Abstract
The hormonal action of abscisic acid (ABA) in plants is controlled by the precise balance between its biosynthesis and catabolism. In plants, ABA 8′-hydroxylation is thought to play a predominant role in ABA catabolism. ABA 8′-hydroxylase was shown to be a cytochrome P450 (P450); however, its corresponding gene had not been identified. Through phylogenetic and DNA microarray analyses during seed imbibition, the candidate genes for this enzyme were narrowed down from 272 Arabidopsis P450 genes. These candidate genes were functionally expressed in yeast to reveal that members of the CYP707A family, CYP707A1–CYP707A4, encode ABA 8′-hydroxylases. Expression analyses revealed that CYP707A2 is responsible for the rapid decrease in ABA level during seed imbibition. During drought stress conditions, all CYP707A genes were upregulated, and upon rehydration a significant increase in mRNA level was observed. Consistent with the expression analyses, cyp707a2 mutants exhibited hyperdormancy in seeds and accumulated six-fold greater ABA content than wild type. These results demonstrate that CYP707A family genes play a major regulatory role in controlling the level of ABA in plants.
Keywords: abscisic acid, Arabidopsis, catabolism, P450
Introduction
Abscisic acid (ABA) is a sesquiterpene phytohormone that controls numerous aspects of plant life cycle, including seed dormancy, germination and adaptive responses to environmental stresses (Zeevaart and Creelman, 1988). Endogenous ABA content is the determinant of these physiological processes and therefore ABA-deficient mutants exhibit reduced seed dormancy and wiltiness (McCarty, 1995). ABA content increases during seed development and when a plant is subjected to various stresses such as dehydration. Conversely, ABA content rapidly decreases during seed germination and when the plant is recovering from stress. ABA content in plants is determined by the balance between its biosynthesis and catabolism. When high levels of ABA are maintained, both ABA biosynthesis and catabolism are active. Consequently, ABA catabolite levels continue to increase (Harrison and Walton, 1975; Zeevaart, 1980; Pierce and Raschke, 1981). Accordingly, constitutive expression of an ABA biosynthetic gene in transgenic plants leads to higher accumulation of the catabolites compared to the moderate increase observed in ABA content (Qin and Zeevaart, 2002). In recent years, considerable progress has been made in the identification and characterization of ABA biosynthetic genes (Seo and Koshiba, 2002; Schwartz et al, 2003). However, the molecular mechanisms underlying ABA catabolism are still poorly understood.
ABA is catabolized into inactive forms either by oxidation or conjugation (Figure 1) (Milborrow, 1969, 1975; Walton and Sondheimer, 1972; Sondheimer et al, 1974; Xu et al, 2002; see the review Cutler and Krochko, 1999). The oxidative pathway is predominant in ABA catabolism and it is triggered by hydroxylation at C-8′ to produce 8′-hydroxy ABA. The 8′-hydroxy ABA is then spontaneously isomerized to form phaseic acid (PA) (Cutler and Krochko, 1999). Therefore, the major regulatory step during inactivation is likely to be the 8′-hydroxylation of ABA to PA. This reaction has been shown to be catalyzed by a cytochrome P450 monooxygenase (P450) (Gillard and Walton, 1976; Krochko et al, 1998). However, despite many reports that describe the kinetics of PA accumulation before and after stress treatments, the gene encoding ABA 8′-hydroxylase is yet to be identified. The identification of this gene is necessary to gain a more complete understanding of the molecular mechanisms controlling ABA levels during various stages of the plant life cycle.
Figure 1.
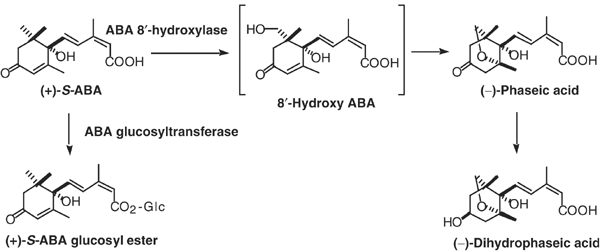
Catabolic pathway of ABA in plants.
P450s constitute a large family of enzymes that catalyze the oxidation of various low-molecular weight compounds. In mammals, P450s play a central role in drug metabolism and have been a major focus of research in the pharmaceutical field. In plants, P450s participate in numerous metabolic processes including phytohormone biosynthesis and catabolism, and secondary metabolism (Schuler, 1996; Chapple, 1998). Completion of Arabidopsis genome sequencing revealed at least 272 P450 genes (Werck-Reichhart et al, 2002). More recently, completion of rice genome sequencing revealed approximately 450 P450 genes (http://drnelson.utmem.edu/rice.html). These numbers reflect the diverse evolution of P450 genes and their important role in the plant life cycle. The genome-sequencing efforts were successful in generating vast amounts of sequence information; however, it remains a major challenge to deduce the function of each gene. Elucidation of gene function will be a major task in the post genomic era and will require novel approaches in addition to the systematic analysis of genes. P450s are especially challenging, because in most cases the substrate of the enzyme cannot be easily predicted. Furthermore, the number of steps in metabolic pathways that P450s participate in is largely unknown.
We have attempted to identify the gene encoding ABA8′-hydroxylase in Arabidopsis from the 272 P450 genes that were revealed by genome sequencing. Identification of the gene encoding ABA 8′-hydroxylase could facilitate the fine-tuning of ABA levels in plants and ultimately lead to improved drought tolerance and/or the prevention of precocious germination in crops. Such modifications would have an enormous impact on the agricultural industry. Our extensive and systematic prediction has led to the first successful identification of CYP707A family members as ABA 8′-hydroxylase genes. Expression and genetic analyses demonstrated that CYP707A genes play a regulatory role in vivo in defining ABA levels during seed imbibition and drought stress conditions.
Results
Phylogenetic and comparative analyses of Arabidopsis P450s
In Arabidopsis, the 272 P450 genes identified have been grouped into A type and non-A type (Durst and Nelson, 1995). The A type represents a more plant-specific branch, whereas the non-A type appears more closely related to P450s from other organisms. These genes are further divided into smaller families according to the CYP number. The two sequences will share the same CYP number when more than 40% identity is seen, while further classification into subfamilies is based on the sequence identity of more than 55% (Schuler and Werck-Reichhart, 2003). In Arabidopsis there are 45 CYP families, within which the number of genes varies considerably. CYP71 and CYP705 are the largest families in Arabidopsis and contain 54 and 33 genes, respectively. Initially, we concentrated on P450s that are involved in phytohormone pathways, as the location of these genes in the phylogenetic tree should point us towards the gene encoding ABA 8′-hydroxylase.
In Arabidopsis, 25 P450s have been functionally identified to date (Werck-Reichhart et al, 2002; Schuler and Werck-Reichhart 2003; see Supplementary data 1, available at The EMBO Journal Online). Among these, 12 have been identified, which participate in phytohormone biosynthesis and catabolism. Interestingly, with the exception of CYP701A3 and CYP79B2/B3 (Werck-Reichhart et al, 2002), all of these genes belong to the non-A-type P450 branch, although CYP701A3 is located in the upper half of the phylogenetic tree, close to non-A-type genes (Supplementary data 2). In contrast, the A-type branch contains many P450s that participate in secondary metabolism and defense-related processes (Supplementary data 1). Moreover, their substrates appear to be mostly aromatic compounds, such as flavonoids and indole derivatives. Therefore, it is possible that ABA 8′-hydroxylase is also located in the non-A-type branch of P450s.
P450s that act on terpenoid substrates are mainly located in the non-A-type branch, except for several CYP71s, which are involved in terpenoid phytoalexin biosynthesis. This is further supported by the fact that two P450s involved in biosynthesis of diterpenoid taxol, taxane 10β-hydroxylase (Schoendorf et al, 2001) and 13α-hydroxylase (Jennewein et al, 2001), show the highest degree of similarity to the non-A-type families CYP716 and CYP718. Furthermore, the mammalian retinoic acid (RA) catabolizing P450, CYP26 (Fujii et al, 1997), also shows the highest degree of similarity to the same families (Supplementary data 2). Both RA and ABA share some degree of similarity in terms of their chemical structure and biosynthesis (Kushiro et al, 2003). This similarity is particularly interesting and may provide a clue as to the nature of the ABA 8′-hydroxylase gene.
In addition, we took advantage of the information from the recently completed rice genome sequencing project. By comparing the P450s from Arabidopsis and rice, it was found that some families are present in one, but not the other species (Supplementary data 3). Clearly, an essential gene that functions in a phytohormone pathway should be highly conserved in both plant species. Furthermore, all of the P450s involved in phytohormone pathways belong to small families that typically contain only one or two members.
DNA microarray analysis on P450s during seed imbibition
With the aforementioned predictions taken into account and the notion that ABA 8′-hydroxylase plays a major role in seed imbibition, we conducted DNA microarray analyses on Arabidopsis P450 genes during seed imbibition using Affymetrix GeneChip ATH1 GenomeArrays (Supplementary data 4). During seed imbibition, the ABA 8′-hydroxylase gene is expected to be highly expressed, causing inactivation of ABA, leading to breaking of seed dormancy. Accordingly, we have observed that the level of ABA in seed decreased dramatically following imbibition and reached the basal level after 12 h (see below).
Of the 272 Arabidopsis P450 genes, 217 were represented on the microarray and their expression data were analyzed comparatively. Among the potential candidate genes we identified in the non-A-type branch, CYP97A3, CYP97B3, CYP97C1, CYP704A2, CYP707A1, CYP707A2, CYP707A3, CYP709B2, CYP714A1 and CYP721A1 were all significantly expressed during seed imbibition, indicating that they are potential candidate genes for ABA 8′-hydroxylase (Supplementary data 3). However, the mRNA for ABA 8′-hydroxylase should also be present in dry seed in order to homeostatically maintain the appropriate ABA levels. This would correlate with our detection of mRNA in dry seed for ABA biosynthetic enzymes such as zeaxanthin epoxidase, nine-cis-epoxycarotenoid dioxygenases (NCEDs) and abscisic aldehyde oxidase 3 (data not shown). Moreover, the level of ABA decreased dramatically immediately after imbibition and reached half the normal level within about 6 h (see below). The target P450 gene would also be expected to follow this trend. By applying these lines of reasoning, CYP709B2 and CYP721A1, whose mRNA was not detected in dry seed but was highly abundant following imbibition, were eliminated as potential target genes. Therefore, these data led us to propose that CYP97A3, CYP97B3, CYP97C1, CYP704A2, CYP707A1, CYP707A2, CYP707A3 and CYP714A1 are the most likely candidate P450s for the ABA 8′-hydroxylase gene.
Functional expression of candidate P450 genes in yeast
Full-length cDNAs for CYP97A3, CYP97C1, CYP707A1, CYP707A3 and CYP714A1, which were available at the RIKEN Bio Resource Center (BRC) (Seki et al, 2002), were functionally expressed in yeast. The yeast expression system has been used successfully in the past to express functionally several plant P450s (Urban et al, 1997; Bishop et al, 1999). The microsomal fractions of the transformants were used for in vitro enzyme assays where they were incubated with 0.5 mM NADPH and 38 μM (+)-S-ABA at 22°C, and analyzed by HPLC and GC-MS. The expression of CYP97A3, CYP97C1 and CYP714A1 failed to produce a peak other than that of the starting material, (+)-S-ABA. Conversely, expression of CYP707A1 and CYP707A3 under identical conditions produced a peak that was equal in retention time with authentic PA (Figure 2A). The peak was not seen in the plasmid only control. These samples were further analyzed by GC-MS after methylation to confirm that the compound corresponding to the peak was indeed PA (Figure 2B). [1,2-13C2]-(±)-ABA (Asami et al, 1999) was incubated with the microsomal fraction from CYP707A1- and CYP707A3-expressing yeast. The product exhibited +2 mass unit increase in molecular ion peak at m/z 296 upon GC-MS analysis, further confirming that the product obtained was PA (data not shown). The production of PA was not seen when NADPH was omitted from the incubation mixture (data not shown). In our assay system, we were unable to detect other peaks that could correspond to 8′-hydroxy ABA (a direct oxidation product of the P450 reaction), which then spontaneously cyclizes to PA. Furthermore, we were also unable to detect 7′-hydroxy ABA (a known minor catabolite) (Hampson et al, 1992).
Figure 2.
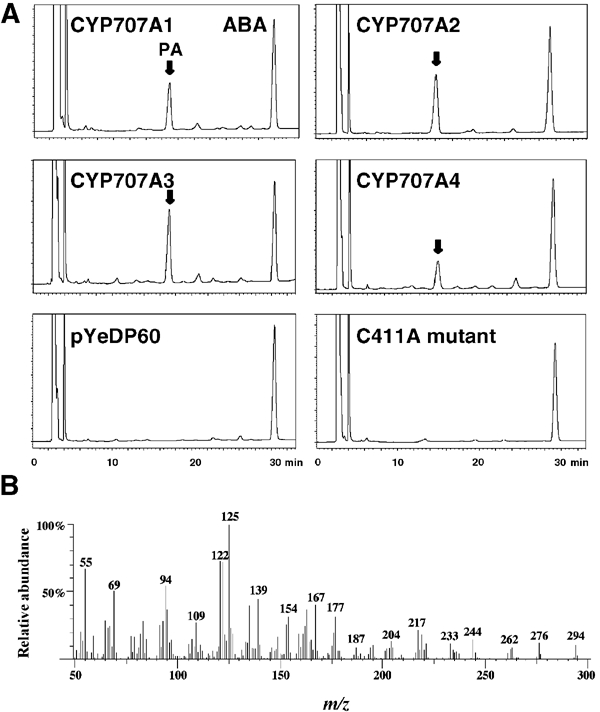
Functional expression of CYP707A genes in yeast. (A) HPLC profiles of reaction products on incubating (+)-S-ABA with 2 μg of microsomal protein. Retention time is given in min, while the vertical axis indicates UV absorbance at 254 nm. The positions of authentic ABA and PA (with an arrow) under the same conditions are labeled. (B) Mass spectra of the reaction product from (+)-S-ABA by CYP707A1.
To test whether the remaining two members of the CYP707A family would also catalyze the same reaction, CYP707A2 and CYP707A4 cDNAs were prepared from dry seeds and siliques, respectively. Under identical conditions, both clones produced a peak corresponding to PA as shown in Figure 2A and, similarly, production of PA was confirmed by GC-MS (data not shown).
(−)-R-ABA, an enantiomer of natural (+)-S-ABA, exhibits ABA-like activity and has previously been shown to be slowly metabolized into minor catabolites in plant cells and cell-free systems (Balsevich et al, 1994a, 1994b). We were interested in observing whether the cloned CYP707A family would catalyze the conversion of (−)-R-ABA into any hydroxylated products. However, we failed to detect products of (−)-R-ABA from four of these clones. Balsevich et al (1994a) reported that in maize suspension cells 7′-hydroxylation was catalyzed at the cell surface, while ABA 8′-hydroxylase activity was located inside the cell. This indicates that minor catabolites produced in feeding experiments are likely to be formed by another enzyme rather than by ABA 8′-hydroxylase. Our results support the notion that ABA 8′-hydroxylase is unable to use (−)-R-ABA as a substrate. However, this cannot be ruled out, as it is possible that the levels of minor catabolites produced are below the detection limit of our system.
To study the biochemical properties of Arabidopsis ABA 8′-hydroxylase, the effect of an inhibitor of this enzyme was investigated on CYP707A1. Tetcyclacis, which was originally developed as an inhibitor of GA biosynthesis, has also been shown to inhibit ABA 8′-hydroxylase (Rademacher, 2000). This inhibitor was added to our microsomal preparation. The production of PA was significantly inhibited by the addition of 10 μM tetcyclacis, whereas 1 μM tetcyclacis reduced the amount of PA production to about 65% of the control nontreated sample (Figure 3). We also tested the effect of another P450 inhibitor metyrapone (Sato et al, 1978). In this case, 10 μM metyrapone failed to inhibit the production of PA (Figure 3), indicating that the enzyme-active site can discriminate between two different types of known P450 inhibitors.
Figure 3.
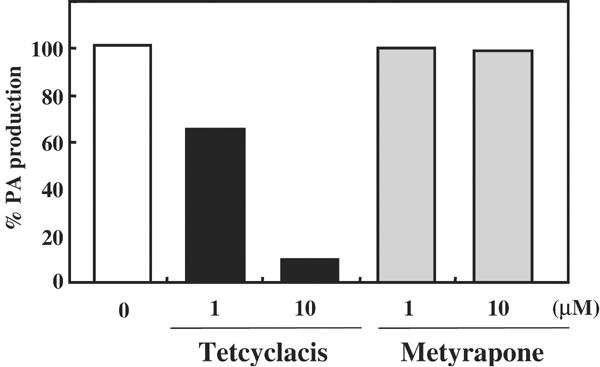
Effect of P450 inhibitors on ABA 8′-hydroxylase activity in vitro. The vertical axis shows the percentage of PA production compared to control in the absence of the inhibitor.
Finally, to confirm that our expressed CYP707A1 does indeed encode a P450, we mutated the highly conserved cysteine residue (PFGNGTHSCPG), which is the putative heme iron ligand. Cys411Ala mutation of CYP707A1 completely abolished the production of PA (Figure 2A). Therefore, in accordance with previous studies (Wu and Chung, 1991), this cysteine residue appears to be essential for catalysis.
Expression analysis of CYP707A genes during seed imbibition and drought stress conditions
To examine whether members of the CYP707A family have different regulatory roles during seed imbibition and drought stress conditions (where ABA is known to play a major role), the following expression analyses were conducted.
Initially, the expression of each gene was measured in different tissues by quantitative reverse transcription–PCR (QRT-PCR) (Figure 4). CYP707A genes were expressed in all tissues, although the relative abundance differed between these genes. In particular, CYP707A2 mRNA predominantly accumulated in dry seed. This observation was consistent with our previous microarray analysis. On the other hand, the highest expression level of CYP707A1 was seen in silique.
Figure 4.
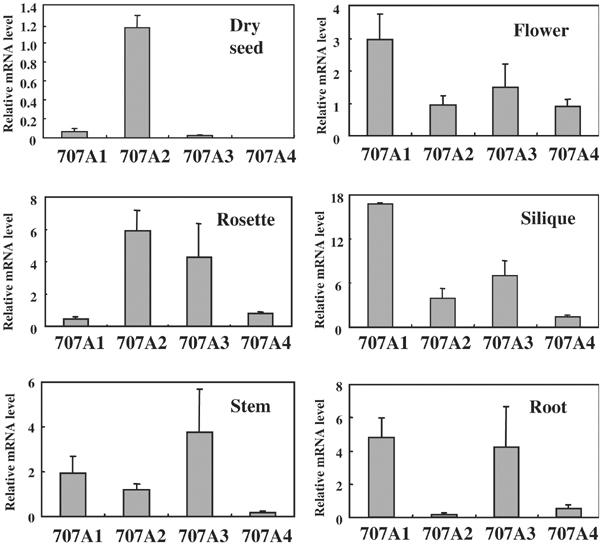
Expression of CYP707A genes among various tissues. QRT–PCR was performed using dry seed, rosette, stem, flower, silique and root. Transcript levels were normalized using 18S rRNA as an internal control. Results from triplicate samples in three independent experiments are shown with error bars.
It has been reported in several plant species that ABA levels decrease during seed imbibition (Yoshioka et al, 1998; Grappin et al, 2000; Jacobsen et al, 2002). To compare the changes in ABA levels to those in CYP707A gene expression during Arabidopsis seed imbibition, we measured the levels of ABA, PA and dihydrophaseic acid (DPA) in dry and imbibed seeds. As shown in Figure 5A, the level of ABA decreased immediately after seed imbibition, reaching the basal level after 12 h. This pattern is consistent with those reported in other plants, indicating that ABA catabolism is active during this period. In support of this observation, the total level of both PA and DPA increased following seed imbibition (Figure 5A), indicating that the major catabolic pathway operates through ABA 8′-hydroxylation. It should be noted that certain amounts of ABA and its catabolites leaked into the medium (see Supplementary data 5) and therefore, for accurate estimation of the catabolites, the level of both endogenous and medium leaked fraction was added.
Figure 5.
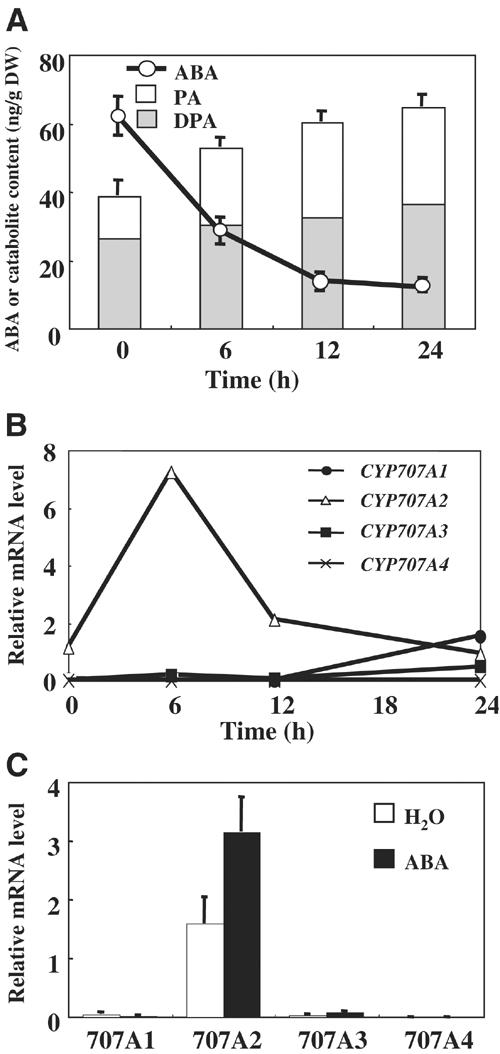
Changes in the level of ABA, PA and DPA, and expression of CYP707A genes during seed imbibition. (A) Quantification of total ABA, PA and DPA levels including both the endogenous and medium leaked fraction during seed imbibition. Levels of ABA, PA and DPA were measured by GC-MS from both seeds and medium, imbibed under continuous light at designated time points. An average from five independent experiments is shown with error bars. DW indicates dry weight. (B) The level of CYP707A transcripts during seed imbibition. QRT–PCR was performed using dry and imbibed seed at the designated time points. Transcript levels were normalized using 18S rRNA as an internal control. An average from duplicate experiments is shown. (C) Induction of CYP707A transcript levels by exogenous ABA (30 μM) at 12 h after seed imbibition. An average from three independent experiments is shown with error bars.
We then examined the temporal patterns of CYP707A gene expression during seed imbibition. QRT–PCR for each gene was carried out from seed samples collected at 0, 6, 12 and 24 h after imbibition (Figure 5B). CYP707A2 mRNA was highly abundant in dry seed and was upregulated immediately after imbibition, reaching a maximum at 6 h and decreasing thereafter. Furthermore, the expression of CYP707A2 was induced by addition of exogenous (+)-S-ABA (Figure 5C). This rapid increase in CYP707A2 mRNA levels correlated strongly with a sharp decrease in ABA levels, supporting the notion that this gene should be responsible for the rapid ABA catabolism during seed imbibition. On the other hand, CYP707A1 and CYP707A3 were upregulated after 12 h, while no significant expression was seen for CYP707A4 during this period.
It has been shown that the levels of ABA and its catabolites change dramatically in response to dehydration and subsequent rehydration (Pierce and Raschke, 1981). Therefore, we analyzed the temporal pattern of the levels of ABA and its catabolites, and CYP707A gene expression patterns during dehydration. In a separate experiment, after 6 h of drought treatment, the plantlet was rehydrated and the expression was further measured. The ABA level increased in response to dehydration and a sharp decrease in ABA level was observed immediately after the plant was rehydrated (Figure 6A). This dramatic decrease in ABA levels took place within 1 h, after which, they decreased gradually. PA and DPA levels increased similarly during dehydration; however, upon rehydration, a significant increase in PA level was observed (Figure 6A). The increase in PA levels correlated strongly with the sharp decrease in ABA levels.
Figure 6.
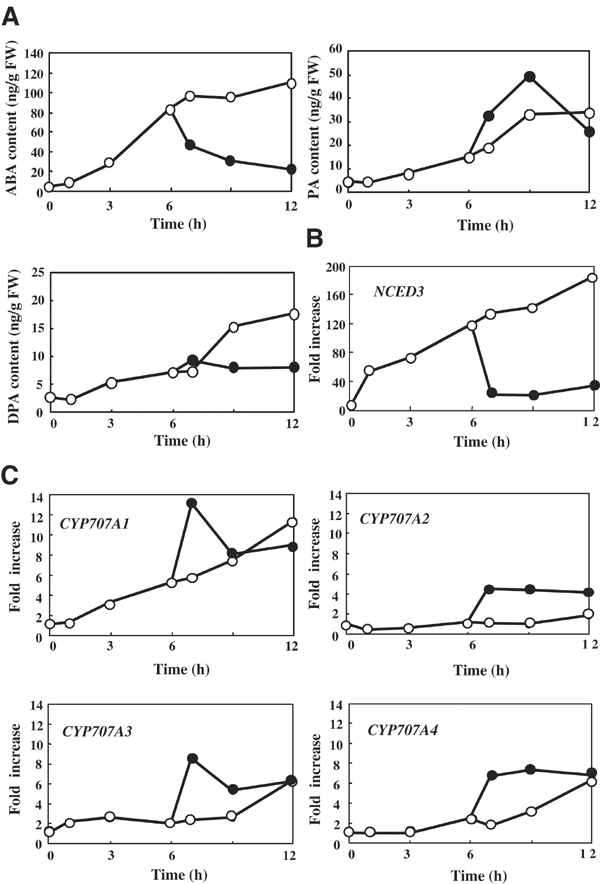
Changes in ABA, PA and DPA levels, and CYP707A gene expression during dehydration and rehydration. (A) Quantification of endogenous ABA, PA and DPA levels during dehydration and rehydration. Endogenous levels of these metabolites were measured by GC-MS from 2-week-old plants subjected to dehydration (open circle). Rehydrated plants were watered after 6 h of dehydration (filled circle). An average from at least duplicate samples in two independent experiments is shown. FW indicates fresh weight. (B) Induction of the NCED3 expression during dehydration and rehydration. QRT–PCR was performed using plants subjected to dehydration (open circle) and rehydration after 6 h of dehydration (filled circle) at the designated time points. Transcript levels were normalized using 18S rRNA as an internal control. Induction of NCED3 expression is indicated as fold increase. An average from duplicate experiments is shown. (C) Induction of CYP707A gene expression during dehydration and rehydration. Induction of CYP707A gene expression is indicated as fold increase.
The expression of CYP707A1 was gradually induced following drought conditions, while CYP707A2, CYP707A3 and CYP707A4 also showed moderate induction (Figure 6C). When water was supplied after 6 h, a dramatic increase in CYP707A1 mRNA levels was observed. Levels reached about 2.5-fold within 1 h and decreased sharply thereafter. CYP707A3 expression also exhibited a temporal pattern similar to that of CYP707A1. This expression pattern during rehydration is opposite to that of NCED3 expression (Iuchi et al, 2001; Figure 6B) and clearly demonstrates that ABA biosynthesis and catabolism are synergistically regulated to control the level of ABA during water stress conditions. Similarly, expression of CYP707A2 was induced significantly upon rehydration and the maximum expression level was maintained even after 6 h of rehydration. A similar pattern was also observed for CYP707A4.
We also tested the effect of exogenous (+)-S-ABA on expression of these genes in the vegetative plant. Plantlets (2-week old) were supplied with either water alone or 30 μM (+)-S-ABA. Again, the levels of PA and DPA, and the expression levels of CYP707A genes, were measured. A significant increase in PA level was observed upon ABA application, indicating that rapid catabolism of applied ABA had occurred, while DPA levels increased only moderately (Figure 7A). Significant induction of expression immediately after ABA application was observed for CYP707A1, while CYP707A2, CYP707A3 and CYP707A4 exhibited moderate inductions (Figure 7B). Induction of these genes was transient and decreased gradually thereafter.
Figure 7.
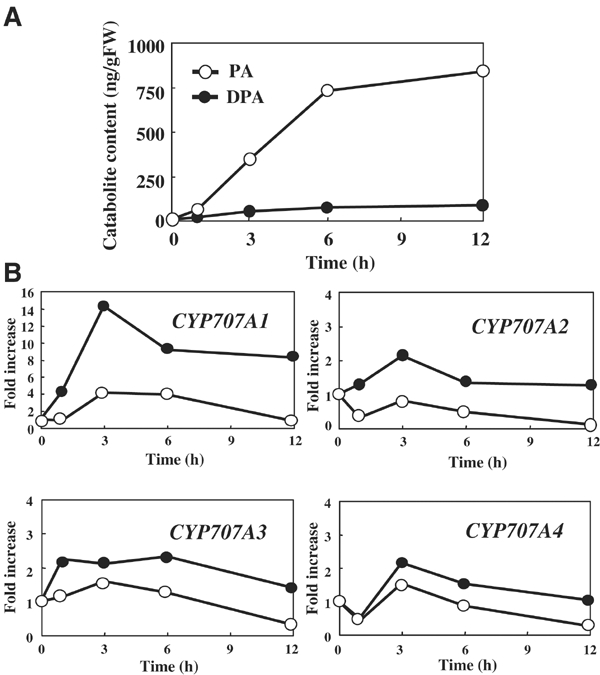
Changes in PA and DPA levels and induction of CYP707A gene expression upon ABA treatment. (A) Quantification of endogenous PA and DPA levels upon ABA treatment. (B) QRT–PCR was performed using plants subjected to 30 μM (+)-S-ABA (filled circle) or water control (open circle) at the designated time points. Transcript levels were normalized using 18S rRNA as an internal control. Induction of CYP707A gene expression is indicated as fold increase. An average from duplicate samples in two independent experiments is shown.
Phenotypic analysis of CYP707A knockout lines
In order to evaluate the in vivo function of CYP707A genes, we analyzed their corresponding insertion mutants. Our expression analyses demonstrated that multiple CYP707A genes are expressed in each tissue. Therefore, it is possible that the loss-of-function of a single CYP707A gene might not confer an apparent phenotype. However, CYP707A2 is expressed predominantly during the early stage of seed imbibition and loss-of-function of this gene could lead to a phenotype. Several insertion lines for CYP707A2 and CYP707A3 were available at the Arabidopsis Stock Center (ABRC) (Alonso et al, 2003), and we have produced and analyzed the homozygous lines (Figure 8A).
Figure 8.
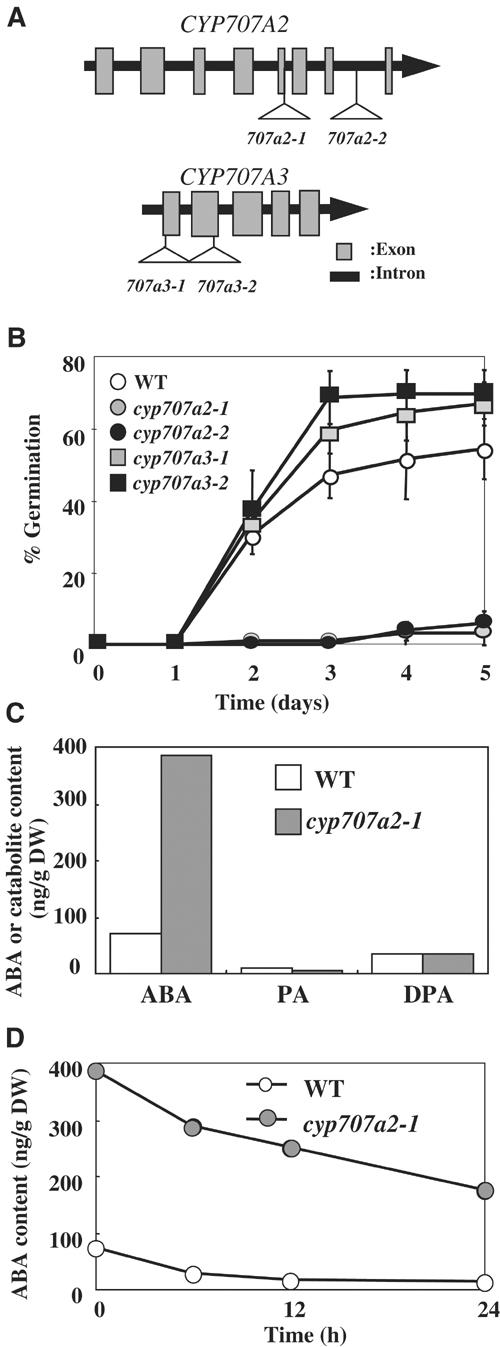
Phenotypic analysis of the cyp707a2 and cyp707a3 mutants. (A) T-DNA insertion in the CYP707A genes. The T-DNA insertion in the cyp707a2-1 mutant is located at the junction between the fifth exon and fifth intron, which disrupts the splicing site, while the cyp707a2-2 mutant contains the T-DNA at the seventh intron. The cyp707a3-1 and cyp707a3-2 mutants harbor the T-DNA at the first exon and second exon, respectively. (B) Dormancy of the cyp707a mutants. Freshly harvested seeds were sown on filter paper moistened with water. Radical emergence was scored as a criterion for germination. Triplicate experiments were performed and the averages are shown with error bars. Wild type, open circle; the cyp707a2-1 mutant, gray circle; the cyp707a2-2 mutant, filled circle; the cyp707a3-1 mutant, gray square; and the cyp707a3-2 mutant, filled square. (C) The endogenous level of ABA, PA and DPA in dry seeds of wild type and the cyp707a2-1 mutant. An average from duplicate experiments from independent samples is shown. (D) The change in the level of endogenous ABA in wild type and the cyp707a2-1 mutant during seed imbibition.
To assess whether CYP707A2 plays a predominant role in controlling seed dormancy and germination, we performed germination tests using wild type, cyp707a2 and cyp707a3 mutants. Seed for the cyp707a2-1 and cyp707a2-2 mutants exhibited significantly delayed germination when it was sown without stratification (Figure 8B). Hyperdormancy of the cyp707a2 mutants was rescued by stratification for 4 days (data not shown). Germination of the cyp707a2 mutants was inhibited by application of 0.5 μM ABA after stratification (data not shown). In contrast, seed for the cyp707a3-1 and cyp707a3-2 mutants germinated similarly to wild type, with or without stratification (Figure 8B). This is consistent with our gene expression analysis which showed CYP707A2 mRNA to be highly abundant in dry seeds, and suggests that it plays a major role in the rapid decrease in ABA content during the early stage of seed imbibition.
In contrast to seed dormancy, the cyp707a2 and cyp707a3 mutants display only subtle phenotypes during other developmental stages in both turgid and dehydrated plants. Once more, this is consistent with our gene expression analyses and indicates that the physiological role of the CYP707A is probably redundant.
Finally, the ABA content of the cyp707a2 mutant was measured during seed imbibition. As shown in Figure 8C, this mutant accumulated significant levels of ABA in dry seeds (six times that of wild type) and high levels of ABA were maintained even at 24 h after imbibition (Figure 8D). These data clearly demonstrate that CYP707A2 is the major ABA 8′-hydroxylase that determines ABA levels in seeds.
Discussion
Identification of the candidate gene for ABA 8′-hydroxylase
The work presented here has demonstrated that members of the Arabidopsis P450 CYP707A family, CYP707A1–CYP707A4, code for ABA 8′-hydroxylase. This enzyme plays a key role in ABA catabolism and in controlling ABA levels in various aspects of the plant life cycle. The gene encoding ABA 8′-hydroxylase has long been sought after. We have taken advantage of the available sequences and phylogenetic relationship for the 272 Arabidopsis P450 genes and narrowed down the number of possible candidates. These candidates were cloned and identified through biochemical analyses. In agreement with our prediction, CYP707A genes were located in the non-A-type branch, in which most of the P450s involved in phytohormone pathways are located. Moreover, the CYP707A family belongs to the same clade as the CYP88A family, which codes for the ent-kaurenoic acid oxidases involved in the early steps of GA biosynthesis (Werck-Reichhart et al, 2002). The CYP85 and CYP90 families from brassinosteroid (BR) biosynthesis are also closely related. Interestingly, many P450s that act on terpenoid substrates are also clustered in this area, and inhibitors of GA biosynthesis were additionally shown to cross-inhibit P450s involved in other phytohormone pathways. Tetcyclacis inhibits ABA 8′-hydroxylase, and paclobutrazol inhibits BR biosynthesis (Rademacher, 2000). These findings indicate that active site structures of these P450s share a high degree of similarity and may have an evolutionary relationship.
The phylogenetic tree representing the CYP707A family has rather long branches and a relatively simple branching pattern. These characteristics were important in evaluating the candidate gene, as essential P450s have presumably remained unchanged for a long period of time.
Phylogenetic analysis has placed the mammalian RA-catabolizing enzyme CYP26 (Fujii et al, 1997), close to the CYP707A family (Supplementary data 2). There is a close resemblance between mammalian RA and ABA in terms of structure and biosynthesis. For example, mammalian β,β-carotene-15,15′-dioxygenase that cleaves a carotenoid substrate to produce RA was cloned through sequence analogy to the NCED gene in plants (Lintig et al, 2000). It appears likely that RA and ABA also share similar catabolic processes.
The CYP707A family appears to be highly conserved throughout different plant species. Two CYP707A family genes have been identified in the rice genome, CYP707A5 and CYP707A6 (http://drnelson.utmem.edu/rice.html). CYP707A homologues have also been found in tomato and soybean (http://drnelson.utmem.edu/CytochromeP450.html). Our criteria requiring that the ABA 8′-hydroxylase gene should be equally conserved in different plants were highly effective in narrowing down the number of candidate genes. Combining the information from the genome sequences of other plants (as they become available) will greatly facilitate the identification of target genes of interest.
After considering the above point for narrowing down the candidate genes, DNA microarray analysis during Arabidopsis seed imbibition was highly effective. In addition to the CYP707A family, a number of genes were also expressed during seed imbibition. These genes may participate in other pathways that function during seed germination and would be the targets of future studies.
CYP707As play a regulatory role in ABA catabolismin vivo
ABA is catabolized into an inactive form either by oxidation or conjugation. It has been generally assumed that ABA 8′-hydroxylation plays a prominent role in ABA catabolism as PA or its derivatives are the predominant catabolites reported in most ABA-related physiological processes. In comparison to ABA, bioassays have shown that PA exhibits substantially reduced biological activities. We have demonstrated that all four members of CYP707A encode ABA 8′-hydroxylase. Moreover, to elucidate the regulatory role of CYP707As in vivo, we showed that expression of these genes was tightly regulated both developmentally and environmentally, such as during seed imbibition, dehydration and subsequent rehydration of the plantlet. Expression levels also correlated closely with endogenous ABA levels. Furthermore, the hyperdormant phenotype exhibited by cyp707a2 mutants clearly renders this gene crucial in defining the ABA content of seeds.
The expression of all the four CYP707A genes is responsive to dehydration and subsequent rehydration (Figure 6C). As upregulation in response to dehydration and strong induction in response to rehydration of CYP707A gene expression were observed in all CYP707A genes, it is likely that the function of CYP707A genes might be partially redundant in these processes. In contrast, CYP707A2 mRNA accumulates predominantly in dry seed and it increases dramatically immediately after initiation of seed imbibition (Figure 5B). The prominent role of CYP707A2 during this process was confirmed by the hyperdormant phenotype of the cyp707a2 seeds (Figure 8B). Furthermore, the cyp707a2 mutants accumulate and maintain significant amounts of ABA in dry and imbibed seeds (Figure 8C and D). These findings clearly demonstrate that CYP707As play a predominant role in vivo in ABA catabolism.
It has been shown that pretreatment of exogenous ABA enhanced PA production in cultured Arabidopsis and potato cells (Windsor and Zeevaart, 1997; Krochko et al, 1998). As ABA 8′-hydroxylation is presumed to be the regulatory step leading to PA production, ABA 8′-hydroxylase may also be activated by ABA. In the present study, we have demonstrated that expression of all the four CYP707A genes, in particular CYP707A1, in vegetative plants was rapidly induced by exogenous ABA (Figure 7B). Interestingly, the induction of CYP707A1 by ABA application was significantly reduced in abi1-1 mutant (Hoth et al, 2002).
Although our results indicate that CYP707A plays a predominant role in ABA catabolism during seed imbibition and drought stress response, several other ABA catabolic pathways might also play a role in determining the ABA content under specific conditions or in different plant species. For instance, ABA glucosylation appears to be the predominant pathway in ABA catabolism in lettuce seed germination (Chiwocha et al, 2003). Further genetic analyses involving the construction of multigenic cyp707a mutants, in combination with studying additional pathways, are necessary to fully understand the role of ABA catabolism and to fine-tune the ABA content during particular physiological processes.
Balance between ABA biosynthesis and catabolism
When ABA levels are increased and maintained upon dehydration, PA levels also increase (Zeevaart, 1980; Pierce and Raschke, 1981). Therefore, it has been argued that ABA levels are maintained by the balance between its biosynthesis and catabolism, rather than simply by biosynthesis alone. Our results demonstrated that expression of CYP707A genes was indeed activated upon dehydration (Figure 6C), although their induction was slower than that of NCED3 expression (Figure 6B). This difference in induction kinetics may define the accumulation profile of stress-induced ABA (Figure 6A). Release from the stressed condition by rehydration greatly stimulated CYP707A gene expression within 1 h. Conversely, expression of NCED3 decreased in response to rehydration. As a result of this difference in gene expression, the balance between biosynthesis and catabolism might be disrupted, causing an abrupt reduction in ABA content.
During seed imbibition, CYP707A1 and CYP707A3 were moderately induced after 12 h (Figure 5B). Interestingly, several NCED genes were also induced after 12 h (unpublished data). Hence, it is likely that an equilibrium between ABA biosynthesis and catabolism is established during this period and prior to germination.
The present work has identified genes encoding the key enzyme in ABA catabolism, ABA 8′-hydroxylase, in Arabidopsis. Once identified, we were able to study the role of these genes during seed imbibition and drought stress conditions. Furthermore, we have demonstrated the synergistic relationship between biosynthesis and catabolism with respect to the control of ABA levels. Our present findings will lay the framework for more detailed and extensive studies into the role and regulation of ABA action during various stages of the plant life cycle.
Materials and methods
Plant materials and growth conditions
Plant materials used in this study were Arabidopsis thaliana (L.) Heynh of ecotype Columbia. Plants were grown under continuous light at 22°C. Nonsterilized plants were grown in pots containing a 1:1 mixture of vermiculite:Jiffy mix (Sakatanotane). To isolate total RNA from various organs, flowers and stems were harvested from plants grown under nonsterilized conditions for 6 weeks. Siliques were harvested at 10 days after flowering, while dry seeds stored for 4 weeks were used. To harvest rosette leaves and roots, plants were grown for 2 weeks on 0.8% agar plates containing 1/2 MS salts. For gene expression analysis during seed imbibition, dry seeds were washed with 0.04% Triton X-100, rinsed with water several times, sown and imbibed on filter papers moistened with water. For gene expression analysis during dehydration and rehydration, 2-week-old plants grown on agar medium were transferred onto the dried filter paper in a sealed container. In a separate experiment, after 6 h of dehydration, water was added to the filter paper. For ABA induction experiments, 2-week-old plants grown on agar medium were transferred onto the filter paper moistened with either water or 30 μM (+)-S-ABA solution. For ABA quantification, the same plant materials as those for RNA isolation were used. For germination tests of cyp707a2 and cyp707a3 mutants, approximately 30 seeds were used in triplicate experiments. The seeds were imbibed on filter papers moistened with water.
Chemicals
(+)-S-ABA was kindly provided by Toray Co Ltd. (−)-R-ABA was prepared from a racemic mixture of (±)-ABA (Sigma) by HPLC separation using Chiralcel OD (20 mm i.d. × 250 mm, Daicel Chemical Industries) (flow rate 20 ml/min, detection with Multiwavelength detector (JASCO MD-910)) with hexane:isopropanol (85:15), 1% acetic acid as a solvent. The retention times for (+)-S-ABA and (−)-R-ABA were ca. 8.5 and 11 min, respectively. [1,2-13C2]-(±)-ABA was prepared according to Asami et al (1999). Tetcyclacis was kindly provided by Dr W Rademacher (BASF, Limburgerhof, Germany). Metyrapone was purchased from Aldrich. Standard and deuterium-labeled PA and DPA were prepared as reported (Hirai et al, 2003).
DNA microarray analysis
Total RNA was prepared from dry or imbibed seeds using RNAqueous columns with Plant RNA isolation aid (Ambion) according to the manufacturer's protocol. The RNA was purified through precipitations with 20% isopropanol containing 0.24 M sodium citrate and 0.16 M sodium chloride, and with 2 M lithium chloride. An aliquot of total RNA (7.5 μg) was used for microarray analysis (GeneChip ATH1 GenomeArray, Affymetrix). cDNA synthesis, biotin-labeled cRNA synthesis, hybridization, washing and staining were carried out according to the manufacturer's instruction. Data analyses were performed using MicroArray Suite (Affymetrix) software.
Functional expression in yeast
Full-length cDNA of candidate P450 genes were obtained from RIKEN BRC Experimental Plant Division, Arabidopsis full-length cDNA collection (Seki et al, 2002). cDNAs were cloned into a yeast expression plasmid, pYeDP60 (Pompon et al, 1996). The resulting plasmids were transformed into Saccharomyces cerevisiae strain WAT11 (Pompon et al, 1996). Transformants were grown in SGI medium for 24–36 h, transferred to SLI medium and induced by galactose for 12 h. The cells were collected, resuspended in 0.1 M potassium phosphate buffer (pH 7.6) and passed through a French press (20 000 psi). Microsomal fractions were suspended in 0.1 M potassium phosphate buffer (pH 7.6). For assaying of ABA 8′-hydroxylase, (+)-S-ABA was incubated with 2 μg of microsomal protein (in a 100 μl volume) containing NADPH, at 22°C for 12–15 h. The amount of protein was measured by the Bradford method. For the P450 inhibitor assay, the inhibitor was added to the reaction mixture at the indicated concentration, incubated and assayed using the same condition as described above. The reaction was stopped by adding 1 N HCl, extracted with EtOAc and analyzed by HPLC using a PEGASIL-B ODS (4.6 mm i.d. × 250 mm, Senshu Scientific) column (flow rate 1.0 ml/min, UV detection at 254 nm) with the following gradient condition at ambient temperature: (A) 10% MeOH, 0.1% acetic acid, (B) 60% MeOH, 0.1% acetic acid, 0–3 min, 50% B, 3–33 min 50–100% B linear gradient. Retention times for PA and ABA were 16 and 30 min, respectively.
cDNAs for CYP707A2 and CYP707A4 were PCR amplified from cDNAs prepared from dry seeds and siliques as described above. C411A mutant clone of CYP707A1 was constructed using an appropriately mutated primer.
Identification of PA by GC-MS
For GC-MS analysis, samples were treated with ethereal diazomethane at room temperature for 5 min to obtain methyl esters of PA. GC-EIMS (JMS-Automass SUN, JEOL) was carried out with DB-1 column (250 μm i.d. × 30 m, film thickness of 0.25 μm, J&W Scientific) with a helium carrier (flow rate 1 ml/min). The column temperature was maintained at 80°C for 1 min, followed by the program, step 1: 80–220°C, 20°C/min; step2: 220–240°C, 5°C/min; step3: 240–300°C, 40°C/min and finally 300°C for 5 min. The retention time for PA methyl esters was 11 min under these conditions.
Quantitative reverse transcription–PCR
The procedures for total RNA isolation from seeds or siliques were described above. To isolate total RNA from other tissues, TRIZOL Reagent (Invitrogen) was used. Total RNA (2 μg) was treated with RQ1 RNase-free DNase (Promega) to eliminate genomic DNA contamination. First-strand cDNA was synthesized with random hexamers using a Superscript first-strand synthesis system according to the manufacturer's instruction (Invitrogen). Quantitative real-time PCR with Taq-Man technology (Holland et al, 1991) was carried out using the first-strand cDNA as a template on a sequence detector system (model 7000, Applied Biosystems). The mean value of three replicates was normalized using a 1000 times diluted 18S rRNA as the internal control. Nucleotide sequences of gene-specific primers and Taq-Man probes were as follows. CYP707A1: forward primer 5′-CTCACTCTCTTCGCCGGAAG-3′, reverse primer 5′-TTCCAAACTCCCACTCCCTCC-3′, TaqMan Probe 5′ FAM-TGTCTAATCTCTCAGCGCCGCTTTGG-TAMRA 3′. CYP707A2: forward primer 5′-CGTCTCTCACATCGAGCTCCTT-3′, reverse primer 5′-CCAAAAGTCCATCAACACCCTC-3′, TaqMan Probe 5′FAM-TCCTCCAAACCCTTTCCTCTTGGACG-TAMRA 3′. CYP707A3: forward primer 5′-CTCTGTTTCTCTGTTTACTCCGATTTA-3′, reverse primer 5′-TGCAGCAAAACAGAGAAGATACG-3′, TaqMan Probe 5′ FAM-CCGCCGTAGCTCCTCCACGAAAC-TAMRA 3′. CYP707A4: forward primer 5′-CCTGAAACCATCCGTAAACTCAT-3′, reverse primer 5′-TTCCTTACAATCTTGGGCCAA-3′, TaqMan Probe 5′ FAM-CTGATATCGAGCACATTGCCCTT-TAMRA 3′. NCED3: forward primer 5′-GCTGCGGTTTCTGGGAGAT-3′, reverse primer 5′-ATCGTCTTCTCAAAGCTCCGAC-3′, TaqMan Probe 5′ FAM-CTTGGTGGCAATCATACTCAGCCGC-TAMRA 3′. Individual data from duplicate experiments are available at The EMBO Journal online (see Supplementary data 6).
Measurement of ABA, PA and DPA levels
Samples were harvested, extracted and quantified according to Cheng et al (2002), except that EtOAc was used for extraction from residual aqueous solution. ABA and PA were methylated as described above, while DPA was TMS derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide at 60°C for 5 min. For GC-selected ion monitoring-MS, 297 (deuterated) and 294 (endogenous) peaks were used for PA, while 429 (deuterated) and 426 (endogenous) peaks were used for DPA quantification. Individual data from duplicate experiments are available at The EMBO Journal online (see Supplementary data 6).
Supplementary Material
Supplementary data 1
Supplementary data 2
Supplementary data 3
Supplementary data 4
Supplementary data 5
Supplementary data 6
Acknowledgments
We thank Dr W Rademacher (BASF, Limburgerhof, Germany) for kindly providing tetcyclacis, Toray Co Ltd for the kind gift of (+)-S-ABA, RIKEN BRC for providing full-length cDNAs, ABRC for providing knockout lines and Dr D Pompon (CNRS, Gif-sur-Yvette, France) for providing the yeast expression plasmid pYeDP60, and yeast strain WAT11. We also thank Ms S Shinoda for sequencing of the ABRC knockout lines, and Dr J Preston (RIKEN, Japan) and Ms S Sarker (University of Toronto, Canada) for critical reading of the manuscript.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Horn E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asami T, Sekimata K, Wang JM, Yoneyama K, Takeuchi Y, Yoshida S (1999) Preparation of (±)-[1,2-13C2]abscisic acid for use as a stable and pure internal standard. J Chem Res (Synop) 11: 658–659 [Google Scholar]
- Balsevich JJ, Cutler AJ, Lamb N, Friesen LJ, Kurz EU, Perras MR, Abrams SR (1994a) Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolites. Plant Physiol 106: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsevich JJ, Abrams SR, Lamb N, König WA (1994b) Identification of unnatural phaseic acid as a metabolite derived from exogenously added (−)-abscisic acid in a maize cell suspension culture. Phytochemistry 36: 647–650 [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JDG, Kamiya Y (1999) The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA 96: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C (1998) Molecular genetics analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311–343 [DOI] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR (2003) A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J 35: 405–417 [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE (1999) Formation and breakdown of ABA. Trends Plant Sci 4: 472–478 [DOI] [PubMed] [Google Scholar]
- Durst F, Nelson DR (1995) Diversity and evolution of plant P450 and P450-reductases. Drug Metab Drug Interact 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H (1997) Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J 16: 4163–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard DF, Walton DC (1976) Abscisic acid metabolism by a cell-free preparation from Echinocystis lobata liquid endosperm. Plant Physiol 58: 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M (2000) Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta 210: 279–285 [DOI] [PubMed] [Google Scholar]
- Hampson CR, Reaney MJT, Abrams GD, Abrams SR, Gusta LV (1992) Metabolism of (+)-abscisic acid to (+)-7′-hydroxyabscisic acid by bromegrass cell cultures. Phytochemistry 31: 2645–2648 [Google Scholar]
- Harrison MA, Walton DC (1975) Abscisic acid metabolism in water-stressed bean leaves. Plant Physiol 56: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai N, Kondo S, Ohigashi H (2003) Deuterium-labeled phaseic acid and dihydrophaseic acids for internal standards. Biosci Biotechnol Biochem 67: 2408–2411 [DOI] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus. Proc Natl Acad Sci USA 88: 7276–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez J-P, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Jennewein S, Rithner CD, Williams RM, Croteau RB (2001) Taxol biosynthesis: Taxane 13α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 98: 13595–13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krochko JE, Abrams GD, Loewen MK, Abrams SR, Cutler AJ (1998) Abscisic acid 8′-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol 118: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Nambara E, McCourt P (2003) The key to signaling. Nature 422: 122. [DOI] [PubMed] [Google Scholar]
- Lintig JV, Vogt K (2000) Filling the gap in vitamin A research. J Biol Chem 275: 11915– 11920 [DOI] [PubMed] [Google Scholar]
- McCarty DR (1995) Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol 46: 71–93 [Google Scholar]
- Milborrow BV (1969) Identification of ‘Metabolite C' from abscisic acid and a new structure for phaseic acid. Chem Commun 966–967 [Google Scholar]
- Milborrow BV (1975) The absolute configuration of phaseic and dihydrophaseic acids. Phytochemistry 14: 1045–1053 [Google Scholar]
- Pierce M, Raschke K (1981) Synthesis and metabolism of abscisic acid in detached leaves of Phaseolus vulgaris L. after loss and recovery of turgor. Planta 153: 156–165 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhanced drought tolerance. Plant Physiol 128: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Sato H, Ashida N, Suhara K, Itagaki E, Takemori S, Katagiri M (1978) Properties of an adrenal cytochrome P-450 (P-450 11β) for the hydroxylations of corticosteroids. Arch Biochem Biophys 190: 307–314 [DOI] [PubMed] [Google Scholar]
- Schoendorf A, Rithner CD, Williams RM, Croteau RB (2001) Molecular cloning of a cytochrome P450 taxane 10β-hydroxylase cDNA from Taxus and functional expression in yeast. Proc Natl Acad Sci USA 98: 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA (1996) Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci 15: 235–284 [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, Muramatsu M, Hayashizaki Y, Kawai J, Carninci P, Itoh M, Ishii Y, Arakawa T, Shibata K, Shinagawa A, Shinozaki K (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Sondheimer E, Galson EC, Tinelli E, Walton DC (1974) The metabolism of hormones during seed germination and dormancy. Plant Physiol 54: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Walton DC, Sondheimer E (1972) Metabolism of 2-14C-(±)-abscisic acid in excised bean axes. Plant Physiol 49: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochromes P450. In The Arabidopsis Book, Somerville CR, Meyerowitz EM (eds) Rockville, MD: American Society of Plant Biologists doi/10.1199/tab.0028, http://www.aspb.org/publications/arabidopsis [Google Scholar]
- Windsor ML, Zeevaart JAD (1997) Induction of ABA 8′-hydroxylase by (+)-S-, (−)-R- and 8′,8′,8′-trifluoro-S-abscisic acid in suspension cultures of potato and Arabidopsis. Phytochemistry 45: 931–934 [DOI] [PubMed] [Google Scholar]
- Wu DA, Chung BC (1991) Mutations of P450c21 (steroid 21-hydroxylase) at Cys428, Val281, and Ser268 result in complete, partial, or no loss of enzymatic activity, respectively. J Clin Invest 88: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z-J, Nakajima M, Suzuki Y, Yamaguchi I (2002) Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from Adzuki bean seedlings. Plant Physiol 129: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Endo T, Satoh S (1998) Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol 39: 307–312 [Google Scholar]
- Zeevaart JAD (1980) Changes in the levels of abscisic acid and its metabolites in excised leaf blades of Xanthium strumarium during and after water stress. Plant Physiol 66: 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1
Supplementary data 2
Supplementary data 3
Supplementary data 4
Supplementary data 5
Supplementary data 6


