing cells in uninfected and infected mice. (A) Total live splenocytes in N67- and N67C-infected and uninfected mice from days 2–16; dash line, averaged cell counts from uninfected mice. (B–E) Representative cell counts for F4/80 positive populations (macrophages) in uninfected (B) N67-infected (C), and N67C-infected (D) mouse spleens day 4 postinfection. (E) Average F4/80 positive cell counts and SDs from three independent experiments. (F) Flow cytometry staining of IFN-α/β of representative day-2 B220−CD11c+ cell populations from spleens of uninfected, N67-, or N67C-infected mice. (G) The same as in F but for B220+CD11c+ cell populations at day 6 postinfection. (H) Fold changes in IFN-α/β positive cell populations (compared with uninfected) carrying B220−CD11c+ and B220+CD11c+ markers in the spleens of N67- and N67C-infected mice from day 0 to day 16. The numbers were averages of three independent measurements.
We also stained splenocytes from N67- and N67C-infected mice with antibodies specific for dendritic cells that are known to produce IFN-I (39) and showed that, in the N67-infected mice, the proportions of B220−CD11c+ conventional DCs (cDCs) expressing IFN-α/β were much higher than in uninfected mice (∼12-fold higher) or those infected with N67C (∼5-fold higher) 2 d postinfection. In contrast, the proportion of B220+CD11c+ plasmacytoid DCs (pDCs) expressing IFN-α/β peaked on day 6 and returned to baseline on day 9 postinfection (Fig. 5 F–H). After peaking on day 2, the proportion of IFN-α/β-expressing B220−CD11c+ cells declined gradually, but was still higher than the uninfected controls even at day 16 (Fig. 5H). The late production of IFN-α/β by B220+CD11c+ pDCs suggested alternative signaling, such as by TLR7/9-mediated pathways because RIG-I/MAD5 pathways are known to be operative in cDCs but have not been demonstrated in pDCs (40).
N67-Infected RBCs Stimulated Strong Phagocytosis.
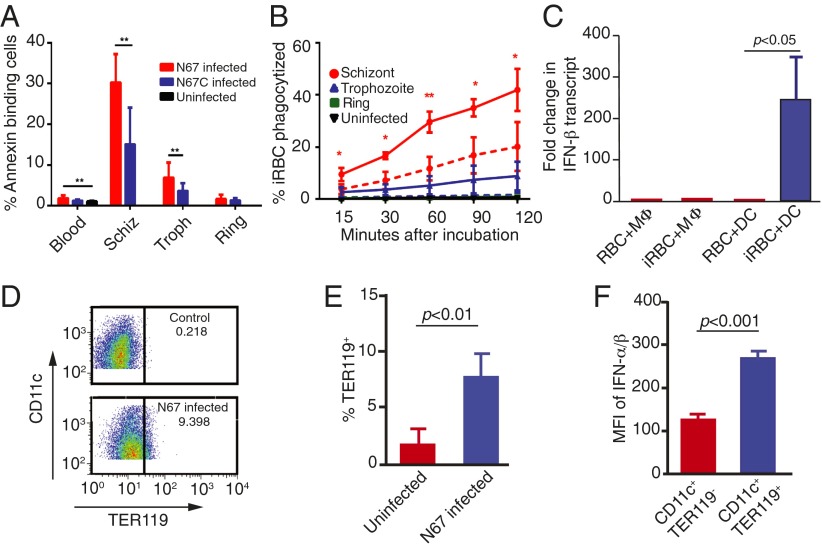
The presence of a large number of MΦs and active phagocytosis of iRBCs in the spleen likely contributed to the day-5 parasitemia decline in the N67 infection (Fig. 1). To investigate the potential mechanism of this enhanced phagocytosis, we measured the levels of PS on the iRBC surface known to trigger phagocytosis by MΦs in vitro. The percentage of late stage iRBCs with surface PS (annexin V-binding cells) was significantly higher in the N67-infected mice than those infected with N67C; ∼30% of N67 schizont-infected RBCs and ∼15% of N67C schizont-infected RBCs were positive for surface PS (Fig. 6A). Consequently, larger numbers of N67 schizont- (significantly more) and trophozoite-infected RBCs were phagocytized by bone-marrow–derived MΦs (BMDMs) compared with those infected with N67C (Fig. 6B). Consistent with the in vivo observations, incubation of N67 schizont-infected RBCs with bone-marrow–derived MΦs and splenic DC cells in vitro showed increased IFN-β transcript in DC cells incubated with iRBCs, but not with uninfected RBCs. No significant changes in the IFN-β level in MΦs incubated with either uninfected RBCs or iRBCs were found (Fig. 6C). Furthermore, DC cells from mice infected with N67 showed significantly increased phagocytosis of RBCs compared with those of uninfected mice on day 4 postinfection (Fig. 6 D and E). The DC cells with phagocytized RBCs also expressed significantly higher levels of IFN-α/β than those without phagocytized RBCs (Fig. 6F), suggesting a link between phagocytosis and IFN-α/β production in DC cells. Phagocytosis and subsequent degradation of iRBCs by DC cells and/or MΦs could promote the release of parasite ligands, leading to activation of IFN-I that in turn stimulates stronger immune responses.
Fig. 6.
Surface phosphatidylserine (PS) exposure on infected red blood cells (iRBCs) and in vitro phagocytosis of iRBCs and IFN-I production. (A) Percentage of infected RBCs that bind to annexin V. Blood, whole infected blood; Schiz, schizont-infected RBCs; Troph, trophozoite-infected RBCs; ring, ring-infected RBCs. (B) Percentages of iRBCs phagocytized by bone marrow-derived macrophages (BMDMs) over time compared with phagocytosis of uninfected RBCs. Solid and dotted lines represent data sets from N67 and N67C, respectively. *P < 0.05; **P < 0.01. (C) Changes in IFN-β transcript levels in BMDMs or splenic dendritic cells after incubation with uninfected RBCs or with RBCs infected with N67 schizonts in vitro. Different stages of the parasites were obtained using a Percoll purification protocol described previously (56). (D and E) Frequencies of splenic CD11c+TER119+ cells (DC cells containing RBCs) from N67-infected and -uninfected mice. D shows representative cell counts, and E shows averaged percentages and SDs of TER119+ DC cells from four mice in each group. (F) DC cells containing RBCs (blue, TER119+) expressed significantly higher IFN-α/β levels than those without RBCs (red, TER119−); MFI, mean fluorescence intensity averaged from four mice each. All tests were performed using t test.
Critical Role of CD14/TLRs and p38 in Inflammatory Response to N67C Infection.
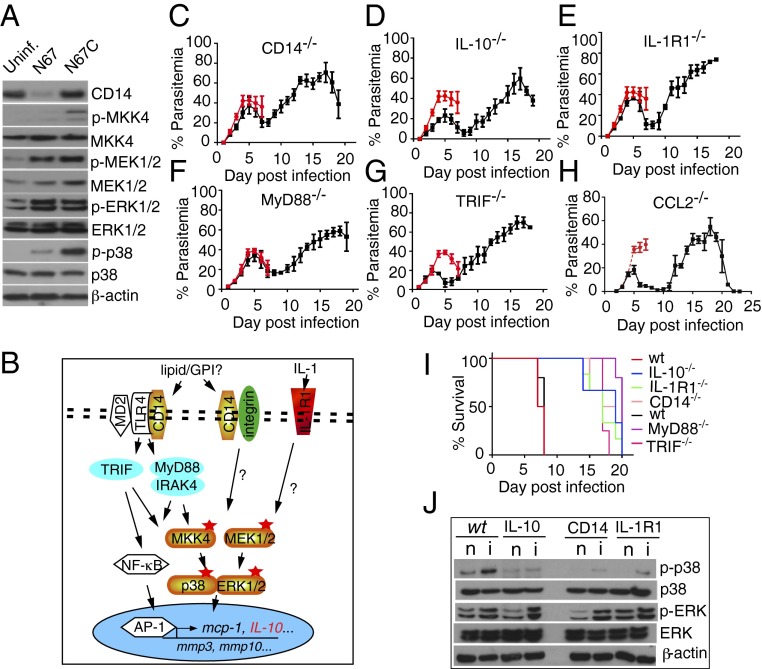
Our microarray analysis suggested that higher levels of proinflammatory responses mediated through CD14 and IL-1R1 in the N67C-infected mice (Fig. S2) likely contributed to early host death. To investigate the signaling pathways associated with these responses, we measured the levels of protein expression and/or phosphorylation of key kinases in inflammation pathways and showed higher levels of CD14 expression and phosphorylation of MKK4, MEK1/2, and p38 kinases in the N67C-infected mice than those infected with N67 (Fig. 7A). These results suggested inflammatory response signaling through CD14/TLR4 and MKK4 and p38 kinase pathways (Fig. 7B).
Fig. 7.
Inflammatory responses and signaling pathways mediated by CD14, TLRs, and IL-1R1 in mice infected with N67C parasite. (A) Protein expression and/or phosphorylation of CD14 and selected kinases in the spleens of uninfected (uninf.), N67- or N67C-infected mice. (B) Putative pathways involved in host inflammatory responses after infection with N67C. (C–H) Parasitemia of genetic knockout mice infected with N67C. Mice were infected with 106 infected red blood cells (iRBCs) (except H, which were infected with 105 iRBCs). Parasitemias were counted daily after Giemsa staining. Red represents parasitemia of wild-type mice. (I) Survival curves of WT and various KO mice infected with N67C. (J) Western blot detection of p38 and ERK kinase expression and phosphorylation in wild-type, IL-10−/−, CD14−/−, and IL-R1−/− mice. β-Actin was used as protein loading control. n, naïve; i, infected.
To further confirm the roles of the key molecules in the inflammatory response, we infected CD14−/−, IL-10−/−, IL-1R1−/−, MyD88−/−, TRIF−/−, and CCL2−/− mice with the N67C parasite. Compared with WT mice, the CD14−/−, IL-10−/−, IL-1R1−/−, MyD88−/−, TRIF−/−, and CCL2−/− mice survived the early N67C infection with minimal symptoms, producing parasitemia curves similar to those infected with N67, whereas most of the N67C-infected WT mice died on days 6–7 (Fig. 7 C–I). Additionally, the TRIF−/−, IL-10−/−, and CCL2−/− mice had lower peak parasitemia than those of WT, suggesting that these molecules also played a role in controlling parasitemia. To further verify that activation of IL-1R1 and CD14 was signaled through p38 kinase, we measured the levels of p38 and ERK protein expression and phosphorylation in IL-10−/−, IL-1R1−/−, and CD14−/− mice with or without infection of N67C. Compared with WT mice, reduced p38 phosphorylation signals were detected in the CD14−/−, IL-1R1−/−, and IL-10−/− mice (Fig. 3J). These data showed that knockout of CD14, IL-1R1, and IL-10 could indeed reduce the levels of p38 activation and established connections between CD14 or IL-R1 binding and p38 activation in response to N67C infection, although more work is necessary to completely understand the inflammation pathways in response to malaria infection.
Discussion
In this study, we systematically compared host transcriptional responses to infections with two isogenic P. yoelii strains (34) that produced different patterns of parasitemia and mortality in C57BL/6 mice. We supported our microarray results with data from qPCR, protein expression and/or phosphorylation, and infection of KO mice, providing functional verifications to the observations. We demonstrated that different innate pathways were stimulated when inbred mice were infected with the two isogenic parasite strains. In our previous study, we showed that out of 21 microsatellites, only one (Py2000 on chromosome 8) was polymorphic between the genomes of N67 and N67C parasites, and sequencing of the gene encoding PyEBL found only one nucleotide substitution leading to an amino acid change at position 741 (C to Y) (34). These results demonstrate that dramatic differences in disease phenotypes can be produced when mice are infected with different parasite strains, even with parasites of similar genomic backgrounds. The results also suggest that conclusions from studies of host–parasite interaction require consideration of variations in both host and parasite genetic backgrounds.
The intriguing question was why the isogenic parasite pair N67/N67C with almost identical genomes produces such different disease outcomes; mice infected with N67C die 6–7 d after infection, whereas those infected with N67 are able to control parasitemia from day 5 (Fig. 1). Our analyses of mRNA transcripts, protein expression, phosphorylation, and infections of KO mice showed up-regulation to high levels of many PICCs and related receptors in mice infected with N67C, which likely resulted in early death of N67C-infected mice. These proinflammatory responses were mainly mediated by ligand binding to CD14 and TLR4 complex, and possibly IL-1R1, followed by signaling through p38 MAPK and ERK kinase pathways because the expression (or protein phosphorylation) of these receptors and kinases were highly up-regulated in the N67C-infected mice. Indeed, induction of proinflammatory response by P. falciparum GPI and signaling through various MAP kinases were reported previously (29, 41). Survival of CD14−/−, IL-R1−/−, MyD88−/−, TRIF−/−, and IL-10−/− mice also supported the notion that PICC production mediated through these molecules played a significant role in the pathology of mice infected with N67C. Interestingly, the N67C parasitemia curves in the KO mice were similar to those of WT mice infected with N67 parasites, suggesting modulation or suppression of inflammatory responses mediated by these molecules may change the balance of host response and promote host survival (Fig. S5). Additionally, the suppression of pathways and genes involved in heme metabolism and hematopoiesis in N67C-infected mice suggests that anemia may have some impact on mortality. Because our current study mainly focuses on the IFN-I activation pathway in N67-infected mice, more experiments are necessary to completely dissect the molecular signaling events in host responses to N67C infection, which may suggest new approaches for managing the care of patients with severe malaria.
Another interesting question was how the mice infected with N67 control early parasitemia. IFN-I and -II responses and the related pathways have been shown to play an important role in controlling malaria parasite infection (12, 42), but the exact effects of the IFN on malaria infection remain to be clearly defined (22, 43–45). Daily injections of a recombinant human IFN-α prevented death by PbANKA cerebral malaria (45), and the majority of ifnar1−/−, ifr3−/−irf7−/−, or tbkΔ/Δ mice survived (or lived) longer than the WT in lethal PbANKA infection (22, 46, 47). Variations at IFN-α promoter (17470-G/G and L168V-G/G genotypes) or receptor IFNAR1 were associated with protection against severe malaria (43, 46), whereas variation at IFNA2-173 and IFNA8-884 reduced IFN-α production and increased susceptibility to severe malarial anemia and mortality (44). Here we showed that the IFN responses to two strains of P. yoelii infection were dramatically different, with mice infected with N67 producing higher IFN-I response than those infected with N67C, whereas mice infected with N67C had higher IFN-γ transcript levels. In addition to the high levels of IFN-I–related molecules, such as IRF7, IFI35, IFIT2, IFIT3, ISG15, and ISG20, transcription factors involved in IFN signaling (STAT1 and STAT2) were also translated or phosphorylated to higher levels in N67-infected mice. A recent report also showed that IRF3/IRF7 and related signaling molecules, STING and TBK1, were key players in P. falciparum DNA (motif ATTTTAC)-mediated response (22). Our results demonstrate that a strong IFN-I response mediated by RNA pol III- and MDA5-sensing parasite DNA/RNA could suppress early parasitemia. Although more studies are necessary to completely elucidate the relationship between the increased phagocytosis and induction of the IFN-I pathway in the N67-infected mice, our data suggest that recognition of iRBCs and phagocytosis by DCs and/or MΦs (releasing parasite materials) could represent one of the mechanisms in promoting the IFN-I response (Fig. S5). Indeed, the larger differences in parasitemia observed in the RAG1−/− and RAG2−/− mice than those of MDA5−/− and MAVS−/− mice suggest T- or B cells may play a role in suppressing parasitemia.
The effect of IFN-I on parasitemia control and/or host pathology is likely dependent on parasite genetic background (species/strain), and the timing and levels of INF-I are also critical. It is possible that early IFN-I response can protect against malaria infection as seen in mice infected with N67; however, prolonged elevated IFN-I level could be harmful to the host. Recent data from viral infection suggested that chronic high levels of IFN-I could lead to immunosuppression and prolonged infection (48, 49). The rebound of parasitemia after day 8 and the death of N67-infected mice after day 15 could be caused by low, but elevated IFN-I levels and immunosuppression (Fig. 2B). The survival and reduced symptoms of experimental cerebral malaria in ifnar1−/−, ifr3−/−irf7−/−, or tbkΔ/Δ mice infected with P. berghei (compared with lethal infection in wt mice) suggest that IFN-I may also contribute to malaria pathology in late stages of infection (22, 46, 47). The relationship of IFN-I response dynamics and protection or pathology in malaria infection requires further investigation.
What are the differences between the N67 and N67C parasites that triggered the dramatically different host responses? Genome sequencing and careful comparison of the parasite DNA sequences may reveal additional unknown variations that may contribute to the differences in virulence, particularly the gene families at subtelomeric regions (50). Immunoprecipitation using antibodies against RIG-I has been used to pull-down viral RNAs bound to the receptor (51); the same technique may be used to identify parasite RNAs bound to MDA5. The amino acid substitution (C741Y) in EBL (34) may also contribute to the differences in pathology between the mice infected with the two parasites by tipping the balance of immune responses. Proteins with DBL domains can bind to the Duffy antigen/chemokine receptor (DARC) expressed on endothelial cells and trigger host innate immune responses (52). DARC is a receptor for chemokines of both the C-C and C-X-C families, including IL-8, MGSA, RANTES, and MCP-1, when expressed on endothelial and other cell types. The C741Y substitution in N67 may change the localization of the EBL protein in the parasites, as reported in another pair of isogeneic P. yoelii parasites (17X and 17XL) (53) and alter the dynamics of protein secretion and its interaction with DARC and/or other unknown receptors. The level of EBL protein in the host blood could affect the binding and activation of DARC (or other receptors) and therefore host immune responses.
Our results also showed that infection with the parasites suppressed components of many host innate immune response pathways (Dataset S2). Expression of genes involved in pathways of antigen processing, immune cell activation, and migration was down-regulated in mice infected with parasites. On the other hand, the expression of genes involved in DNA replication, cell division, and host chemokine response were up-regulated. Down-regulation of components of immune response related pathways could represent a mechanism that parasites use to evade host killing, whereas higher gene expression levels for those involved in the activation and division of cells, including cells in host immune response and hematopoiesis, could represent active host responses. Our analyses clearly demonstrated that there are many strain-specific pathways of host innate immune responses. Although these results were from a mouse malaria model, they provide important information for future studies of pathogenesis in human malaria infection and potentially for better disease intervention strategies.
Materials and Methods
Malaria Parasites and Infection of Mice.
The parasite P. yoelii nigeriensis N67 (N67) and N67C were initially obtained from the Malaria Research and Reference Reagent Resource Center (MR4; www.mr4.org/) and was described previously (34). Freshly thawed parasites were injected into naïve C57BL/6 mice to initiate infection. An inoculum containing 1 × 106 iRBCs suspended in 100 µL phosphate buffer saline (PBS), pH 7.4, from the donor mice was injected i.v. into experimental mice. Naïve mice receiving an equivalent number of uninfected RBCs served as a negative control group. Parasitemia was monitored daily by examination of Giemsa-stained thin tail blood smears. Inbred female C57BL/6 mice, aged 6–8 wk old, or various gene KO mice were purchased from Charles River Laboratories, Jackson Laboratory, or obtained from the NIAID/Taconic repository. All mouse-related procedures were performed according to protocol LMVR11E approved by the Animal Care and Use Committee in NIAID, National Institutes of Health, or a protocol approved by the Animal Care and Welfare Committee at The Methodist Hospital Research Institute.
Immunohistochemistry and Microscopy.
Animals were killed using CO2 inhalation and necropsied according to a standard protocol. For hematoxylin and eosin (H&E) staining, tissue sections were fixed by immersion in 10% (vol/vol) neutral buffered formalin, embedded in paraffin, sectioned at 4 μm, and stained with H&E. Immunostaining by the avidin-biotin peroxidase complex (ABC) method was performed using a Vectastain Elite ABC kit (Vector) as described previously (54). Antibody against F4/80 was purchased from Santa Cruz Biotechnology. For apoptosis detection, a standard TUNEL assay was performed. Briefly, following deparaffinization and rehydration, tissue sections were treated with proteinase K and H2O2/PBS to block the endogenous peroxidase activity and then incubated with TdT (New England Biolabs) before detection using antidigoxigenin-peroxidase (Roche Diagnostics) and DAB. The samples were counterstained with hematoxylin before microscopy.
RNA Samples for Microarray Hybridization.
Four days after parasite injection, spleens from infected or uninfected mice were removed and preserved in RNA Later solution (Qiagen). RNA was isolated using an RNeasy mini kit following the manufacturer's protocol (Qiagen).
Microarray Hybridization and Analysis.
Spleen RNA from six mice per group was individually amplified and labeled from 500-ng input using the Illumina TotalPrep RNA Amplification kit (Applied Biosystems). Biotinylated cDNA was hybridized to Illumina MouseRef-8 v2.0 Expression BeadChip (GEO accession no. GPL6885) having 25,697 unique probes using reagents provided. Amplification and labeling of RNA samples were performed using the Illumina TotalPrep RNA Amplification kit (Applied Biosystems). Illumina HiScan was used for chip imaging and Genome Studio software for data extraction. Quantile normalization, P value adjustments, and gene set enrichment analysis (35) were performed using JMP/Genomics software (SAS Institute). Gene sets were derived from Gene Ontology term assignments available from National Center for Biotechnology Information.
Western Blot and Kinase Phosphorylation Assay.
Mouse spleens were removed 4 d after infection and immediately frozen in liquid nitrogen. Total proteins were extracted using the Zoom 2D Protein Solubilizer kit (Invitrogen) supplemented with Protease Inhibitor Mixture, Phosphatase Inhibitor Mixture (Sigma), and Protease Inhibitor Tablets (Roche Applied Sciences). The proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS/PAGE), transferred to a PVDF Western blotting membrane (Roche Diagnostics), and probed with the indicated primary antibodies and corresponding secondary antibody before chemiluminescent detection (SuperSignal West Pico Chemiluminescent Substrate; Pierce).
Antibody Treatment.
To block the IFN-I receptor, anti-IFNAR1antibodies (Leinco Technologies) in the amount of 500 mg in PBS on days 2, 3, and 5, and then 250 mg on days 7, 9, and 11 postinfection, were injected i.p. into each of five mice, and the parasitemias were monitored by daily Giemsa-stained blood smears.
In Vitro Transfection of Murine RAW264.7 Cell Line and RNAi.
siRNA oligonucleotides for MDA5, RIG-I, RNA Pol III, and control (scrambled SiRNA) were purchased from Invitrogen and nucleotransfected into Raw264.7 cells using Amaxa nucleofector kit V following the manufacturer’s instruction (Lonza) for 48 h before stimulation with 1 μg Plasmodium DNA or RNA using Lipofectamine 2000 reagent (Invitrogen) at the indicated time points. Total RNAs were extracted using TRIzol reagent (Invitrogen), and complementary cDNAs were generated using reverse superscriptase III (Invitrogen).
qPCR.
RNA samples from the splenic tissues or transfected cell lines were prepared using an RNeasy mini kit following the manufacturer's protocol (Qiagen). cDNA was prepared from the total RNA using SuperScript III First-Strand cDNA synthesis kit (Invitrogen) and oligo dT primer. All quantifications were normalized to an endogenous beta actin (Actb) or glyceraldehyde 3-phosphate dehydrogenase (gapdh) control. The relative quantification value for each target gene compared with the calibrator for that target is expressed as 2-(Ct-Cc) (Ct and Cc are the mean threshold cycle differences after normalizing to Actb or gapdh). For qPCR reactions, iQ SYBR Green Supermix or the SYBR GreenER qPCR Super Mix Universal (Invitrogen) was used according to the manufacturer's instructions. The primer sequences for the PCR are listed in Dataset S3.
RNA Polymerase III Inhibitor Treatment and ELISA.
The RNA polymerase III inhibitor ML-60218 was purchased from EMD Millipore. Raw264.7 cells were treated with RNA polymerase III inhibitor (25 μM) in dimethyl sulfoxide (DMSO) for 24 h and then stimulated with 1 μg Plasmodium gDNA for the indicated times. The expression of IFN-β mRNA or protein was measured using qPCR and ELISA, respectively. For ELISA, supernatants were collected 6 h after DNA stimulation, and IFN-β was measured by using VerKine Mouse IFN Beta ELISA kit following the manufacturer's instructions.
Preparation of BMDMs and DC Cells and Phagocytosis Assay.
Bone marrow cells were collected from the femur and tibia of C57BL/6 mice and cultured in RPMI medium 1640 with 10% FBS, 0.1 mM nonessential amino acids, 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM Hepes, and 20% L929 conditioning medium (the source of M-CSF) for 7 d. Carboxyfluorescein succinimidyl ester (CFSE)-labeled iRBCs were mixed with the BMDM cells at the ratio of 10:1 and examined at various time points. After incubation, BMDM cells were washed and detached with Accutase (Innovative Cell Technologies). The cells were treated with ACK lysis buffer (Lonza) to remove the nongulfed iRBCs and analyzed using FACSort flow cytometer (BD Bioscience). The fluorescence of CFSE was monitored as an indicator of RBC engulfment. The cell population with CFSEhigh was regarded as the cells that engulfed the iRBCs. Data acquisition and analysis were done using FACSort flow cytometer (BD Bioscience) with Cell Quest software (v3.3), and analyzed using FlowJo (v7.6; Tree Star).
Cell suspensions of splenocytes were prepared from naïve mice, and DC cells were enriched using the EasySep Mouse Pan-DC Enrichment kit (Stemcell Technologies), seeded into each well (1.0 × 105) of a 24-well plate, and incubated with 1.0 × 106 N67 schizont-infected or uninfected RBCs. Eight hours after incubation, mRNA were extracted for qPCR analysis of IFN-β transcript.
Splenocyte Staining.
Mouse splenocytes were stained at 4 °C in PBS containing 2% FBS after FcγRII/III blocking with anti-mouse CD16/CD32 (clone 93; eBiosciences). Surface staining was performed with antibodies purchased from either BD Pharmingen or eBioscience (anti-CD11c, clone N418; anti-CD11b, clone M1/70; anti-B220; clone RA3-6B2; anti-TER119, clone TER-119). For intracellular IFN-α/β staining, cells were incubated at 37 °C for 6 h with Brefeldin A (BD Bioscience) before cell surface staining. After fixation and permeabilization (BD; Fix & Perm), the cells were stained with anti-IFNα (clone RMMA-1, 10 µg/mL, PBL) and anti-IFNβ (clone 7F-D3, 10 µg/mL; Abcam) followed by staining with biotin anti-rat IgG1 (clone MRG1-58, 10 µg/mL; BioLegend) and streptavidin-APC (eBiosciences). Stained cells were analyzed by flow cytometry. DC cells were also stained for intracellular TER119 using the procedures described (55). For Annexin V binding assay, iRBCs were incubated with APC-conjugated Annexin V (eBiosciences) in binding buffer (eBiosciences) at room temperature for 15 min. After washing with binding buffer, cells were analyzed in a flow cytometer.
Supplementary Material
Acknowledgments
We thank Drs. Alan Sher and David Sacks for advice and National Institute of Allergy and Infectious Diseases (NIAID) intramural editor Brenda Rae Marshall for assistance. This work was supported by the Intramural Research Program of the Division of Intramural Research at the NIAID, National Institutes of Health (NIH) and in part by grants from the National Cancer Institute, NIH R01CA090327 and R01CA101795 (to R.W.). X.Y. is a recipient of The China Scholarship Council (CSC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.Z. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the NCBI database (accession no. GSE51329).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316467111/-/DCSupplemental.
References
- 1.WHO 2012. World Malaria Report 2012. Available at www.who.int/malaria/publications/world_malaria_report_2012/en/index.html. Accessed December 31, 2013.
- 2.Metcalf D. Hematopoietic cytokines. Blood. 2008;111(2):485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liehl P, Mota MM. Innate recognition of malarial parasites by mammalian hosts. Int J Parasitol. 2012;42(6):557–566. doi: 10.1016/j.ijpara.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Gazzinelli RT, Denkers EY. Protozoan encounters with Toll-like receptor signalling pathways: Implications for host parasitism. Nat Rev Immunol. 2006;6(12):895–906. doi: 10.1038/nri1978. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4(3):169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas K, Tongren JE, Riley EM. The war between the malaria parasite and the immune system: Immunity, immunoregulation and immunopathology. Clin Exp Immunol. 2003;133(2):145–152. doi: 10.1046/j.1365-2249.2003.02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley EM, Wahl S, Perkins DJ, Schofield L. Regulating immunity to malaria. Parasite Immunol. 2006;28(1-2):35–49. doi: 10.1111/j.1365-3024.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 8.Ockenhouse CF, et al. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74(10):5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther M, et al. Innate immune responses to human malaria: Heterogeneous cytokine responses to blood-stage Plasmodium falciparum correlate with parasitological and clinical outcomes. J Immunol. 2006;177(8):5736–5745. doi: 10.4049/jimmunol.177.8.5736. [DOI] [PubMed] [Google Scholar]
- 10.Coban C, et al. Toll-like receptor 9 mediates innate immune activation by the malaria pigment hemozoin. J Exp Med. 2005;201(1):19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coban C, Ishii KJ, Horii T, Akira S. Manipulation of host innate immune responses by the malaria parasite. Trends Microbiol. 2007;15(6):271–278. doi: 10.1016/j.tim.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Franklin BS, et al. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci USA. 2009;106(14):5789–5794. doi: 10.1073/pnas.0809742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Gowda NM, Kumar S, Gowda DC. Protein-DNA complex is the exclusive malaria parasite component that activates dendritic cells and triggers innate immune responses. J Immunol. 2010;184(8):4338–4348. doi: 10.4049/jimmunol.0903824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erdman LK, Finney CA, Liles WC, Kain KC. Inflammatory pathways in malaria infection: TLRs share the stage with other components of innate immunity. Mol Biochem Parasitol. 2008;162(2):105–111. doi: 10.1016/j.molbiopara.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Rosanas-Urgell A, et al. Expression of non-TLR pattern recognition receptors in the spleen of BALB/c mice infected with Plasmodium yoelii and Plasmodium chabaudi chabaudi AS. Mem Inst Oswaldo Cruz. 2012;107(3):410–415. doi: 10.1590/s0074-02762012000300017. [DOI] [PubMed] [Google Scholar]
- 16.Lepenies B, et al. Induction of experimental cerebral malaria is independent of TLR2/4/9. Med Microbiol Immunol (Berl) 2008;197(1):39–44. doi: 10.1007/s00430-007-0057-y. [DOI] [PubMed] [Google Scholar]
- 17.Togbe D, et al. Murine cerebral malaria development is independent of toll-like receptor signaling. Am J Pathol. 2007;170(5):1640–1648. doi: 10.2353/ajpath.2007.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dostert C, et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS ONE. 2009;4(8):e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shio MT, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5(8):e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimer T, et al. Experimental cerebral malaria progresses independently of the Nlrp3 inflammasome. Eur J Immunol. 2010;40(3):764–769. doi: 10.1002/eji.200939996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kordes M, Matuschewski K, Hafalla JC. Caspase-1 activation of interleukin-1β (IL-1β) and IL-18 is dispensable for induction of experimental cerebral malaria. Infect Immun. 2011;79(9):3633–3641. doi: 10.1128/IAI.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35(2):194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGilvray ID, Serghides L, Kapus A, Rotstein OD, Kain KC. Nonopsonic monocyte/macrophage phagocytosis of Plasmodium falciparum-parasitized erythrocytes: A role for CD36 in malarial clearance. Blood. 2000;96(9):3231–3240. [PubMed] [Google Scholar]
- 26.Patel SN, et al. CD36 mediates the phagocytosis of Plasmodium falciparum-infected erythrocytes by rodent macrophages. J Infect Dis. 2004;189(2):204–213. doi: 10.1086/380764. [DOI] [PubMed] [Google Scholar]
- 27.Nebl T, De Veer MJ, Schofield L. Stimulation of innate immune responses by malarial glycosylphosphatidylinositol via pattern recognition receptors. Parasitology. 2005;130(Suppl):S45–S62. doi: 10.1017/S0031182005008152. [DOI] [PubMed] [Google Scholar]
- 28.Parroche P, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc Natl Acad Sci USA. 2007;104(6):1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Krishnegowda G, Gowda DC. Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: The requirement of extracellular signal-regulated kinase, p38, c-Jun N-terminal kinase and NF-kappaB pathways for the expression of proinflammatory cytokines and nitric oxide. J Biol Chem. 2005;280(9):8617–8627. doi: 10.1074/jbc.M413539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh AP, et al. Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell. 2007;131(3):492–504. doi: 10.1016/j.cell.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Mavoungou E, Held J, Mewono L, Kremsner PG. A Duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. J Infect Dis. 2007;195(10):1521–1531. doi: 10.1086/515579. [DOI] [PubMed] [Google Scholar]
- 32.McCormick CJ, Craig A, Roberts D, Newbold CI, Berendt AR. Intercellular adhesion molecule-1 and CD36 synergize to mediate adherence of Plasmodium falciparum-infected erythrocytes to cultured human microvascular endothelial cells. J Clin Invest. 1997;100(10):2521–2529. doi: 10.1172/JCI119794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longley R, et al. Host resistance to malaria: Using mouse models to explore the host response. Mamm Genome. 2011;22(1-2):32–42. doi: 10.1007/s00335-010-9302-6. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii. Proc Natl Acad Sci USA. 2011;108(31):E374–E382. doi: 10.1073/pnas.1102261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SY, Volsky DJ. PAGE: Parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sexton AC, et al. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J Infect Dis. 2004;189(7):1245–1256. doi: 10.1086/382596. [DOI] [PubMed] [Google Scholar]
- 37.Schaecher K, Kumar S, Yadava A, Vahey M, Ockenhouse CF. Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes. Infect Immun. 2005;73(9):6091–6100. doi: 10.1128/IAI.73.9.6091-6100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim CC, et al. Splenic red pulp macrophages produce type I interferons as early sentinels of malaria infection but are dispensable for control. PLoS ONE. 2012;7(10):e48126. doi: 10.1371/journal.pone.0048126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald-Bocarsly P, Dai J, Singh S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 2008;19(1):3–19. doi: 10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, et al. MAPK-activated protein kinase 2 differentially regulates plasmodium falciparum glycosylphosphatidylinositol-induced production of tumor necrosis factor-alpha and interleukin-12 in macrophages. J Biol Chem. 2009;284(23):15750–15761. doi: 10.1074/jbc.M901111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCall MB, Sauerwein RW. Interferon-γ—central mediator of protective immune responses against the pre-erythrocytic and blood stage of malaria. J Leukoc Biol. 2010;88(6):1131–1143. doi: 10.1189/jlb.0310137. [DOI] [PubMed] [Google Scholar]
- 43.Aucan C, et al. Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun. 2003;4(4):275–282. doi: 10.1038/sj.gene.6363962. [DOI] [PubMed] [Google Scholar]
- 44.Kempaiah P, et al. Reduced interferon (IFN)-α conditioned by IFNA2 (-173) and IFNA8 (-884) haplotypes is associated with enhanced susceptibility to severe malarial anemia and longitudinal all-cause mortality. Hum Genet. 2012;131(8):1375–1391. doi: 10.1007/s00439-012-1175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vigário AM, et al. Recombinant human IFN-alpha inhibits cerebral malaria and reduces parasite burden in mice. J Immunol. 2007;178(10):6416–6425. doi: 10.4049/jimmunol.178.10.6416. [DOI] [PubMed] [Google Scholar]
- 46.Ball EA, et al. IFNAR1 controls progression to cerebral malaria in children and CD8+ T cell brain pathology in Plasmodium berghei-infected mice. J Immunol. 2013;190(10):5118–5127. doi: 10.4049/jimmunol.1300114. [DOI] [PubMed] [Google Scholar]
- 47.Palomo J, et al. Type I interferons contribute to experimental cerebral malaria development in response to sporozoite or blood-stage Plasmodium berghei ANKA. Eur J Immunol. 2013;43(10):2683–2695. doi: 10.1002/eji.201343327. [DOI] [PubMed] [Google Scholar]
- 48.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340(6129):207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlton JM, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419(6906):512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 51.Baum A, Sachidanandam R, García-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010;107(37):16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hadley TJ, Peiper SC. From malaria to chemokine receptor: The emerging physiologic role of the Duffy blood group antigen. Blood. 1997;89(9):3077–3091. [PubMed] [Google Scholar]
- 53.Otsuki H, et al. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proc Natl Acad Sci USA. 2009;106(17):7167–7172. doi: 10.1073/pnas.0811313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi CF, et al. Expression of the cyclin-dependent kinase inhibitor p27 and its deregulation in mouse B cell lymphomas. Leuk Res. 2006;30(2):153–163. doi: 10.1016/j.leukres.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 55.Ohyagi H, et al. Monocyte-derived dendritic cells perform hemophagocytosis to fine-tune excessive immune responses. Immunity. 2013;39(3):584–598. doi: 10.1016/j.immuni.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 56.Bettiol E, Carapau D, Galan-Rodriguez C, Ocaña-Morgner C, Rodriguez A. Dual effect of Plasmodium-infected erythrocytes on dendritic cell maturation. Malar J. 2010;9:64. doi: 10.1186/1475-2875-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.