Abstract
Ubiquitin (Ub) functions in many different biological pathways, where it typically interacts with proteins that contain modular Ub recognition domains. One such recognition domain is the Npl4 zinc finger (NZF), a compact zinc-binding module found in many proteins that function in Ub-dependent processes. We now report the solution structure of the NZF domain from Npl4 in complex with Ub. The structure reveals that three key NZF residues (13TF14/M25) surrounding the zinc coordination site bind the hydrophobic ‘Ile44' surface of Ub. Mutations in the 13TF14/M25 motif inhibit Ub binding, and naturally occurring NZF domains that lack the motif do not bind Ub. However, substitution of the 13TF14/M25 motif into the nonbinding NZF domain from RanBP2 creates Ub-binding activity, demonstrating the versatility of the NZF scaffold. Finally, NZF mutations that inhibit Ub binding by the NZF domain of Vps36/ESCRT-II also inhibit sorting of ubiquitylated proteins into the yeast vacuole. Thus, the NZF is a versatile protein recognition domain that is used to bind ubiquitylated proteins during vacuolar protein sorting, and probably many other biological processes.
Keywords: NMR spectroscopy, NZF domain, structure, ubiquitin, vacuolar protein sorting
Introduction
Protein ubiquitylation performs essential functions in a variety of biological processes, including protein degradation, DNA repair, transcription, translation, signal transduction, cell cycle control, organelle assembly/disassembly, protein trafficking, and viral budding (Hershko and Ciechanover, 1998; Varshavsky et al, 2000; Pickart, 2001; Schnell and Hicke, 2003). Ubiquitylated proteins are commonly recognized by other proteins that contain modular ubiquitin (Ub)-binding domains (Buchberger, 2002; Katzmann et al, 2002). These domains typically function together with additional localization and recognition signals that help to recruit and retain ubiquitylated proteins in the appropriate biological pathway, and with effector domains that perform specific biological functions (Di Fiore et al, 2003).
The Npl4 zinc fingers (NZFs) are a widespread family of zinc-binding modules that appear to function primarily by binding other macromolecules, including Ub (Nakielny et al, 1999; Yaseen and Blobel, 1999; Meyer et al, 2002; Wang et al, 2003). The Ub-binding activity of NZF domains was first discovered in studies of mammalian nuclear protein localization 4 (Npl4) protein (Meyer et al, 2002), which functions together with the ubiquitin fusion degradation 1 (Ufd1) protein to form an adaptor complex for the AAA ATPase, p97/Cdc48. The Ufd1–Npl4 complex mediates several different biological processes, including endoplasmic reticulum (ER)-associated degradation (ERAD), regulated ubiquitin-dependent processing (RUP), nuclear envelope closure, and mitotic spindle disassembly (Cao et al, 2003; Hetzer et al, 2001; reviewed in Tsai et al, 2002). Although the intact Ufd1–Npl4 complex can bind ubiquitylated proteins via the NZF domain of Npl4, the biological relevance of this interaction is not yet clear because the single NZF domain of the mammalian Npl4 proteins is not conserved in the yeast Npl4 protein (Meyer et al, 2002; Ye et al, 2003). Moreover, deletion of the mammalian Npl4 NZF domain does not measurably affect the retrotranslocation of polyubiquitylated substrates from the ER in vitro (Ye et al, 2003). Indeed, the functional importance of Ub binding has not yet been established for any NZF domain.
General searches for proteins containing NZF domains have revealed that more than 100 proteins contain NZF domains that conform to the consensus sequence: x(4)-W-x-C-x(2)-C-x(3)-N-x(6)-C-x(2)-C-x(5) (where x represents any residue), and many more that contain recognizable NZF variants (Falquet et al, 2002; Wang et al, 2003). The four conserved Cys residues coordinate a single zinc ion, and the Trp and Asn residues stabilize the domain core (Wang et al, 2003). NZF domains are frequently found in proteins that function in Ub-dependent pathways, and several isolated NZF domains have been shown to bind mono- and polyubiquitin in vitro (Meyer et al, 2002; Wang et al, 2003). However, not all NZF domains can bind Ub (Meyer et al, 2002), and there is therefore a need to identify the determinants that dictate Ub binding and to understand the NZF/Ub interaction in molecular detail. The current studies were undertaken with the goals of determining the structure of the NZF/Ub complex, establishing the determinants of Ub recognition, and testing the functional relevance of Ub/NZF interactions.
Results
Spectroscopic characterization of the Npl4 NZF/Ub complex
The 1:1 complex formed between Npl4 NZF and Ub was characterized spectroscopically and the structure of the complex was determined using NMR spectroscopy (see Figure 1A for sequences and secondary structures of the two proteins). Atomic absorption spectroscopy was used to confirm that the NZF domain contained a single zinc ion, and EXAFS analyses of the Npl4 NZF/Ub complex showed that the zinc was coordinated in a tetrahedral S4 coordination site, as previously observed for the uncomplexed Npl4 NZF domain (Wang et al, 2003) (see Experimental Procedures). Nearly complete resonance and stereospecific assignments for both Npl4 NZF and Ub were obtained using standard 1H/13C/15N heteronuclear NMR experiments, and intermolecular NOEs were identified in complementary half-filtered NOESY spectra from complexes containing a single labeled component (e.g., Figure 1B). The final structure of the NZF/Ub complex was calculated using a total of 2072 NOE restraints (109 intermolecular NOEs), and is of high quality, as reflected by the good agreement between the 20 lowest penalty structures (backbone atom rmsd=0.35 Å over NZF residues 5–30 and Ub residues 1–70), good Ramachandran statistics, a lack of restraint violations, and low final energies (see Table I and Figure 2A).
Figure 1.
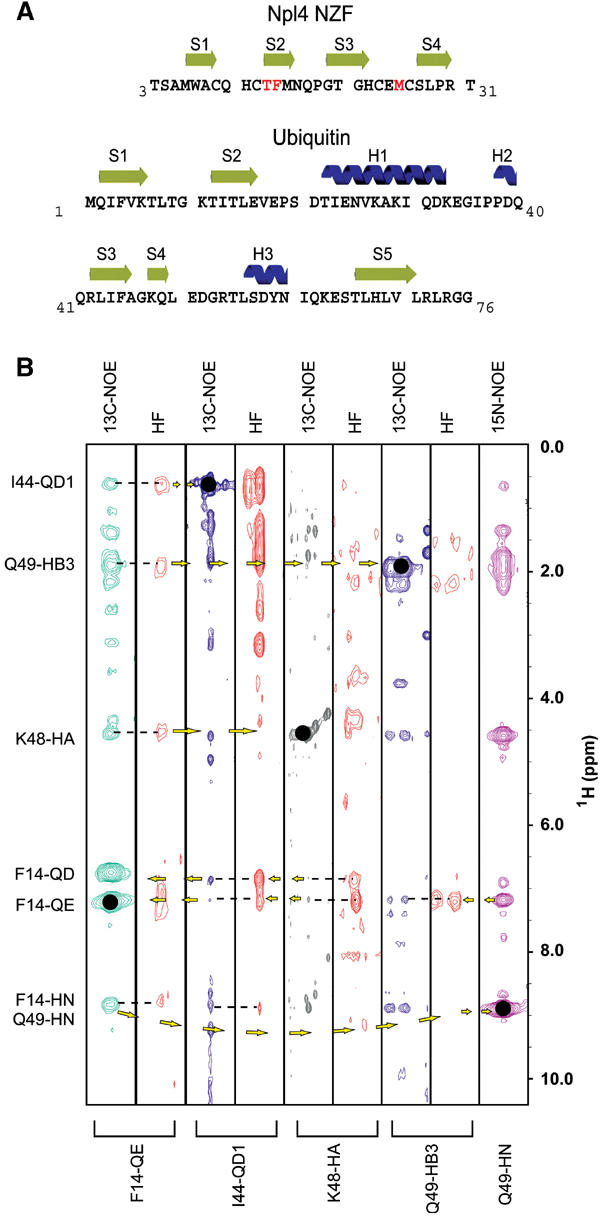
Secondary structures and intermolecular contacts in the Npl4 NZF/Ub complex. (A) Sequences, numbering schemes, and secondary structures of the rat Npl4 NZF and human Ub proteins. The Ub-binding TF/M motif of Npl4 NZF is highlighted in red. Note that rat and human Npl4 NZF proteins differ by a single S-to-A substitution at residue 4. (B) Matched pairs of unfiltered and half-filtered (HF) NOESY strips, aligned to illustrate reciprocal intermolecular NOE contacts between NZF residue Phe14 (strip 1) and Ub residues Ile44, Lys48, and Gln49 (strips 2–5). Resonances on the diagonal are denoted with solid black dots, and their identities are given below each strip. Dashes represent intermolecular NOEs, and arrows denote forward and return NOEs. The intermolecular NOEs from NZF F14 to Ub are labeled at left and the proton indirect chemical shift scale is shown at right (ppm).
Table 1.
Structure statistics for the Npl4 NZF/Ub complex
| 〈TAD〉a | 〈CNS〉a | |
|---|---|---|
| NOE distance restraintsb (Å) | 2072 | 2072 |
| Npl4 NZF | 478 | 478 |
| Ub | 1485 | 1485 |
| Intermolecular | 109 | 109 |
| Intraresidue | 400 | 400 |
| Sequential (∣i−j∣=1) | 532 | 532 |
| Medium range (2⩽∣i−j∣⩽5) | 388 | 388 |
| Long range (∣I−j∣>5) | 677 | 677 |
| Zinc coordination restraints | 14 | 14 |
| Hydrogen bond distance restraintsc (Å) | 104 | 104 |
| Hydrogen bondsd | 48(4) | 48(4) |
| Dihedral restraintsd | 112(33) | 112(33) |
| DYANA target function (Å2)e | 0.8±0.09 | NA |
| CNS energy (kcal/mol)e | ∼300f | 134.7±4.9 |
| Residual distance restraint violations | ||
| Number of violations ⩾0.1 Å | 0±0 | 0.0 |
| Sum of violations (Å or kcal/mol)e | 1.7±0.1 | 27.0±1.0 |
| Maximum violation (Å) | 0.14 | <0.1 |
| Residual dihedral restraint violations | ||
| Number of violations ⩾1° | NA | 0.0 |
| Sum of violations (deg or kcal/mol)e | NA | 1.29±0.8 |
| Maximum violation (deg) | NA | <2 |
| Van der Waals violations | ||
| Number ⩾0.1 Å | 0±0 | 0.0 |
| Sum of violations (Å or kcal/mol)e | 0.7±0.1 | 34.5±1.7 |
| Maximum violation (Å) | 0.09 | <0.1 |
| Ramachandran statistics (Ub)g | ||
| Favored | 87.3(94.6)% | 84.5(93.2)% |
| Allowed | 11.3(3.8)% | 14.4(5.1)% |
| Generously allowed | 1.3(1.5)% | 1.1(1.7)% |
| Disallowed | 0.0(0.0)% | 0.0(0.0)% |
| Ramachandran statistics (Npl4 NZF)g | ||
| Favored | 59.8% | 68.6% |
| Allowed | 34.5% | 31.4% |
| Generously allowed | 2.6% | 0.0% |
| Disallowed | 3.1% | 0.0% |
| rmsds from the average coordinatesh (Å) | ||
| Secondary elements (backbone, Ub)i | 0.23±0.06 | 0.29±0.08 |
| Secondary elements (heavy atoms, Ub) | 0.85±0.13 | 0.82±0.12 |
| Backbone (complex)j | 0.31±0.07 | 0.35±0.09 |
| Heavy atoms (complex) |
0.92±0.12 |
0.92±0.12 |
| a〈TAD〉 is the ensemble of 40 lowest-penalty structures calculated using the program DYANA. 〈CNS〉 is the same ensemble after 1000 steps (15 ps each) of simulated annealing at 25 K, 1000 slow-cooling steps to 0 K, and 10 000 steps of restrained Powell minimization in Cartesian space (anneal.inp protocol). | ||
| bOnly meaningful and nonredundant restraints as determined by the DYANA CALIBA function. | ||
| cTwo upper-limit distance restraints were used to define each hydrogen bond. | ||
| dHydrogen-bonding and dihedral restraints are reported separately for Ub (no parentheses) and NZF (parentheses). | ||
| eViolation energies from DYANA have units of (Å2) or deg, while energies from CNS are in kcal/mol. | ||
| fEnergies for structures input into CNS (from DYANA) were estimated within the generate_easy.inp program after the first regularization without restraints. | ||
| gDetermined using PROCHECK-NMR. | ||
| hSuperposition and overall rmsds were calculated using the program MOLMOL and PROCHECK_NMR. | ||
| iSecondary elements in Ubiquitin (2–6, 12–16, 22–35, 37–39, 41–45, 48–49, 56–60, and 66–71). | ||
| jAll nondisordered residues (NZF: 5–30; Ub: 1–71). | ||
| NA: not applicable. | ||
Figure 2.
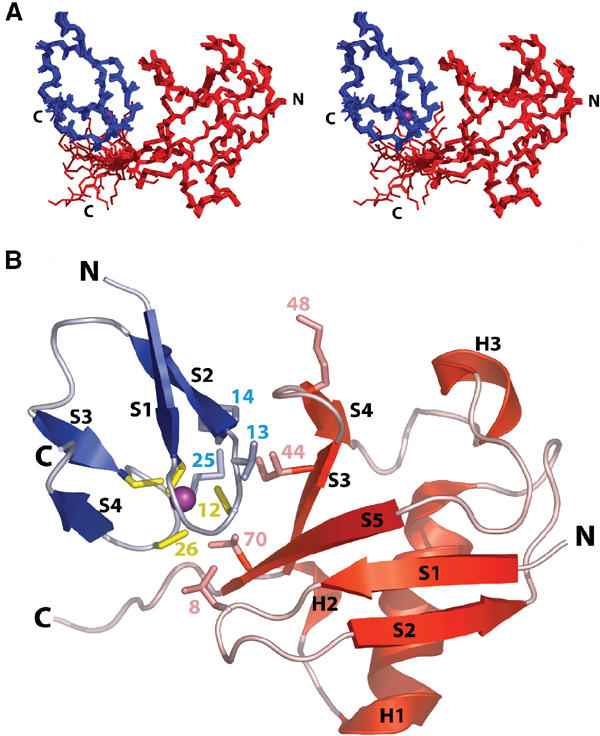
Structure of the Npl4 NZF/Ub complex. (A) Stereoview showing an overlay of the 20 lowest penalty complexes of Npl4 NZF and Ub. Ub is shown in red, the NZF domain is shown in blue, and the Zn2+ ion in purple. (B) Ribbon diagram of the Npl4 NZF/Ub complex. Color coding is the same as in (A), with the secondary structural elements in both proteins labeled, and key interface residues from both proteins shown explicitly. The Ub Lys48 residue is also shown to help orient the reader.
Structure of the Npl4 NZF/Ub complex
The structure of the Npl4 NZF/Ub complex is shown in Figures 2 and 3. Neither the Npl4 NZF domain nor Ub changes structure dramatically upon complex formation, although slight adjustments are evident in the flexible loops of both proteins. Npl4 NZF is composed of four antiparallel strands organized about a rubredoxin-like Zn(Cys)4 metal-binding site (Wang et al, 2003) (Figure 2B). Ordered backbone heavy atoms in the free and complexed Npl4 NZF proteins overlay with an rmsd of 0.78 Å (residues 5–30), with the largest changes occurring in the loop that connects strands 3 and 4 (Wang et al, 2003). Ub forms a five-stranded mixed β-sheet with helices between strands 2/3 and 4/5, and the structures of free and complexed Ub also overlay well (rmsd=0.71 Å for the heavy atoms in residues 1–71; Vijay-Kumar et al, 1985). The biggest changes occur in the strand 3/4 loop (e.g., at Lys48), which is pulled away from the body of Ub and closer to the NZF domain. Several residues in the C-terminal tail of Ub also increase in order as judged by changes in their order parameters (see Supplementary figure).
Figure 3.
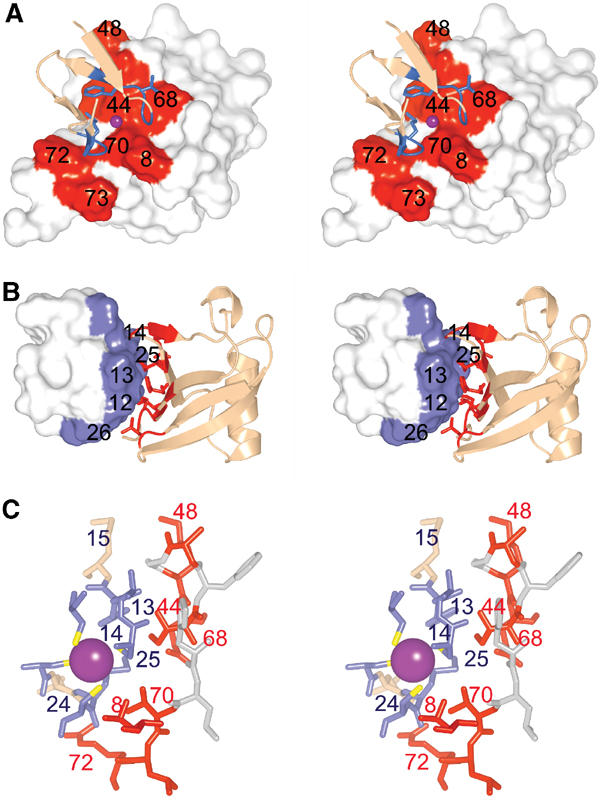
Npl4 NZF/Ub interaction surfaces. (A) Stereoview of Npl4 NZF (ribbon) bound to Ub (surface). Ub residue numbers are shown. (B) Stereoview of Ub (ribbon) bound to Npl4 NZF (surface). NZF residue numbers are shown. (C) Expanded stereoview showing the key interface residues (numbered) from Npl4 NZF (blue) and Ub (red).
The two metal-binding loops of Npl4 NZF form ‘rubredoxin knuckles' and contact the hydrophobic Ub surface defined by the three C-terminal β-strands and the strand 1/2 loop (the ‘I44' surface). The NZF zinc ion is centered between Ub residues Ile44 and Val70 (Figures 2B and 3). The core of the interface is hydrophobic on both sides, and includes Ub residues Leu8, Ile44, Val70 and Leu73, which interact with NZF residues Cys12, Phe14, Met25, and Cys26. The hydrophilic side chains of several other Ub residues (Arg42, Lys48, Glu49, His68, and Arg72) form a ring of hydrophilic interactions that surrounds the core of the interface.
In the complex, the two knuckles of NZF clamp over a ridge formed by the Ub Val70 and Leu8 side chains (Figure 3A). The C-terminal edge of the first knuckle (residues 13–15) crosses a bulge formed by Ile44, with NZF residues Thr13 and Phe14 straddling the Ile44 side chain. Loop residues of the second NZF knuckle (Met25 and Cys26) fill a cleft lined by Ub residues Val70, Arg72, and Leu73. To complete the interface, the tip of the Ub strand 3/4 loop (including Lys48) contacts the β-strands of the first NZF knuckle. NZF residues Thr13 and Phe14 appear to play the most important roles in Ub recognition. Specifically, the carbonyl oxygen of Thr13 makes an intermolecular hydrogen bond with the Ub Gly47 amide backbone, the Thr13 hydroxyl group points toward the face of the imidazole ring of Ub His68, where it may hydrogen bond with one of the ring nitrogens, and the Thr13 methyl group makes a close contact with the Ub Ala46 methyl group. The NZF Phe14 ring makes intramolecular packing interactions with Met25 and Glu24, and buttresses one side of the Ub Ile44 side chain.
Determinants of Ub recognition
To help characterize the determinants of Ub/NZF recognition, biosensor experiments were used to quantitate the Ub binding of five different representative NZF domains (termed Npl4, Vps36-1, Vps36-2, TAB2, and RanBP2). The NZF sequences and Ub dissociation constants are provided in Table II, together with a brief description of each protein (see footnote a). Previous studies have established that some, but not all, NZF domains can bind free Ub (Meyer et al, 2002; Wang et al, 2003), and this was again confirmed by our experiments, which showed that Npl4, Vps36-1, and TAB2 all bound Ub with dissociation constants between 100 and 400 μM, whereas RanBP2 and Vps36-2 did not bind Ub significantly (Kd>5000 μM). Although these binding affinities are modest, most other Ub recognition domains also bind Ub with micromolar dissociation constants (Raiborg et al, 2002; Shekhtman and Cowburn, 2002; Fisher et al, 2003; Kang et al, 2003; McKenna et al, 2003). It is therefore likely that these domains cooperate with other recognition elements to achieve the affinities and specificities necessary for biological function.
Table 2.
Ub-binding affinities of different NZF domains
| NZF domainsa | NZF sequenceb | Kd (μM)c |
|---|---|---|
| Npl4 (wt) | 580TSAMWACQHCTFMNQPGTGHCEMCSLPRT | 126±26d |
| Npl4 (T13L,F14V) | 580TSAMWACQHCLVMNQPGTGHCEMCSLPRT | >5000e |
| Vps36-1 (wt) | 177VNSENICPACTFANHPQIGNCEICGHRLP | 182±19d |
| Vps36-1 (T13G) | 177VNSENICPACGFANHPQIGNCEICGHRLP | 1900(100)e |
| Vps36-1 (T13G,F14S) | 177VNSENICPACGSANHPQIGNCEICGHRLP | >5000e |
| TAB2 (wt) | 664EGAQWNCTACTFLNHPALIRCEQCEMPRH | 338(1)f |
| VPS36-2 (wt) | 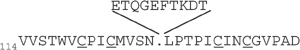 |
>5000e |
| RanBP2 (wt) | 1479KEGQWDCSACLVQNEGSSTKCAACQNPRK | >5000e |
| RanBP2 (L13T,V14F) | 1479KEGQWDCSACTFQNEGSSTKCAACQNPRK | 976(3)f |
| RanBP2 (L13T, V14F,A25M) | 1479KEGQWDCSACTFQNEGSSTKCAMCQNPRK | 92(2)f |
| RanBP2 (L13T,V14F,A24E,A25M) |
1479KEGQWDCSACTFQNEGSSTKCEMCQNPRK |
85±2g |
| aNpl4 (nuclear protein localization gene 4; NCBI NP_542144) forms a heterodimer with Ufd1 that recognizes ubiquitylated target proteins and acts as a cofactor for Cdc48/p97 in the processes of ERAD, RUP, and nuclear envelope closure (Hetzer et al, 2001; Meyer et al, 2002; Tsai et al, 2002). The metazoan Npl4 proteins have a single NZF domain, which is also found in some but not all fungal Npl4 proteins. | ||
| Vps36 (vacuolar protein sorting protein 36; NCBI NP_013521) is a component of the yeast ESCRT-II complex and is required for the sorting of ubiquitylated protein cargos into the multivesicular body (Babst et al, 2002; Katzmann et al, 2002). Yeast Vps36 contains two NZF domains, one of which binds Ub (Vps36-1) and the other which does not (Vps36-2). Note that these domains are not conserved in the mammalian EAP45/Vps36 protein. | ||
| TAB2 (TAK1-binding protein 2; NCBI NP_055908) is a subunit of the heterotrimeric TRIKA2 kinase complex, a Ub-dependent MKK and IKK kinase involved in IL-1 receptor signaling (Wang et al, 2001). The mammalian and amphibian proteins have a single NZF domain. | ||
| RanBP2 (Ran-binding protein 2/Nup358; NCBI NM_006267) is a Ran-GDP-binding subunit of the nuclear pore complex (Nakielny et al, 1999; Yaseen and Blobel, 1999). This NZF domain corresponds to the third of eight NZF domains in human RanBP2. | ||
| bSubscripts denote the starting position of each NZF domain within the relevant protein. Underlined residues coordinate zinc, and residues mutated from the wt sequence are shown in bold. | ||
| cKd values were determined in BIACORE biosensor measurements of soluble Ub binding to immobilized GST-NZF fusion proteins at 20°C in a buffer of 25 mM Tris–HCl pH 7.0, 2 μM ZnCl2, 0.005% P20, 50 μg/ml bovine serum albumin, and 2 mM DTT. Note that several of these measurements were reported previously (Wang et al, 2003), but are also included here for completeness. | ||
| dDissociation constant is the mean of four or more independent measurements and the error is the standard deviation in the measurements. | ||
| eExtrapolated from less than 50% saturation binding at 1.5 mM free Ub protein concentration. | ||
| fDissociation constant and error were estimated from a statistical fit of a single binding isotherm derived from triplicate measurements at seven different Ub concentrations (2.1–1500 μM). Values in parentheses are statistical errors estimated from the fit of the data to a simple 1:1 binding model. | ||
| gDissociation constant is the mean of two independent measurements and the error is the range in the two measurements. | ||
Notably, all three Ub-binding NZF domains contained the 13Thr-Phe14 dipeptide sequence that makes critical contacts in the Npl4 NZF/Ub complex, whereas the two nonbinding NZF domains lacked this dipeptide sequence. Moreover, mutation of the 13Thr-Phe14 sequences of Npl4 NZF (to Leu-Val) reduced Ub binding to undetectable levels, demonstrating the importance of this sequence motif for Ub recognition. More generally, our structural and binding studies indicated that NZF domains may form a structurally defined scaffold that can orient as few as five key residues in the appropriate geometry for Ub binding (see Figure 3C). Two of these residues are conserved metal-binding ligands (Cys12 and Cys26), two are the key variable residues displayed in the first zinc-binding knuckle (Thr13 and Phe14), and one is a hydrophobic amino acid displayed in the second knuckle (Met25 in Npl4 NZF, although other hydrophobic aliphatic residues (Φ) can occur at position 25 in other Ub-binding NZF domains). Thus, we suggest that the 13TF14/Φ25 motif might serve as the ‘signature' for the subset of NZF domains that bind Ub.
To test the validity of this simplified model for Ub recognition, we attempted to introduce Ub-binding activity into the NZF domain of RanBP2. Importantly, the wild–type (wt) RanBP2 NZF domain does not bind Ub, and exhibits little sequence similarity to the Ub-binding NZF domains of Npl4, Vps36-1, and TAB2, beyond the six conserved structural residues that identify it as an NZF domain (see Table II and Figure 4A). As shown in Figure 4, a minimal L13T,V14F substitution in the RanBP2 NZF introduced weak, but significant, Ub-binding activity (Kd=973 μM). A further Ala25 to Met substitution increased Ub-binding affinity more than 10-fold, to a level that was actually slightly tighter than the wt Npl4 and Vps36-1 NZF domains (Kd=92 μM). Ub-binding activity was not enhanced further by an Ala24 to Glu substitution (Kd=85 μM), indicating that although the Glu24 side chain of Npl4 NZF contacts Ub, it contributes little to the overall binding energy. These data demonstrate that the 13TF14/M25 surface can support Ub binding when transplanted onto a highly diverged NZF domain, indicating that the three-dimensional structure of different NZF cores is conserved and can be used to display a small number of variable loop residues for specific ligand binding.
Figure 4.
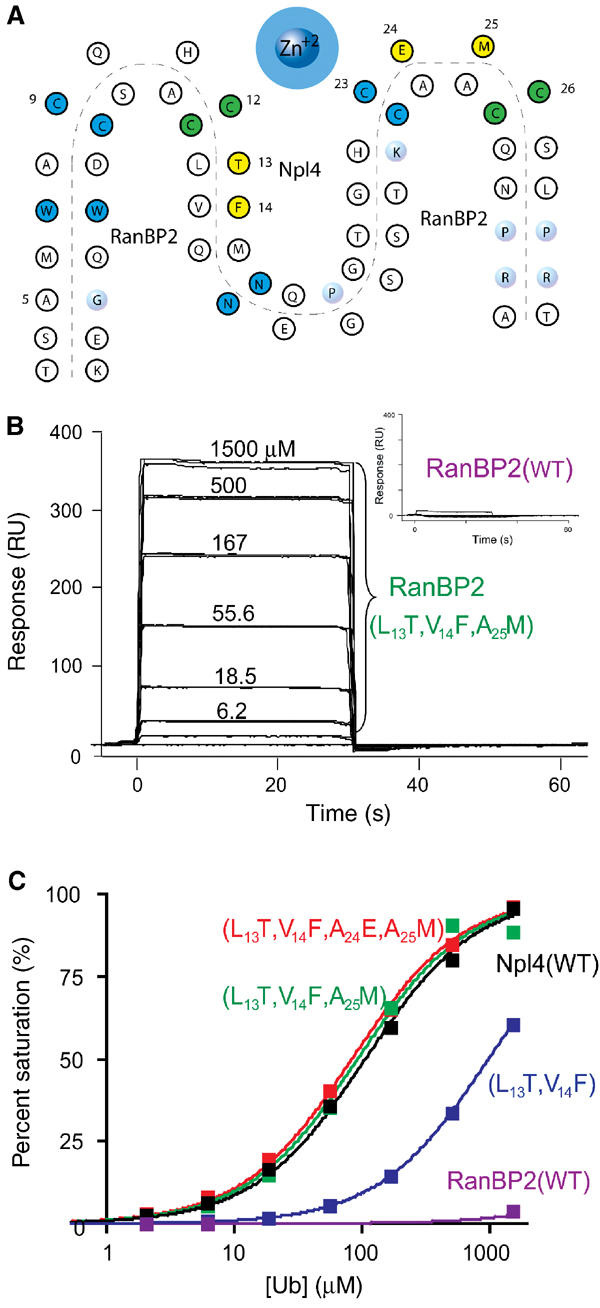
Transferability of the NZF 13TF14/M25 Ub-binding motif. (A) Schematic alignment of the Npl4 and RanBP2 NZF sequences. Dark blue: highly conserved NZF residues (>50%); light blue: moderately conserved NZF residues (>20%); yellow: residues that contact Ub; and green: residues that are both highly conserved and contact Ub. NZF residue conservation was defined as in Wang et al (2003). (B) Ub binding by wt (inset) and mutant RanBP2 (L13T,V14F,A25M) NZF domains. Ub was injected in triplicate at concentrations of 0–1500 μM over GST-RanBP2 NZF proteins captured on anti-GST surfaces. Note that Ub binding by the wt RanBP2 NZF domain (inset) was negligible, even at 1500 μM Ub. (C) Binding isotherms for the NZF domains of Npl4 (positive control, black), wt RanBP2 (negative control, purple), RanBP2 (L13T,V14F) (blue), RanBP2 (L13T,V14F,A25M) (green), and RanBP2 (L13T,V14F,A24E,A25M) (red). Kd values are given in Table II.
Biological relevance of NZF/Ub interactions
The biological relevance of Ub binding by the 13TF14/Φ25 class of NZF domains has not yet been established in any system. We therefore tested the functional consequences of mutating the Vps36-1 NZF domain on vacuolar protein sorting in yeast. Vacuolar protein sorting is an attractive system for such studies because the sorting of membranes and membrane-bound proteins like carboxypeptidase S (CPS) into the lumen of the yeast vacuole is a Ub-dependent process (Katzmann et al, 2002), and is closely related to the Ub-dependent release of many enveloped RNA viruses, including HIV (Pornillos et al, 2002). Yeast vacuolar protein sorting requires the sequential action of a series of soluble protein complexes, termed the Hrs/Vps27 complex, ESCRT-I, -II, and -III (endosomal sorting complexes required for transport). Vps36 is an essential subunit of the yeast ESCRT-II complex (Babst et al, 2002; Katzmann et al, 2002).
As noted above, one of the two NZF domains of Vps36 (Vps36-1) contains a 13TF14/Φ25 motif and binds Ub (Kd=182 μM), whereas the other NZF domain (Vps36-2) lacks this motif and does not bind Ub (Table II). As expected, point mutations in the 13TF14 dipeptide of the isolated Vps36-1 NZF motif reduced the Ub-binding affinity substantially (Table II and Figure 5). In particular, the T13G,F14S double mutation reduced binding more than 30-fold (although Ub binding was not completely abolished by this mutation).
Figure 5.

Ub binding by Vps36 NZF domains. (A) Surface plasmon resonance biosensor analyses of the wt Vps36-1 NZF/Ub interaction. Ub was injected in triplicate at concentrations of 0–1500 μM over GST-Vps36-1 NZF captured on an anti-GST surface. The inset depicts the much weaker binding responses obtained for the same concentrations of Ub injected over a mutant GST-Vps36-1 (13TF14 to 13GS14) captured on an anti-GST surface. (B) Isotherms for Ub binding to wt and mutant NZF domains from Vps36p: wt Vps36-1 NZF (black), Vps36-1 (T13G) NZF mutant (red), Vps36-1 (13TF14 to 13GS14) NZF mutant (blue), and wt Vps36-2 NZF (green). Full sequences of the NZF domains and estimated dissociation constants are given in Table II.
We next tested whether the intact yeast ESCRT-II complex exhibited Ub-binding activity. A vps36 allele expressing a Myc-tagged protein was integrated in place of wt vps36 at the normal chromosomal locus, and the Myc-tagged protein fully complemented the activity of the wt Vps36 protein. Detergent-solubilized extracts were prepared from yeast cells expressing the Myc-tagged Vps36 protein, and incubated with either GST or Ub-GST beads. Unbound proteins were washed away, and bound proteins were analyzed by Western blotting with anti-Myc antiserum. As shown in Figure 6A, the wt Vps36 (ESCRT-II) bound to Ub-GST, but not to GST alone, demonstrating that yeast Vps36/ESCRT-II exhibits Ub-binding activity. The Vsp36-1 NZF domain was responsible for most, if not all, of the Ub-binding activity because a mutation in the Vps36-1 NZF TF dipeptide motif (Vps36T187G,F188S) reduced Vps36 binding significantly (∼10-fold; see Figure 6A). The low levels of residual binding seen in this experiment likely reflected the fact that this mutation does not eliminate Ub binding entirely (see Figure 5B). The composition and stability of the ESCRT-II complex was unaffected by the Vps36T187F,G188S mutation, as demonstrated in control immunoprecipitations in which a second ESCRT-II subunit (Vps22) was detected. These control experiments also showed that both wt and mutant Vps36 bound Ub as a component of the intact ESCRT-II protein complex (data not shown). To test whether the Ub binding was direct, the ESCRT-II complex was also expressed in Escherichia coli and then tested for Ub-binding activity. As shown in Figure 6B, ESCRT-II (as detected by Vps22-HA) bound to Ub-GST, but not to GST alone. Taken together, these studies demonstrated that the intact ESCRT-II complex can bind directly to Ub, and that the Vps36-1 NZF domain of Vps36 plays an important role in this interaction.
Figure 6.
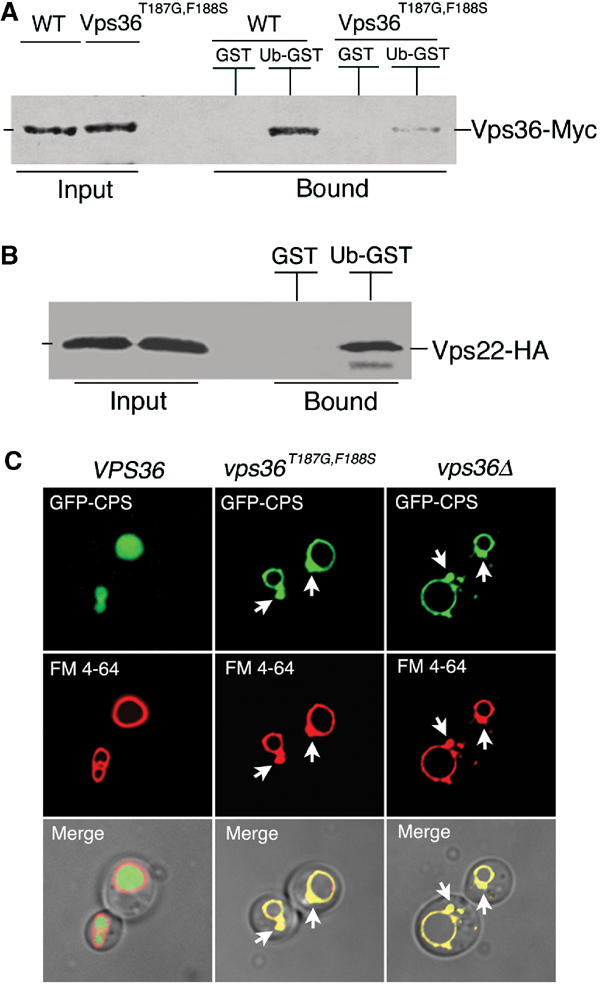
A Vps36 NZF mutant is defective in Ub binding and MVB sorting. (A) Extracts were prepared from vps36Δ(MBY30) yeast cells expressing the indicated VPS36 alleles and incubated with immobilized GST (negative control) or Ub-GST. Bound, Myc-tagged Vps36 proteins were visualized by Western blotting with anti-Myc antisera. (B) Extracts were prepared from E. coli expressing all three ESCRT-II genes (pMB202) and incubated with immobilized GST (negative control) or Ub-GST. Bound, HA-tagged Vps22 protein (ESCRT-II complex) was visualized by Western blotting with anti-HA antisera. (C) Normarski optics (bottom panels) and fluorescence localization of GFP-CPS (top panels) and FM 4-64 (middle panels) in vps36Δ cells expressing either wt Vps36, Vps36T187G,F186S, or no Vps36 (vps36Δ) at 37°C. Class E compartments are indicated by arrowheads.
We next tested the functional consequences of the Vps36T187G,F188S mutation on vacuolar protein sorting. Wild type and mutant Vps36 proteins were expressed in vps36Δ cells and the multivesicular body (MVB) sorting of a GFP-CPS fusion reporter protein was examined using fluorescence microscopy (Odorizzi et al, 1998). As expected, GFP-CPS was delivered to the lumen of the vacuole via the MVB pathway in cells expressing the wt Vps36 protein and the distribution of GFP did not overlap with staining of the vacuolar membrane by FM 4-64. (Figure 6C, left column of panels). In cells lacking Vps36 (vps36Δ), however, GFP-CPS was mislocalized to the limiting membrane of the vacuole and to aberrant endosomal structures (Figure 6C, right column). Importantly, cells expressing Vps36T187G,F188S were also defective in GFP-CPS sorting (Figure 6C, middle column). GFP-CPS colocalized with FM 4-64 on the limiting membrane of the vacuole and in the class E compartment (prevacuolar endosome), similar to vps36Δ cells. This defect was exaggerated at higher temperatures, even though control experiments demonstrated that the mutant ESCRT-II complex was stable at all temperatures tested (not shown). To quantify the class E phenotype, 50 cells of each genotype were evaluated by their fluorescence staining. As expected, 96% of the wt cells displayed lumenal GFP-CPS staining, whereas 92% of the vps36Δ cells and 94% of the Vps36T187G,F188S-expressing cells, respectively, displayed vacuolar membrane and class E compartment staining. None of the mutant cells exhibited CPS staining of the vacuolar lumen. Thus, we conclude that the Ub-binding activity of the Vps36 NZF domain is required for efficient sorting of ubiquitylated substrates into the MVB.
Discussion
Protein ubiquitylation is a remarkably common, yet complex post-translational modification that helps regulate a broad variety of biological processes. For example, more than 1000 different yeast proteins appear to be covalently linked either to single Ub molecules (monoubiquitylation) or to one of seven different types of polyubiquitin chains (Peng et al, 2003). The diversity and prevalence of protein ubiquitylation suggests that cells must also have a variety of different Ub sensors. Indeed, a number of structurally distinct Ub-binding modules have now been identified, including the UIM (ubiquitin interacting motif), the UBA (ubiquitin-associated domain), the UEV (ubiquitin E2 variant/UBC-like domain), the CUE (coupling of ubiquitin conjugation to ER degradation domain), and the NZF domain (Hofmann and Bucher, 1996; Hofmann and Falquet, 2001; Buchberger, 2002; Katzmann et al, 2002; Madura, 2002; Lima, 2003).
In the NZF Npl4/Ub complex, the zinc-binding knuckles of the NZF domain interact with the hydrophobic I44 surface of Ub. This surface is essential for yeast viability (Sloper-Mould et al, 2001), and has emerged as the most important recognition site for Ub-binding proteins. High-resolution structures of Ub in complex with the CUE (Kang et al, 2003; Prag et al, 2003), UIM (Fujiwara et al, 2003; Mueller and Feigon, 2003; Swanson et al, 2003), UEV (Sundquist et al, 2004), and NZF recognition domains have now been determined, and in every case Ub binds its partner through the I44 surface. The binding of acceptor Ub to the Mms2/Ubc13 complex also involves the I44 surface of Ub (McKenna et al, 2001). The recognition domains themselves adopt very different folds, however, as the CUE and UIM folds are helical, the E2/UEV domains have mixed α/β structures, and the NZF domain is a four-stranded zinc-binding module.
Our studies demonstrate that NZF domains can bind Ub using as few as five key residues that are clustered about the zinc-binding site (Cys12, Thr13, Phe14, Φ25, and Cys26). The two cysteine residues coordinate zinc and are found in all NZF domains, whereas the 13TF14/Φ25 motif is unique to the specialized subset of NZF domains that bind Ub. Remarkably, we find that Ub-binding activity is created when the 13TF14/Φ25 motif is transplanted into a nonbinding NZF domain from RanBP2, which implies that different NZF domains probably adopt very similar core structures and can be used to display different clusters of variable loop residues that recognize different protein surfaces. This appears to occur naturally, as several pairs of residues covary strongly at positions 13 and 14, creating distinct subsets of NZF domains (Wang et al, 2003). The two most striking examples are (1) 13TF14-type NZF domains, which bind Ub, and (2) 13LV14-type NZF domains, which are present in multiple copies in different nuclear pore proteins including RanBP2, and can bind the GDP form of Ran (Nakielny et al, 1999; Yaseen and Blobel, 1999) and exportin 1 (Singh et al, 1999).
The splicing factor ZNF265 also contains two NZF domains, and one of these has been shown to bind RNA (Plambeck et al, 2003). This ZNF265 NZF domain again adopts the same overall fold, but in this case displays a series of basic residues that are used for RNA binding. It appears, however, that the NZF/RNA interface is more complex than the NZF/Ub surface, in part because the first zinc-binding loop of ZNF265 NZF is expanded relative to the first knuckle of Npl4 NZF (Plambeck et al, 2003). The variety of natural applications in which the conserved NZF core structure is used to display variable surface residues for ligand binding suggests that NZF domains would also be useful scaffolds for use in protein design and selection experiments.
Finally, our experiments demonstrate that the yeast ESCRT-II complex can bind Ub through one of the two NZF domains of Vps36, and reveal that this activity is required for the proper sorting of CPS into the lumen of the vacuole. The vacuolar protein sorting pathway represents a particularly striking use of Ub because both cargoes and components of the Vps machinery itself are ubiquitylated (Katzmann et al, 2002; Raiborg and Stenmark, 2002), and at least four distinct proteins or complexes within the pathway have now been shown to bind Ub: Vps9 (Katzmann et al, 2002; Davies et al, 2003; Donaldson et al, 2003; Shih et al, 2003), HRS/Vps27/STAM (HRS complex) (Polo et al, 2002; Raiborg et al, 2002; Shih et al, 2002), TSG101/Vps23 (ESCRT-I complex) (Garrus et al, 2001; Katzmann et al, 2001), and Vps36 (ESCRT-II complex). The HRS, ESCRT-I, and ESCRT-II complexes are recruited sequentially to the endosomal membrane, where they play essential roles in helping to sort ubiquitylated proteins into vesicles that bud into the late endosome. All three complexes recognize the I44 surface of Ub, suggesting sequential (or competitive) binding to this essential surface as protein cargoes are sorted. Our studies reveal precisely how the versatile NZF fold has been adapted to recognize Ub in the yeast vacuolar protein pathway, and indicate that similar NZF/Ub interactions will also occur in other important biological pathways.
Experimental procedures
Protein expression and purification
Expression constructs for wt and mutant NZF domains from rat Npl4, yeast Vps36p, human TAB2, and human RanBP2 were created by cloning annealed oligonucleotides into the BamH1/Xho1 sites of pGEX4T (Amersham Pharmacia), the NdeI/BamHI sites of a modified pGEX2T vector (WISP01-69), or by QuickChange™ mutagenesis of wt constructs (Stratagene). These vectors expressed GST-NZF fusion proteins with either thrombin (pGEX4T) or TEV (WISP01-69) cleavage sites between the GST and NZF domains.
Unlabeled, 15N-labeled, and 13C/15N-labeled human Ub and NZF domains were expressed in BL21(DE3) E. coli and purified as described previously (Wang et al, 2003). These procedures typically yielded 4–5 mg of pure NZF domain and 40 mg Ub per liter culture. Purified NZF proteins were characterized using SDS–PAGE (GST-NZF), N-terminal sequencing and MALDI mass spectrometry (NZF alone). NZF domains were aligned and numbered following previous conventions (Meyer et al, 2002; Wang et al, 2003) (see Table II and Figures 1A and 4A).
Zn and EXAFS analyses
Unbound zinc was removed from the Npl4 NZF domain prior to atomic absorption and EXAFS analyses by gel filtration (Superdex-75) in a degassed buffer lacking zinc (20 mM potassium phosphate pH 5.5, 150 mM NaCl, and 2 mM DTT). The stoichiometry of zinc:mole Npl4 NZF (0.93) was obtained by determining the protein concentration using optical spectroscopy and the zinc concentration by flame atomic absorption spectroscopy (Wang et al, 2003). XAFS spectra of the 1:1 Npl4 NZF/Ub complex (with excess Ub present) were collected and analyzed as described previously for the Npl4 NZF domain alone (Wang et al, 2003). The best-fit model to the transformed EXAFS spectrum yielded a zinc coordination sphere of four sulfur atoms (n=4.33), and an average Zn–S bond length of 2.33 Å, with a Debye–Waller factor of 0.0043 Å. These data are consistent with a slightly distorted tetrahedral geometry about Zn2+ and are very similar to data for the free Npl4 NZF domain (Wang et al, 2003).
NMR spectroscopy
Sample preparation. Samples for structure determinations were either 1.0 mM in labeled Npl4 NZF and 2.0 mM in unlabeled Ub, or 1.0 mM in labeled Ub and 2.0 mM unlabeled NZF in NMR buffer (20 mM sodium phosphate, 50 mM NaCl, 2 mM D10-DTT, 10 μM ZnCl2, pH 5.5 in 90% H2O/10% D2O). Samples were degassed and flame-sealed under argon in NMR sample tubes to reduce cysteine oxidation.
Data collection and resonance assignments
All NMR spectra were collected at 18°C on a Varian Inova 600 MHz spectrometer equipped with a triple-resonance 1H/13C/15N probe and z-axis pulsed field gradient capability. The Npl4 NZF/Ub complex was in fast exchange, as is typical for complexes with dissociation constants in the 100 μM range. Backbone and side-chain assignments were made using a standard suite of triple-resonance experiments as described previously (Wang et al, 2003), with 2D versions of the HNCACB (Wittekind, 1993) and HN(CA)CO (Yamazaki et al, 1994) experiments used for NZF assignments and 3D versions of these experiments used for Ub assigments. Side-chain assignments were completed using 3D-H(CCO)NH-TOCSY, 3D-(H)C(CCO)NH-TOCSY (Grzesiek et al, 1993), and 3D-15N-edited TOCSY experiments. Aromatic resonances were assigned using a combination of 1H,13C HSQC and 13C-edited NOESY experiments centered on the aromatic carbon resonances (125 ppm), together with a 13C-edited NOESY experiment centered on the aliphatic region. Stereospecific assignments for β-methylene protons (and estimation of χ1 dihedral angles) were accomplished using a combination of HNHB, HN(CO)HB (Bax et al, 1994b), 15N-edited TOCSY, and NOESY data (Powers et al, 1993). Stereospecific assignments of side-chain methyl groups and qualitative determination of χ1 and χ2 dihedral propensities were evaluated using long-range carbon–carbon and carbon–proton couplings observed in LRCC (Bax et al, 1994a) and LRCH (Vuister et al, 1993) experiments. 3D 15N-edited NOESY-HSQC (Zhang et al, 1994; Mori et al, 1995) and 3D 13C-edited NOESY-HSQCs (Muhandiram et al, 1993; Pascal et al, 1994) (100 ms mixing times) were used to generate distance restraints. Interdomain NOEs between NZF and Ub were identified using half-filtered NOESY experiments (Ikura and Bax, 1992; Ogura et al, 1996; Zwahlen et al, 1997; Ferentz and Wagner, 2000) (e.g., see Figure 1B). Three-bond coupling constants (3JHNHA) were obtained from a 3D HNHA experiment (Kuboniwa et al, 1994) and φ/ψ dihedral restraints were derived from backbone chemical shift evaluation within the program TALOS (Cornilescu et al, 1999). All spectra were processed in FELIX 97 (MSI), and referenced indirectly to DSS (Wishart et al, 1995).
Structure determination. Backbone and side-chain correlations were assigned and NOE intensities were integrated using tools in SPARKY (TD Goddard and DG Kneller, University of California, San Francisco). The solution structure for the Npl4 NZF/Ub complex was refined using torsion angle dynamics (TAD) in DYANA and CYANA (Guntert et al, 1997; Herrmann et al, 2002) and regularized in CNS (Brunger et al, 1998) through a gentle simulated annealing with 2072 interproton NOE restraints (including 109 intermolecular NOEs), 104 hydrogen-bonding restraints, and 145 total dihedral restraints (φ, ψ, χ1, and χ2; see Table I for details). Initial rounds of refinement included only NOE data to define the general fold of the two proteins and their relative juxtaposition, and to determine the stereochemistry about the coordinated zinc ion. Final refinements added restraints for zinc–sulfur distances, cysteine-Sγ-to-Sγ distances, Zn-to-Cβ distances (total 14), hydrogen-bonding restraints, 3JHNHA coupling constant data, and dihedral restraints. Structures were analyzed using PROCHECK-NMR (Laskowski et al, 1996), INSIGHT II (MSI) (Table I), and the validation programs supplied at the PDB deposition site (http://deposit.pdb.org/adit/). All figures were created with PYMOL (DeLano Scientific).
Ub binding
Binding of purified Ub to GST-NZF peptides captured on anti-GST antibody surfaces was detected using a Biacore biosensor, and the equilibrium data were fit to simple 1:1 interaction models to obtain dissociation constants (Garrus et al, 2001). All measurements were performed at 20°C in 25 mM Tris–HCl pH 7.0, 2 mM DTT, and 10 μM ZnCl2.
Plasmid construction and yeast strains
The VPS36 sequence was amplified from genomic DNA and cloned into pRS414 to create pMB131. The T187G, F188S mutations were introduced into pMB131 by Quickchange™ mutagenesis (Stratagene) to create pSJ001. DNA encoding a Myc epitope was introduced just before the stop codon in the chromosomal copy of VPS36 by the method of Longtine et al (1998). VPS36-Myc sequence was then amplified from genomic DNA. The SphI/SacII-digested PCR product was ligated with SphI/SacII-digested pMB131 and pSJ001 vector, respectively, to create pSJ004 and pSJ008. pGO45 (GFP-CPS) has been described (Odorizzi et al, 1998).
The following yeast strains were used: SEY6210 (MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901lys2-801 suc2-Δ9) (Wurmser and Emr, 1998); MBY30 (SEY6210; vps36Δ1(VPS36::HIS3)) (Babst et al, 2002).
In vitro Ub-binding studies
Preparation of GST sepharose affinity matrices. Sepharose matrices were prepared according to the manufacturer's instructions (Amersham Biosciences). In brief, glutathione S-transferase (GST) and Ub-GST were expressed in E. coli and purified by affinity chromatography on glutathione agarose. Reduced glutathione used to elute the proteins was removed by a PD-10 desalting column, and equivalent amounts of the pure proteins were covalently linked to cyanogen bromide-activated sepharose.
Yeast extract preparation. Cellular extracts were prepared essentially as described previously (Katzmann et al, 2001). In brief, 100 OD600 units of cells were lysed with glass beads in lysis buffer (20 mM Hepes–KOH, pH 6.8, 50 mM potassium acetate, 2 mM EDTA plus protease inhibitors: 1:50 diluted complete, EDTA-free (Roche), 1 μg/ml pepstatin A, 1 μg/ml chymostatin, and 1 mM AEBSF) plus 5 mM N-ethylmaleimide (NEM) plus 0.5% Triton X-100.
E. coli sample preparation. The ESCRT-II complex was expressed in BL21(DE3)pLysS (Novagen) from plasmid pMB202, which contained all three ESCRT-II genes (VPS22, VPS25, and VPS36) (Babst et al, 2002). Protein expression was induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 1 h at 37°C (OD600=0.8). The soluble cytoplasmic fraction was isolated with BugBuster Protein Extraction Reagent according to the manufacturer's instructions (Novagen). In brief, cells from 25 ml cultures were harvested and lysed in 2.5 ml of BugBuster plus the same protease inhibitors as used for the yeast samples (see above).
In vitro Ub-binding assays. Yeast or E. coli cell lysates were cleared at 20 000 g and passed over immobilized GST or Ub-GST. For yeast samples, the sepharose affinity matrices were washed three times with Hepes lysis buffer plus 0.5% Triton X-100, once with Hepes lysis buffer plus 0.05% Triton X-100, and once with Hepes lysis buffer alone. For E. coli samples, the sepharose affinity matrices were washed three times with BugBuster, once with Hepes lysis buffer plus 0.05% Triton X-100, and once with Hepes lysis buffer alone. Bound proteins were eluted with 0.5 M (pH 3.4) acetate buffer, TCA precipitated, acetone washed, and resolubilized in SDS sample buffer. Yeast samples for the input (1 OD600 equivalent of cell extract) and bound fractions (20 OD600 equivalent of cell extract) were subjected to SDS–PAGE and Western blotting with anti-Myc antisera. E. coli samples for the input (0.25 ml equivalent of cell culture) and bound fractions (5 ml equivalent of cell culture) were subjected to SDS–PAGE and Western blotting with anti-HA antisera.
Microscopy
Living yeast cells expressing GFP-CPS were harvested at an OD600 of 0.4–0.6 and labeled with FM 4-64 at 37°C as described (Vida and Emr, 1995). GFP and FM 4-64 fluorescence was visualized using a fluorescence microscope (Axiovert S1002TV; Carl Zeiss MicroImaging, Inc.) equipped with FITC and rhodamine filters, captured with a digital camera (Photometrix), and deconvolved using Delta Vision software (Applied Precision Inc.). GFP and FM 4-64 images were merged using Adobe Photoshop 7.0 (Adobe Systems, Inc.). Results presented were based on observations of >120 cells.
Supplementary Material
Supplementary Materials
Acknowledgments
We thank Rebecca Rich and David Myszka for BIACore biosensor analyses, Bob Schackmann for N-terminal protein sequencing, the SLAC staff and Tim Stemmler for EXAFS data collection, Eric Ross for computer support, and Chris Hill for critical reading of the manuscript. WIS and DRD are supported by grants from the NIH, HHM was supported by grants from the NIH and the Human frontiers science project to Graham Warren, and this work was also partially supported by grant CA58689 from the NIH (to SDE) and by the Howard Hughes Medical Institute. SDE is an investigator of the Howard Hughes Medical Institute. The Utah Biomolecular NMR Facility is supported by grants from the NIH and NSF. Structure coordinates and chemical shifts have been deposited in the PDB database (1Q5W).
References
- Babst M, Katzmann D, Snyder W, Wendland B, Emr S (2002) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 [DOI] [PubMed] [Google Scholar]
- Bax A, Delaglio F, Grzesiek S, Vuister GW (1994a) Resonance assignment of methionine methyl groups and c3 angular information from long-range proton–carbon and carbon–carbon J correlation in a calmodulin–peptide complex. J Biomol NMR 4: 787–797 [DOI] [PubMed] [Google Scholar]
- Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G (1994b) Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol 239: 79–105 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Buchberger A (2002) From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol 12: 216–221 [DOI] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115: 355–367 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Davies BA, Topp JD, Sfeir AJ, Katzmann DJ, Carney DS, Tall GG, Friedberg AS, Deng L, Chen Z, Horazdovsky BF (2003) Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J Biol Chem 278: 19826–19833 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 4: 491–497 [DOI] [PubMed] [Google Scholar]
- Donaldson KM, Yin H, Gekakis N, Supek F, Joazeiro CA (2003) Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr Biol 13: 258–262 [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferentz AE, Wagner G (2000) NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys 33: 29–65 [DOI] [PubMed] [Google Scholar]
- Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem 278: 28976–28984 [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M (2003) Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: 55–65 [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Anglister J, Bax A (1993) Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of carbon-13 magnetization. J Magn Reson B 101: 114–119 [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319: 209–227 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 3: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci 21: 172–173 [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26: 347–350 [DOI] [PubMed] [Google Scholar]
- Ikura M, Bax A (1992) Isotope-filtered 2D NMR of a protein–peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J Am Chem Soc 114: 2433–2440 [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I (2003) Solution structure of a CUE–ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113: 621–630 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905 [DOI] [PubMed] [Google Scholar]
- Kuboniwa H, Grzesiek S, Delaglio F, Bax A (1994) Measurement of HN-H alpha J couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J Biomol NMR 4: 871–878 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lima CD (2003) CUE'd up for monoubiquitin. Cell 113: 554–556 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Madura K (2002) The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle 1: 235–244 [PubMed] [Google Scholar]
- McKenna S, Hu J, Moraes T, Xiao W, Ellison MJ, Spyracopoulos L (2003) Energetics and specificity of interactions within Ub.Uev.Ubc13 human ubiquitin conjugation complexes. Biochemistry 42: 7922–7930 [DOI] [PubMed] [Google Scholar]
- McKenna S, Spyracopoulos L, Moraes T, Pastushok L, Ptak C, Xiao W, Ellison MJ (2001) Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J Biol Chem 276: 40120–40126 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Abeygunawardana C, Johnson MO, van Zijl PC (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B 108: 94–98 [DOI] [PubMed] [Google Scholar]
- Mueller TD, Feigon J (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J 22: 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhandiram DR, Xy GY, Kay LE (1993) An enhanced-sensitivity pure absorption gradient 4D 15N, 13C-edited NOESY experiment. J Biomol NMR 3: 463–470 [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 18: 1982–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Ogura K, Terasawa H, Inagaki F (1996) Fully 13C-refocused multidimensional 13C-edited pulse schemes using broadband shaped inversion and refocusing pulses. J Magn Reson B 112: 63–68 [DOI] [PubMed] [Google Scholar]
- Pascal SM, Muhandiram DR, Yamazaki T, Forman-Kay JD, Kay LE (1994) Simultaneous acquisition of 15N- and 13C-edited NOE spectra of proteins dissolved in H2O. J Magn Reson B 103: 197–201 [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Plambeck CA, Kwan AH, Adams DJ, Westman BJ, van der Weyden L, Medcalf RL, Morris BJ, Mackay JP (2003) The structure of the zinc finger domain from human splicing factor ZNF265 fold. J Biol Chem 278: 22805–22811 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Pornillos OP, Garrus JE, Sundquist WI (2002) Mechanisms of enveloped RNA virus budding. Trends Cell Biol 12: 569–579 [DOI] [PubMed] [Google Scholar]
- Powers R, Garrett DS, March CJ, Frieden EA, Gronenborn AM, Clore GM (1993) The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry 32: 6744–6762 [DOI] [PubMed] [Google Scholar]
- Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH (2003) Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113: 609–620 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H (2002) Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell Struct Funct 27: 403–408 [DOI] [PubMed] [Google Scholar]
- Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Shekhtman A, Cowburn D (2002) A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochem Biophys Res Commun 296: 1222–1227 [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol 4: 389–393 [DOI] [PubMed] [Google Scholar]
- Shih SC, Prag G, Francis SA, Sutanto MA, Hurley JH, Hicke L (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J 22: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA (1999) The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 274: 37370–37378 [DOI] [PubMed] [Google Scholar]
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L (2001) Distinct functional surface regions on ubiquitin. J Biol Chem 276: 30483–30489 [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Schubert HL, Kelly B, Hill G, Hill CP (2004) Ubiquitin recognition by the human TSG101 protein. Mol Cell, in press [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I (2003) Solution structure of Vps27 UIM–ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J 22: 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA (2002) Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol 3: 246–255 [DOI] [PubMed] [Google Scholar]
- Varshavsky A, Turner G, Du F, Xie Y (2000) Felix Hoppe-Seyler Lecture 2000. The ubiquitin system and the N-end rule pathway. Biol Chem 381: 779–789 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ (1985) Three-dimensional structure of ubiquitin at 2.8 Å resolution. Proc Natl Acad Sci USA 82: 3582–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister GW, Yamazaki T, Torchia DA, Bax A (1993) Measurement of two- and three-bond 13C-1H J couplings to the C delta carbons of leucine residues in staphylococcal nuclease. J Biomol NMR 3: 297–306 [DOI] [PubMed] [Google Scholar]
- Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI (2003) Structure and ubiquitin interactions of the conserved zinc finger domain of npl4. J Biol Chem 278: 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6: 135–140 [DOI] [PubMed] [Google Scholar]
- Wittekind M (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide–proton and nitrogen resonances with the alpha and beta carbon resonances in proteins. J Magn Reson B 101: 201–205 [Google Scholar]
- Wurmser AE, Emr SD (1998) Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J 17: 4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Lee W, Revington M, Mattiello DL, Dahlquist FW, Arrowsmith CH, Kay LE (1994) An HNCA pulse scheme for the backbone assignment of 15N,13C,2H-labeled proteins: application to a 37-kDa Trp repressor–DNA complex. J Am Chem Soc 116: 6464–6465 [Google Scholar]
- Yaseen NR, Blobel G (1999) Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA 96: 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2003) Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang O, Kay LE, Olivier JP, Forman-Kay JD (1994) Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR 4: 845–858 [DOI] [PubMed] [Google Scholar]
- Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lamda N-peptide/boxB RNA complex. J Am Chem Soc 119: 6711–6721 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials


