Significance
Discovering the genetic basis of common diseases, such as diabetes, heart disease, and schizophrenia, is a key goal in biomedicine. Genomic studies have revealed thousands of common genetic variants underlying disease, but these variants explain only a portion of the heritability. Rare variants are also likely to play an important role, but few examples are known thus far, and initial discovery efforts with small sample sizes have had only limited success. In this paper, we describe an analytical framework for the design of rare variant association studies of disease. It provides guidance with respect to sample size, as well as the roles of selection, disruptive and missense alleles, gene-specific allele frequency thresholds, isolated populations, gene sets, and coding vs. noncoding regions.
Keywords: mapping disease genes, power analysis, statistical genetics
Abstract
Genetic studies have revealed thousands of loci predisposing to hundreds of human diseases and traits, revealing important biological pathways and defining novel therapeutic hypotheses. However, the genes discovered to date typically explain less than half of the apparent heritability. Because efforts have largely focused on common genetic variants, one hypothesis is that much of the missing heritability is due to rare genetic variants. Studies of common variants are typically referred to as genomewide association studies, whereas studies of rare variants are often simply called sequencing studies. Because they are actually closely related, we use the terms common variant association study (CVAS) and rare variant association study (RVAS). In this paper, we outline the similarities and differences between RVAS and CVAS and describe a conceptual framework for the design of RVAS. We apply the framework to address key questions about the sample sizes needed to detect association, the relative merits of testing disruptive alleles vs. missense alleles, frequency thresholds for filtering alleles, the value of predictors of the functional impact of missense alleles, the potential utility of isolated populations, the value of gene-set analysis, and the utility of de novo mutations. The optimal design depends critically on the selection coefficient against deleterious alleles and thus varies across genes. The analysis shows that common variant and rare variant studies require similarly large sample collections. In particular, a well-powered RVAS should involve discovery sets with at least 25,000 cases, together with a substantial replication set.
Genomic studies over the last half decade have shed light on the genetic basis of common polygenic human diseases and traits, identifying thousands of loci and revealing key biological pathways. Nonetheless, the genetic variants identified thus far appear to explain less than half of the estimated heritability in most diseases and traits. The sources of the so-called missing heritability remain unclear (1). This article is our second paper exploring the mystery of missing heritability.
In our first paper (2), we explored a methodological issue. We showed that genetic interactions, if present, could account for substantial missing heritability. Still, this is likely to be only a partial explanation. In this paper, we turn to the search for additional genetic variants underlying common human diseases, focusing on rare genetic variants.
The discovery of genes underlying common diseases depends on association studies (except in special cases where Mendelian subtypes of common diseases show clear segregation in large families). Association studies involve testing whether the frequency of a set of one or more alleles differs between cases and a control population, indicating that the set of alleles is associated with the disease. Association studies to date have largely focused on studying individual common variants, because they could be more readily assayed with initial genomic technologies. However, association studies are increasingly being applied to sets of rare variants as well.
Association studies of individual common variants are often referred to as genomewide association studies (GWAS), whereas association studies of sets of rare variants in coding regions are often described as exome-sequencing studies. This nomenclature is unfortunate because it conflates statistical methodology (association testing) and laboratory methodology (DNA sequencing). To highlight the parallelism, we will use the terms common variant association study (CVAS) and rare variant association study (RVAS). Ideally, the term GWAS should encompass both types of association studies.
CVAS.
By common variants, we mean those that occur often enough that it is practical to test each variant individually by estimating its frequency in cases and controls. Given the feasibility of collecting many thousands of cases, common variants will be operationally defined as those with frequency ≥0.5% (one carrier per 100 people).
The theory and practice of CVAS is well advanced. Catalogs of common variants in human populations are nearing completion, and tens of thousands (and in some cases, hundreds of thousands) of cases and controls have been genotyped for many traits and diseases. Studies to date have largely used genotyping arrays and linkage disequilibrium patterns, but will increasingly use inexpensive massively parallel sequencing.
Some missing heritability is clearly due to additional common variants that remain to be discovered. First, CVAS to date has used genotyping arrays with good coverage of variants with frequency 5–50% but poorer coverage of the range 0.5–5%. [The potential importance of these low-frequency common variants, sometimes called “Goldilocks” alleles (3), is demonstrated by nonsense mutations of PCSK9, which have led to new therapeutics for decreasing LDL cholesterol (4, 5).] Second, CVAS has been limited by sample size; studies with larger sample sizes continue to reveal many new loci (6–8). Third, indirect statistical methods indicate that association with common variants can explain at least 30% (and likely more) of the heritability for a number of diseases and traits (9–11).
RVAS.
Rare variants may also make a major contribution to missing heritability, although much less is known at present. The theoretical case for an important role of rare variants is that alleles that strongly predispose to disease are likely to be deleterious and thus kept at low frequencies by purifying selection (12–14). Moreover, rare variants with strong effects are valuable, because they may enable clinical studies in individual patients and reveal protective null alleles that define targets for pharmaceutical intervention.
RVAS differs from CVAS in two key respects. First, rare variants are too numerous to catalog comprehensively and so must be directly enumerated in every sample by DNA sequencing rather than by genotyping known variants. The importance of this distinction is fading, however: with the plummeting cost of DNA sequencing, both CVAS and RVAS will increasingly use direct DNA sequencing. Second and more fundamentally, rare variants occur too infrequently to allow association tests of individual variants. RVAS thus require aggregating rare variants into sets and comparing the aggregate frequency distribution in cases vs. controls (15).
The need to aggregate variants poses a challenge: which variants to aggregate? Ideally, one would aggregate only damaging alleles and ignore benign alleles. Unfortunately, one cannot perfectly distinguish the former from the latter. To enrich for harmful alleles, RVAS typically focuses on (i) nonsynonymous variants in protein-coding regions, ignoring the rest of the genome; and (ii) variants with frequency below a specified threshold T. Even with these limitations, the resulting variants remain a mixture of damaging and benign alleles.
The practice of RVAS is still in its infancy. In pioneering studies of the genetics of obesity, O’Rahilly and colleagues studied unrelated cases with a common disease to look for rare variants in candidate genes related to the disease (16). Hobbs and Cohen subsequently applied this approach to well-established candidate genes for lipid-related phenotypes, showing, for example, that PCSK9, in which gain-of-function alleles caused a Mendelian hypercholesterolemia syndrome, also harbored rare loss-of-function alleles that lowered LDL cholesterol (4).
The challenge, however, has been to move beyond handfuls of candidate gene studies to unbiased gene discovery. Some early efforts organized by the US National Institutes of Health were premised on the notion that rare variants underlying common diseases could be reliably identified in small collections of 50–100 cases. However, results have made clear that RVAS, like CVAS, requires much larger samples.
The few discoveries from RVAS to date have largely emerged from candidate gene studies rather than unbiased surveys and, in some cases, have reached only nominal rather than genomewide significance (17–24). However, exomewide studies of large samples are now underway for several diseases.
Given the early stage of the field, the analytical methodology for RVAS remains in flux. Many authors have proposed a rich collection of possible statistics (reviewed in refs. 25 and 26 and tests compared in ref. 27; SI Appendix, Section 3.4). Our goal here is neither to evaluate their relative merits nor to propose alternatives.
Rather, the goal of this paper is to offer a simple conceptual framework that provides insight into the design of RVAS and to apply it to address some fundamental questions:
-
i.
Choice of variants. What are the relative merits of disruptive alleles (stop, frameshift, and splice-site mutations, which severely disrupt protein structure) vs. missense alleles?
-
ii.
Frequency threshold. What is the optimal threshold T for filtering alleles? Is it constant or does it vary with the properties of each gene?
-
iii.
Sample sizes. How many cases are needed to detect association by RVAS? How does the answer depend on the properties of the gene, the relative risk of harmful alleles, and the strategic choices above?
-
iv.
Other strategies. What approaches might be used to increase power, such as studying special populations, specific gene sets, or de novo mutations?
-
v.
Whole genome analysis. Can RVAS be extended from the exome to include the noncoding portion of the genome?
To gain intuition, we focus on a simple situation. We consider a binary trait (e.g., a disease, such as schizophrenia, or the tail of a quantitative phenotype, such as LDL cholesterol above 200 mg/dL) analyzed with burden tests (which compare the number, or ‘burden’, of variants in cases and controls), where the variants studied are those with frequency below a fixed threshold. We assume a ‘two-class model’, wherein all alleles are either null (abolishing gene function) or neutral (having no effect on gene function). Although the model is simple, it can provide clear insight into the many alternative methods for RVAS.
Below, we show how the answers depend crucially on the selection coefficient of each gene; more modestly on the proportion of missense mutations that are null; and in some cases on population history. Results are summarized in the main text and supported by mathematical formulas, proofs, simulations, graphs and tables in the extensive Supplementary Information.
Results
Designing an Association Study.
A burden test for association with a disease examines whether a class C of alleles in a gene G is enriched or depleted in cases vs. random controls from the general population, with individuals assumed to be unrelated so that events are independent. (Although we consider random population controls, one can alternatively compare cases with disease-free controls.) The class of alleles may be selected in many ways. It could, for example, consist of a single allele (as in CVAS), all nonsense alleles, all missense variants with frequency <1%, or all variants at evolutionarily conserved nucleotides in a given gene.
The power of an association study depends on two key quantities: (i) fC, the combined allele frequency (CAF) of class C, meaning the expected number of alleles present in a haploid genome in the population; and (ii) λC, the excess relative risk of disease conferred by alleles in the class, meaning that a heterozygous carrier has a (1 + λC)-fold higher risk of disease than a random member of the population.† By Bayes’ theorem, (1 + λC)-fold is also the expected enrichment of such alleles in cases vs. the overall population [SI Appendix: Eq. 3.1]. Consequently, λC can be directly estimated from the excess enrichment in cases (SI Appendix, Section 3.1). Enrichment of alleles in cases can be tested with likelihood ratio tests (LRTs) (SI Appendix, Section 3.2).
The number of cases‡ needed to detect an association using an LRT is given by a formula (SI Appendix, Eq. 3.6) that is well approximated, when fC is small, by
 |
Here,  is a simple function§ and
is a simple function§ and  is a constant that controls the false-positive rate a and false-negative rate b. For 90% power (b = 10%),
is a constant that controls the false-positive rate a and false-negative rate b. For 90% power (b = 10%),  = 10.5 when testing a single hypothesis (a = 0.05) and
= 10.5 when testing a single hypothesis (a = 0.05) and  = 35.9 when correcting for multiple hypothesis testing of 20,000 genes (a = 2.5 × 10−6). The values for 50% power are
= 35.9 when correcting for multiple hypothesis testing of 20,000 genes (a = 2.5 × 10−6). The values for 50% power are  = 3.8 and 22.2, respectively. Calculations below assume multiple hypothesis correction.
= 3.8 and 22.2, respectively. Calculations below assume multiple hypothesis correction.
Although discussions of RVAS often contemplate alleles that increase disease risk, the formulas apply equally well to alleles that decrease disease risk—that is, protective alleles. In this case, λC is a negative number between −1 and 0. (For example, protective alleles that decrease risk by fourfold have λC = −0.75.)
To maximize the power of an association study, we want fC, the combined allele frequency, and |λC|, the absolute value of the excess relative risk, to be as large as possible. Unfortunately, these goals sometimes pull in opposite directions. For example, expanding the alleles under study m increases fC but dilutes  .
.
Below, we explore six strategies for RVAS. To lay the foundation, we start by considering several issues related to alleles and their frequencies.
Mutation Rates for Observable Classes.
First, we need to understand the rate at which various types of mutations are born. For protein-coding regions, three classes of mutations can be directly observed: silent (S), missense (M), and disruptive (D, defined as nonsense, splice site, and frameshift changes, which severely disrupt protein structure). For any human gene, we can obtain good estimates of the mutation rates for these observable classes (μS, μM, μD) based on (i) the length and sequence composition of the gene and (ii) the rate and mutational spectrum for point mutations in the local region. (The latter quantities can be estimated from comparative genomics, medical genetics or large-scale human parent-offspring trio sequencing studies.) (SI Appendix, Section 2.6). Below, we will focus on a typical human gene with the median mutation rate and a coding region of 1,500 bp. (One could readily incorporate gene-specific rates, if desired.) For this typical gene, the mutation rates for silent, missense, and disruptive mutations are 5.6 × 10−6, 12.8 × 10−6, and 1.7 × 10−6 per gene copy per generation, respectively (Table 1).
Table 1.
Mutation rates per gamete
| Mutation type | Absolute mutation rate (×10−6) |
|||
| α = 10% | 25% | 33% | 60% | |
| Silent | 5.6 | 5.6 | 5.6 | 5.6 |
| Missense | 12.8 | 12.8 | 12.8 | 12.8 |
| Null | 1.3 | 3.2 | 4.2 | 7.7 |
| Neutral | 11.5 | 9.6 | 8.6 | 5.1 |
| Disruptive | 1.7 | 1.7 | 1.7 | 1.7 |
| Nonsense, splice | 0.9 | 0.9 | 0.9 | 0.9 |
| Frameshift | 0.8 | 0.8 | 0.8 | 0.8 |
| All null | 3.0 | 5.0 | 6.0 | 9.4 |
| Disruptives: All nulls | ||||
| Ratio | 1: 1.8 | 1: 2.8 | 1: 3.5 | 1: 5.5 |
| Percentage | 57% | 34% | 29% | 18% |
For a typical gene as defined in the text, with α being the proportion of newborn missense alleles that are null.
Two-Class Model for Impact of Mutations.
Second, we need to consider the impact of mutations on gene function. We will adopt a simple two-class model in which mutations in protein-coding regions are either (i) neutral, having no effect on function, or (ii) null, abolishing gene function. Null alleles cause excess relative risk λ = λnull ≥ 0 and have selection coefficient s = snull ≥ 0, which is assumed to have been constant.
We will assume that (i) all silent mutations are neutral, (ii) all disruptive mutations are null (which is reasonable, at least if one excludes disruptive mutations occurring late in a coding region), and (iii) missense mutations are a mixture of null and neutral alleles, with proportion α being null and 1 − α being neutral.
Our model is clearly a simplification, because we ignore the possibility of alleles with intermediate effects. Evolutionary studies and analysis of mutations responsible for Mendelian diseases suggest that ∼25% of missense mutations are strongly deleterious (essentially equivalent to disruptive mutations, with selection coefficients in the range of 10−2 and below); ∼50% are weakly deleterious (hypomorphic alleles, with selection coefficients in the range 10−3–10−4); and ∼25% are truly neutral (13, 28–30). [Some studies have suggested slightly different proportions for strongly deleterious mutations (31–33).] In our two-class model, we will ignore the hypomorphic mutations, treating them as being effectively neutral. We also ignore the possibility of countervailing alleles; e.g., protective alleles if null alleles are deleterious or vice versa (4, 5). Discussion elaborates on the reasons for and limitations of the model.
Our analyses below will primarily use the value α = 25%, because it is the average value reported for human genes. Of course, the actual proportion α varies across genes. For example, the depletion of missense alleles relative to silent alleles suggests values α  5% for BRCA2 but 50% for CHD8 (SI Appendix, Section 2.1).
5% for BRCA2 but 50% for CHD8 (SI Appendix, Section 2.1).
To illustrate the impact of varying proportions of null missense alleles, we sometimes compare results for four values: α = 10%, 25%, 33%, or 60%. For these values, Table 1 shows that (i) the mutation rates for all null alleles (disruptive plus missense) are 3.0, 5.0, 6.0, and 9.4 × 10−6; (ii) the ratio of all null alleles to disruptive alleles is 1.8-, 2.8-, 3.5-, and 5.5-fold; and (iii) disruptive alleles comprise 57%, 34%, 29%, and 18% of all null alleles.
CAF.
Third, we need to understand the properties of the CAF. For this purpose, we focus on classes where all alleles have the same selection coefficient s.
The ancestral human population that existed ∼1,000 generations ago is typically modeled as being at equilibrium with a constant effective population size Neq of ∼10,000 (34). For a population at equilibrium, classical population genetics (35–37) provides a precise formula for the expected CAF (SI Appendix, Proposition 2), which is well approximated by
 |
where μC is the rate per chromosome per generation of new mutations in class C.
Modern human populations have undergone massive expansions, interrupted in some cases by bottlenecks. Although no simple formulas are available, the value of f can be found by simulation. We analyzed various population histories (SI Appendix, Section 1.3), including two models of uninterrupted expansions (denoted expansions 1 and 2) and models meant to represent Europe (bottleneck of 1,550 chromosomes occurring 1,250 generations ago), Finland (100 chromosomes, 100 generations), and Iceland (1,000 chromosomes, 50 generations).
Importantly, the expected value of the CAF is essentially unaffected by the vast changes in the size or structure of human populations over the last ∼1,000 generations [Fig. 1A]. The reason is intuitively clear: (i) for alleles under significant selection, the expected CAF is held at μC/s by mutation-selection balance (38), whereas (ii) for nearly neutral alleles, a simple back-of-the-envelope argument shows that the expected CAF increases from the equilibrium value of 4μCNeq at an extremely slow rate.¶ Eq. 2 thus provides a good approximation for the expected CAF in modern human populations. (Note: although the expected CAF is constant, the variance across genes varies across populations; see RVAS strategy 4 below.)
Fig. 1.

Allele frequencies from simulations for various demographic models. (A) CAF (average 50,000 simulations) as a function of selection coefficient s. CAF is not sensitive to demographic history, as noted in the text. (B) Cumulative distribution of IAF as a function of s, for the ancestral population at equilibrium. (C) Cumulative distribution of IAF as a function of s, for the European population. Compared with the ancestral population, the IAF distribution is skewed toward rare alleles for intermediate to strong selection s (SI Appendix, Sections 2.2–2.3).
The CAF depends strongly on the selection coefficient, with the expected value falling precipitously as selection increases: fC ∼ 40,000μC, 13,000μC, 1,000μC, 100μC, and 10μC for s = 0, 10−4, 10−3, 10−2, and 10−1, respectively (see Eq. 2 and SI Appendix, Proposition 2 for exact formula). If their birth rates were identical, neutral alleles would thus outnumber deleterious alleles by 400-fold if s = 10−2 and by 4,000-fold if s = 10−1.
Using the mutation rate for disruptive alleles in our typical gene (1.7 × 10−6) and the approximation in Eq. 2, the expected frequency for disruptive alleles is 0.17%, 0.017%, and 0.0017% for s = 10−3, 10−2, and 10−1, respectively; that is, approximately one heterozygote per 300, 3,000, and 30,000 individuals. For our typical gene with α = 25%, the expected frequency of all null alleles (disruptive plus missense) is ∼2.8-fold higher (Table 1).
Individual Allele Frequencies.
Fourth, we need to understand the individual allele frequency (IAF) distribution, ΨC(x), for a class C of alleles, defined as the probability that an individual allele in C sampled from a random chromosome|| has population frequency ≤ x. The IAF distribution may be calculated from closed-form formulas for populations at equilibrium (35, 36) and from simulations for other models (SI Appendix, Sections 1.2 and 1.3).
The IAF distribution (i) shows when selection is strong enough that the allelic spectrum will consist primarily of rare variants and, (ii) by providing the frequency of null and neutral missense alleles, allows one to calculate the proportion ρC(T) of missense alleles with frequency ≤ T that are null.
Unlike the CAF, the IAF distribution may be significantly shaped by population history. We calculated the IAF distribution for each population model (equilibrium and European model in Fig. 1 B and C; remaining populations in SI Appendix, Section 2.3).
For the ancestral human population, the frequency distribution of null alleles in a gene (sampled from a randomly chosen chromosome) should be roughly equally balanced between common and rare frequencies if s ∼ 10−2.5. If selection is stronger, null alleles will be mostly rare, comprising ≥90% of alleles on randomly chosen chromosomes for s ≥ 10−2. If selection is weaker, null alleles will be mostly common.
The dramatic expansion of the human population has important effects on allele frequencies. Even if the population were to stabilize at current levels, the large modern population size would eventually drive all nonneutral alleles toward extremely low allele frequencies. However, the speed of approach to this new equilibrium varies substantially depending on s (39). Population expansion over the last ∼103 generations has a dramatic effect for alleles with  , but little effect when s
, but little effect when s
 . In the former case, the ancestral alleles are largely eliminated and replaced by new alleles, which are born at a much lower frequency as a result of population expansion. In the latter case, the ancestral alleles continue to dominate and the median remains relatively high (although there is a small excess of rare variants that provides valuable information about population history) (40, 41).
. In the former case, the ancestral alleles are largely eliminated and replaced by new alleles, which are born at a much lower frequency as a result of population expansion. In the latter case, the ancestral alleles continue to dominate and the median remains relatively high (although there is a small excess of rare variants that provides valuable information about population history) (40, 41).
The greater impact of population expansion on alleles under strong selection can be seen by comparing the ancestral population vs. European model: the median allele frequencies are 1.7% vs. 0.6% if s = 10−3; 0.18% vs. 0.005% if s = 10−2; and 0.02% vs. 0.00005% if s = 10−1 (SI Appendix, Table S3). The selection intensity s at which common and rare variants are equally frequent is slightly weaker in Europe than in the ancestral population (10−3 vs. 10−2.5).
Impact of Population Expansion on the Genetic Architecture of Disease.
Various authors have speculated that the dramatic increase in the number of new alleles caused by population expansion has altered the genetic architecture of disease and the explanation for missing heritability (42–44). Specifically, it has been suggested that expansion may have dramatically increased (i) the total frequency of genetic disease by raising mutational load; (ii) the relative importance of new alleles in disease risk; or (iii) the role of rare variants in disease risk.
-
i.
Disease frequency. In fact, population expansion does not increase disease frequency. The increase in the number of distinct alleles caused by population expansion does not translate into a meaningful increase in the combined frequency of alleles, because the vast majority of the newly added alleles are so rare. Although the number of alleles is relevant for studies of population history, disease frequency depends on the CAF, which, as noted above, is largely unaffected by recent human population expansion.
-
ii.
Role of new alleles. Similarly, population expansion does not substantially increase the proportion of disease due to new alleles. For alleles under significant selection, simulations show that the age distribution of a randomly chosen allele is essentially unaltered by population expansion (SI Appendix, Section 2.4).
-
iii.
Role of rare alleles. Population expansion does increase the role of rare variants, although the effect is limited. Expansion shifts the allelic spectrum from predominantly common to predominantly rare only for s in a relatively narrow range (10−3 ≤ s ≤ 10−2.5 for Europe). Outside this range, the allelic spectrum remains either predominantly common or predominantly rare.
Relevant Range of Selection Coefficients.
The analysis above shows that RVAS is important when s > 10−3 and essential when s > 10−2.5 (with ≥90% of variants being rare). We focus below on s in the range of 10−3–10−1.
To understand how this range of selection might relate to common diseases, it is useful to consider the situation of direct selection—where the reduction in fitness caused by a null allele is proportional to the extent to which it increases risk for the disease under study. In this case, s = (λπ)sD, where π is the disease prevalence in the general population and sD is the decreased reproductive fitness of individuals manifesting the disease. Schizophrenia (prevalence ∼1%) has a severe fitness cost, which has been estimated at sD = 50% (45). An allele conferring relative risk of 10-fold (λ = 9) would have s = 0.045 (∼10−1.3), whereas one with 3-fold risk (λ = 2) would have s = 0.01 (∼10−2). Type 2 diabetes has a higher prevalence of ∼10%, but likely has more modest effect on fitness—perhaps sD = 1%. Alleles conferring 10-fold and 3-fold risk would have s = 10−2.0 and s = 10−2.7, respectively. For very late-onset diseases such as Alzheimer’s, the disease itself likely has little impact on reproductive fitness. Direct selection would be weak ( ) and CVAS would likely be the better strategy. Of course, these illustrations assume direct selection. Genes may be under pleiotropic selection for their effect on multiple phenotypes.
) and CVAS would likely be the better strategy. Of course, these illustrations assume direct selection. Genes may be under pleiotropic selection for their effect on multiple phenotypes.
Knowledge of Key Parameters.
Below we typically assume that the values of μ, fD, s, and α for each gene are known. In reality, they must be inferred from data.
-
i.
Mutation rate μ. From local mutation rates, one can estimate the mutation rate μ for observable classes (S, M, D).
-
ii.
Frequency fD. The allele frequency fD for the class D of disruptive alleles can be estimated simply by counting alleles in a large sample, with the precision increasing with sample size. For a coefficient of variation of 20%, about 25 events must be seen (corresponding to 6,250, 62,500, and 625,000 people for s = 10−3, 10−2, and 10−1, respectively).
-
iii.
Selection coefficient s. The selection coefficient for each gene can be obtained from fD = μD/s (Eq. 2), by using the estimated values of fD and μD and solving for s. The estimate’s precision is inherently limited by the fact that the realized value of fD in any given population fluctuates around the expected value due to the stochastic nature of population history; this variation cannot be reduced by increasing the sample size. When selection is strong, s can be estimated to within roughly half an order of magnitude (SI Appendix, Section 12.1). The precision might be improved by combining results across unrelated populations.
-
iv.
Proportion of nulls, α. The fraction α of newborn missense mutations that are null can be estimated from the deficit of missense alleles seen in a population or evolutionary comparisons. However, there are issues with the precision and accuracy of these estimates (SI Appendix, Section 12.2). The best solution may be to use a range of values of α in RVAS analysis.
With these foundations, we now turn to the design of RVAS.
RVAS Strategy 1: Studying Disruptive Alleles Only.
One simple strategy is to study only disruptive (D) variants in each gene: they are all null alleles and confer the same excess relative risk λD (=λnull). Disruptive alleles can thus be lumped into a single meta-allele, whose frequency can be measured in cases and the overall population, with the ratio providing an estimate of (1 + λnull).
For disease-predisposing alleles, Fig. 2A shows the number of cases needed to detect association with 90% power. (To achieve 50% power, one needs ∼62% as many cases.) The expected values are largely independent of population history.
-
i.
For the weakest selection in our range (s = 10−3, where common and rare variants occur with equal frequency), we need 260, 770, 2,700, 8,400, and 28,000 cases where the relative risk (1 + λ) is 20-, 10-, 5-, 3-, and 2-fold, respectively. Current case-control collections used for CVAS for many diseases already have the required sizes for RVAS.
-
ii.
For stronger selection (s = 10−2), the sample size is 10-fold larger: 2,600–280,000 cases. Current collections should suffice to detect strong effects (1 + λ > 5).
-
iii.
For the strongest selection in our range (s = 10−1), the sample size grows another order of magnitude: 26,000–2.8 million. Achieving such numbers is challenging, but should eventually be feasible by aggregating clinical information, with patient consent, across populations.
Fig. 2.
Power to detect association for a typical gene. (A) Number of cases needed to detect association based on excess of disruptive disease-predisposing variants in cases vs. general population, as a function of selection coefficient s (or frequency fD of disruptive alleles). Curves represent various effect sizes 1 + λ. (Values for 90% power and 5% false-positive rate after Bonferroni correction for testing 20,000 genes.) (SI Appendix, Sections 3.2 and 4). (B) Number of cases to detect association based on deficit of disruptive protective variants. Curves represent various values of (1 + λ)−1. The curve for (1 + λ)−1 = 2 corresponds to twofold protection (50% lower disease risk), whereas the curve for (1 + λ)−1 =  corresponds to complete protection. (C) Relative contribution of missense vs. disruptive variants for European model. Curves for various selection coefficients show ratio of expected LOD scores for testing an excess of rare missense variants with frequency below threshold T divided by expected LOD score for disruptive variants. Values are calculated for effect size for null alleles of 1 + λ = 4. For each s, there is an optimal threshold T* at which missense alleles provide maximal relative contribution (typically ∼1.0- to 1.3-fold) (SI Appendix, Section 5). (D) Relative contribution of missense (vs. disruptive) variants at optimal threshold T* as a function of 1 + λ for various populations. Solid lines show values when filtering missense alleles by frequency threshold T*. Dashed lines show values when also filtering to include only missense alleles predicted to be deleterious (by a high-quality predictor with false-positive and false-negative rates of 20%). The functional predictor increases the contribution of missense alleles—e.g., from 1.5-fold to 2.1-fold for genes 1 + λ = 10 in the European population (SI Appendix, Sections 5 and 6).
corresponds to complete protection. (C) Relative contribution of missense vs. disruptive variants for European model. Curves for various selection coefficients show ratio of expected LOD scores for testing an excess of rare missense variants with frequency below threshold T divided by expected LOD score for disruptive variants. Values are calculated for effect size for null alleles of 1 + λ = 4. For each s, there is an optimal threshold T* at which missense alleles provide maximal relative contribution (typically ∼1.0- to 1.3-fold) (SI Appendix, Section 5). (D) Relative contribution of missense (vs. disruptive) variants at optimal threshold T* as a function of 1 + λ for various populations. Solid lines show values when filtering missense alleles by frequency threshold T*. Dashed lines show values when also filtering to include only missense alleles predicted to be deleterious (by a high-quality predictor with false-positive and false-negative rates of 20%). The functional predictor increases the contribution of missense alleles—e.g., from 1.5-fold to 2.1-fold for genes 1 + λ = 10 in the European population (SI Appendix, Sections 5 and 6).
The corresponding sample sizes for detecting protective alleles are shown in Fig. 2B. More samples are needed to detect protective alleles than harmful alleles, because it is harder to detect a deficit (rather than an excess) of null alleles in cases against the already low background in the population. In the extreme case of a completely protective allele (λ = −1), the required sample size is the same as for detecting harmful alleles that increase disease risk by a factor of e
 2.718.
2.718.
Bottom line.
Contrary to some initial expectations, the number of cases needed for a well-powered RVAS is large and similar to that needed for CVAS. To obtain enough cases to detect association, RVAS for disease should thus focus primarily, for the foreseeable future, on sequencing case-control collections rather than cohorts randomly selected from the population.
RVAS Strategy 2: Adding Missense Alleles Filtered by Frequency.
One obvious idea for increasing power is to include null missense alleles in the association study. If we could perfectly recognize the null missense alleles, we could simply count them alongside the disruptive alleles. For a typical gene with α = 25%, the number of events would increase by 2.8-fold (Table 1) and the required sample size would decrease by 2.8-fold.
Given that we lack perfect knowledge, the question is as follows: how close can we come to this best possible case of 2.8-fold reduction in sample size? The inability to distinguish between null and neutral missense alleles creates a serious problem because neutral alleles are much more abundant. The usual solution is to impose a frequency threshold T—that is, to study only missense alleles with frequency ≤ T—with the aim of eliminating the vast majority of neutral alleles (which tend to have higher frequencies) while retaining most null alleles.
Even with this approach, missense alleles may be less useful than disruptive alleles because the null alleles are always diluted by neutral alleles: the proportion ρ(T) of nulls among missense alleles with frequency ≤ T is always lower than the proportion α among newborn alleles (for example, α = 25% for our typical gene). As a consequence, the observed enrichment of missense alleles in cases will always underestimate the true excess enrichment for null alleles: specifically, λM(T) = ρ(T)λnull. It is thus essential that missense variants and disruptive variants be analyzed separately.
The threshold T is often chosen in an ad hoc manner (e.g., 1%, 0.5%, or 0.1%), but it should ideally be chosen to maximize power (SI Appendix, Section 5). In general, the optimal threshold T* turns out to be roughly the frequency where (i) ∼75% of null alleles are retained and (ii) ∼16% of retained missense alleles are null, meaning that the apparent value of λ will underestimate the true value by ∼6.5-fold (SI Appendix, Tables S4–S9). Importantly, the optimal threshold T* depends strongly on the selection coefficient and thus differs across genes.
Using the optimal values of T* for Europe, missense alleles together contribute 0.6- to 1.3-fold as much information as the disruptive alleles, with the value rising as λnull increases from 0 to 10 (Fig. 2 C and D). When disruptive and missense alleles are combined, the sample size thus falls by 1.6- to 2.3-fold.
Achieving this reduction requires using the optimal threshold T*. If the threshold is too high or too low by 10-fold, the contribution of missense alleles can fall by approximately twofold (Fig. 2C). Moreover, using too high a threshold will underestimate the apparent relative risk (possibly dramatically), owing to dilution by neutral alleles.
Bottom line.
Disruptive alleles provide the most robust information and require no knowledge of s and α. Missense alleles should be analyzed as well, but require care in selecting the correct frequency cutoff for each gene (based on its selection coefficient). For typical genes with α = 25%, incorporating missense alleles may decrease sample size by approximately twofold. [The potential improvement in sample size is only ∼1.4-fold for genes where only a small proportion (α = 10%) of missense alleles are null and could be as high as 3-fold for genes where the proportion is unusually high (α= 60%).]
RVAS Strategy 3: Filtering Missense Alleles by Severity.
It may be possible to glean information about whether missense alleles are null or neutral—either from biochemical experiments (24) (where in vitro assays are available) or computational programs, such as PolyPhen-2, SIFT, or MutationTaster (reviewed in ref. 46)—that offer computational predictions of whether a mutation is likely to be damaging. Given the quality of the predictions (proportions, γnull and γneutral, of true null and neutral variants declared to be damaging), one can calculate the optimal threshold T* and the corresponding sample size.
If we use a predictor with γnull = 80% and γneutral = 20% [close to the values reported for PolyPhen-2 (47)] and filter with the optimal threshold T*, missense alleles contribute 1.2- to 2.0-fold as much information as the disruptive alleles (SI Appendix, Section 6) and thus decrease total sample size by 2.2- to 3.0-fold.
Bottom line.
For typical genes with α = 25%, using a high-quality predictor of the mutational impact of missense alleles can yield a decrease of ∼2.5-fold in sample size (vs. twofold when filtering by frequency alone).
Example: LDL Receptor in Early-Onset Myocardial Infarction.
The points above are nicely illustrated by a recent analysis of the association between rare variants in the LDL receptor (LDLR) and early-onset myocardial infarction (MI) (SI Appendix, Section 7.1). The study found that LDLR harbors (i) disruptive variants in 20/2,743 cases and 1/2,465 controls corresponding to a relative risk of 18.1 (or λD = 17.1), and (ii) missense variants with frequency ≤1% in 172 cases and 102 controls, for a dramatically lower relative risk of 1.5 (excess relative risk of λM(1%) = 0.5).
What accounts for the striking difference in the apparent relative risk between disruptive and rare missense alleles? Our framework provides a simple quantitative explanation. The selection coefficient for null alleles can be estimated as s ∼ 10−1.7 [from the equation s = μD/fD, using fD = 1/(2 × 2,465) and the mutation rate for LDLR (μD = 3.8 × 10−6); SI Appendix, Section 2.6]. Given such strong selection, the frequency threshold of 1% used by the authors is expected to result in only a small proportion of missense alleles being null: ρ(0.01) = 0.02. Given this low proportion, the expected excess relative risk is only λM(1%) = (0.02)λnull = 0.35, which is in good agreement with the observed value of 0.5. In fact, the optimal threshold T* given such strong selection is not 0.01 but ∼0.0001. At this much lower threshold, we expect ∼17% of missense alleles to be null and to observe an excess relative risk of 1 + λM(0.01%) = 3.8.
We note that there is a practical problem with using such low thresholds. We assumed above that the frequency fD is known precisely for every allele based on large population surveys. Based on a limited number of samples, we cannot tell precisely which alleles have frequency ≤0.0001. The best solution is to focus only on singleton alleles (SI Appendix, Section 7). Given the sample size, the expected relative risk for singleton alleles is 1 + λ = 3.7 compared with 3.8, when the frequencies are known precisely. It is successful because the sample has ∼10,000 chromosomes, and thus singleton alleles are likely to have a true frequency that is not much greater than 0.0001. If the sample size was much smaller, the apparent effect size (1 + λ) for singletons would be much lower: 2.1 and 1.4 for 1,000 and 100 chromosomes, respectively.**
Bottom line.
The strong effect of null LDLR alleles on early-onset myocardial infarction is evident from the enrichment of disruptive alleles. [Interestingly, the relative risk is similar to that inferred in the classic study of familial hypercholesterolemia (48).] The true effect is much harder to discern from the missense variants, because the apparently strong selection on LDLR causes missense variants to be swamped by neutral alleles even when one imposes a frequency threshold of 1%.
RVAS Strategy 4: Hitting the Jackpot with Isolated Populations.
Another approach to reducing sample size is to hope to be lucky. Although the expected value of the CAF is essentially identical across populations (see above), the variance of the CAF differs substantially. Accordingly, we might select a population in which the CAF for some genes associated with a disease happens, by chance, to be much larger than the expected value of μ/s. If we are lucky enough that the CAF is 10-fold higher than expected, we will be able to detect the gene with a sample size that is 10-fold lower than expected.
Using simulations, we find that the CAF has a tight coefficient of variation for expansion models and Europe but has a fat right tail for isolated populations with recent bottlenecks (Fig. 3 and SI Appendix, Section 8). If the expected CAF is small relative to the reciprocal of the bottleneck size, we expect either 0 or 1 ancestral allele to pass through the bottleneck. In the latter case, the CAF suddenly jumps to a much higher frequency (e.g., 1% for Finland, with a bottleneck of 100 chromosomes) and declines only slowly back to the expected level. This phenomenon explains the high prevalence of dozens of so-called “Finnish diseases” (monogenic disorders found at much higher frequencies than in the rest of Europe). It also explains genetic features in the Ashkenazi Jewish population, including the high prevalence of certain disorders and the simpler allelic spectrum for certain diseases [such as BRCA1 and BRCA2, in which three founder alleles together account for the majority of early-onset breast cancer among Ashkenazi Jewish women (49)].
Fig. 3.

Chances of being lucky. Figures show the right tail of the CAF (fnull) distribution for four selection coefficients s (10−3, 10−2.5, 10−2, 10−1.5) and four demographic models. Curves show probability that the realized value of the CAF (fnull) for all null alleles, (in absolute terms and normalized to the expected value given the selection coefficient) exceeds the value on the x-axis, with results obtained from 50,000 simulations of gene histories for each value of s and demography. Finland and Iceland show heavy right tails (genes with CAF much larger than the expected value), because population bottlenecks scatter allele frequencies. For s = 10−3 in Finland, 3.5% of genes have CAF that is 10-fold higher than expected—making it possible to discover the genes with a 10-fold lower example size than expected. The distributions depend on bottleneck size, number of generations since expansion, mutation rate and selection coefficient (SI Appendix, Section 8). Tighter bottlenecks, as in Finland vs. Iceland, allow fewer alleles to pass, but result in greater proportional increase in allele frequency. (Calculations assume μnull = 5 × 10−6, corresponding to α = 25%.)
For Finland’s tight bottleneck, the “sweet spot” occurs when s ∼ 10−2.5–10−3. For s = 10−3, the CAF for null alleles in Finland will be 5-, 10-, and 20-fold higher than expected for about 6%, 3.5%, and 1% of genes, respectively. For Iceland’s wider bottleneck, the sweet spot occurs for somewhat stronger selection (s ∼ 10−1.5–10−2). For s = 10−2, the CAF for null alleles in Iceland will be 5-, 10-, and 20-fold higher for about 5%, 2%, and 0.5% of genes, respectively. The distributions depend on the bottleneck size, number of generations since expansion, mutation rate of the allelic class, and selection coefficient (SI Appendix, Section 8).
Bottom line.
Tight bottlenecks scatter the relative contributions of genes. Studying recently bottlenecked populations should thus make it much easier to discover some disease-associated genes, although it will be harder to detect other genes (whose ancestral alleles failed to pass through the bottleneck). Although incomplete, such early discoveries may prove especially valuable by providing initial insights into disease pathogenesis. Studying multiple bottlenecked populations may be a powerful strategy.
RVAS Strategy 5: Studying Gene Sets.
Another potentially powerful idea is to focus not just on single genes but on gene sets. Given a gene set G with m genes, the simplest approach is to compare the total number of events (e.g., disruptive alleles) across all genes seen in cases and controls. The analysis is straightforward if each gene in G has the same background frequency fD: the sample size to achieve a given nominal significance level is smaller (relative to detecting a single gene with excess relative risk λ) by a factor of  , where λavg is the average excess relative risk across the set (SI Appendix, Section 9). If 50 genes each confer the same high relative risk, we thus need 50-fold fewer samples to achieve the same nominal significance level for the set as for any individual genes alone. If 20 of 100 genes confer a relative risk of 1 + λ = 11 and the rest have no effect, the sample size required to detect the set is smaller by a factor of ∼8 (=100 g(2)/g(10)) than the size required to detect one of the risk-increasing genes alone.
, where λavg is the average excess relative risk across the set (SI Appendix, Section 9). If 50 genes each confer the same high relative risk, we thus need 50-fold fewer samples to achieve the same nominal significance level for the set as for any individual genes alone. If 20 of 100 genes confer a relative risk of 1 + λ = 11 and the rest have no effect, the sample size required to detect the set is smaller by a factor of ∼8 (=100 g(2)/g(10)) than the size required to detect one of the risk-increasing genes alone.
The analysis is trickier if the background frequency differs across genes. Genes with larger fD will disproportionately affect the variance. On average, these genes will be under weaker selection (because f ≈ μ/s) and thus likely to have smaller effect size λ. More sophisticated LRT methods can be used to improve power. Methods such as overdispersion tests can also allow for both deleterious and protective alleles (50, 51).
The challenge is to select a set likely to be enriched for genes associated with the disease or trait. An obvious set is the genes implicated by CVAS, as genes related to a disease are likely to harbor both common and rare variants. [For example, many genes related to lipid levels harbor both types of variants (7).] CVAS may provide a valuable foundation for RVAS.
Bottom line.
Studying gene sets, especially those identified by CVAS, is likely to be a powerful strategy. Discovering association with a gene set does not reveal precisely which genes are connected with the disease, but the findings can suggest targeted population-based follow-up and laboratory investigation.
RVAS Strategy 6: Studying De Novo Mutations.
An extreme example of a frequency threshold is studying only de novo mutations. The framework above applies directly, with the allele frequency fD,denovo being the mutation rate μD. Focusing only on de novo mutations would seem to be very inefficient, because they are so rare (μD ∼2 × 10−6 for our typical gene). By Eq. 1, detecting a gene in which disruptive alleles increase risk by 20-fold would require ∼100,000 cases.
However, RVAS with de novo mutations can be effective for genes with very large effect sizes. For example, CHD8 was associated with autism with mental retardation (prevalence < 1/300) (52) based on the observation of de novo disruptive mutations in 3 of 1,078 cases (53, 54). Given the mutation rate of CHD8 (μD ∼ 5 × 10−6), this observation is highly improbable (P ∼ 1.7 × 10−6), and the effect size appears to be huge (∼300-fold before correcting for the “winner’s curse”), corresponding to nearly complete penetrance (SI Appendix, Section 2.6 and 10).
The example highlights a subtle advantage of de novo mutations vs. standing variation in the general population. We assumed above that background allele frequencies are known precisely, but they must actually be inferred from data. The observation for CHD8 (three de novo events in cases) is only significant because the extremely low frequency fD,denovo = μD can be estimated with high precision without the need for population studies, based on the ability to infer mutation rates with high precision. A study of CHD8 based on standing variation could be similarly effective given a precise estimate of fD; however, this could require a survey of more than half a million people in the case of strong selection (e.g., s = 0.1).
The comparative advantage for de novo mutations is thus greatest for genes with the largest selection coefficient. The proportion of null alleles that are de novo mutations is largest for such genes. Indeed, under mutation selection balance, the expected proportion of null alleles that are de novo is precisely s.
The gene set approach discussed above can be applied to de novo mutations, with the nice feature that the genes contribute more evenly because the frequency fD,denovo does not depend on the selection coefficient. For example, the set of all 20,000 protein-coding genes shows an excess of de novo variants for autism, indicating that many genes likely contribute (53, 54).
Bottom line.
RVAS with de novo mutations can be valuable for genes with huge effects and large values of s. However, the design is not well suited for finding genes that increase risk by merely 20-fold or selection coefficients of merely 1%. Notably, the approach does not require prior knowledge of population frequencies, which can be a significant advantage.
Prospects for RVAS in Noncoding Regions.
Whereas the methodology for CVAS applies identically to both coding and noncoding regions, extending RVAS to noncoding regions poses major new challenges. The problem is that RVAS requires selecting the genomic regions across which to aggregate variants. The choice is problematic.
-
i.
If one focuses on a single regulatory element, such as a promoter or enhancer, the target size is so small that the CAF is tiny and the required sample size is huge. For example, focusing on a 50-base regulatory element (vs. a 1,500-base coding region) necessitates a 30-fold larger sample.
-
ii.
On the other hand, if one combines all variants across a large intergenic region, most sites will be functionally unimportant; this dilutes the apparent value of λ and thereby inflates the sample size. If ∼5% of intergenic DNA is functional, the signal will be diluted by 20-fold and the sample size will be inflated by 20- to 400-fold (depending on the value of λ).
-
iii.
Outside of coding regions, there are no classes analogous to disruptive variants, which can provide a pure signal largely undiluted by neutral mutations, or silent mutations, which can be discarded as benign.
In addition, scanning the entire genome requires imposing a more stringent threshold for statistical significance, which increases the sample size by ∼50% (55) (SI Appendix, Section 11.1).
Finally, it is unclear whether noncoding regions will harbor many signals to be found by RVAS. Although common variants with moderate effects found by CVAS occur predominantly in noncoding regions, there is reason to think that most rare variants of large effect may lie in coding regions. This expectation is based on the fact that single-base changes may have dramatic effects in coding regions but rarely obliterate the function of noncoding elements. Consistent with this, the inferred selection coefficient in coding regions is about 10-fold higher than in evolutionarily conserved noncoding sequences (56, 57).
Despite these challenges, it is appropriate to explore whole-genome RVAS in certain well-chosen situations. A recent paper (58) reported whole-genome sequence from 962 individuals and searched for an association of HDL cholesterol levels with both common and rare variants. The rare variant analysis involved studying alleles with frequency ≤1% in sliding windows of 4 kb across the genome. A handful of peaks with low P values was identified, although none reach statistical significance.††
Bottom line.
To perform RVAS with reasonable sensitivity in noncoding regions, it will be important to have fairly precise knowledge of the functionally important regulatory sequences related to each human gene to aggregate them together. Without such information, the sample sizes required to detect association will be larger by one to two orders of magnitude. At present, it makes more sense to deploy resources primarily toward whole-exome, rather than whole-genome, sequence. This approach will maximize the number of samples that can be analyzed for coding regions, where the power is currently vastly greater, where the effect sizes are expected to be larger, and where the discoveries are likely to be more immediately actionable.
Variance Explained.
Finally, the analysis above relates directly to the search for missing heritability. For a disease with prevalence π, the proportion of phenotypic variance (on the observed scale) explained by the null alleles in a gene G is
 |
The case of LDLR in early-onset MI discussed above is unusual in that it explains 1.8% of the phenotypic variance (or ∼3.6% of the genetic variance). In contrast, most RVAS findings thus far explain a tiny proportion the phenotypic variance. For example, rare variants in SLC12A3 (also known as NCCT), SLC12A2 (also known as NKCC2), and KCNJ1 (also known as ROMK1) each explain an average of <0.17% of the phenotypic variance in risk of hypertension, whereas rare variants in MTNR1B similarly explain <0.1% of the phenotypic variance in risk of type 2 diabetes (18, 21) (SI Appendix, Section 13.3).
From Eqs. 1 and 4, one can calculate the number of cases needed to detect genes explaining a given fraction of the phenotypic variance. To detect a gene explaining 1% of the phenotypic variance based on studying risk-increasing disruptive alleles, the required number of cases should be ∼2,800, ∼4,700, and ∼6,500 for diseases with prevalences of 1%, 5%, and 10%, respectively (SI Appendix, Section 13.2). After reaching this sample size, one can reasonably conclude that the remaining genes each explain less than 1% of the variance. (For 0.1% of the variance, the sample size should be 10-fold larger.)
Discussion
Some early RVAS efforts were premised on the notion that searching for rare variants in small numbers of patients could reveal the genes underlying disease. However, it is now clear that rare variant studies, like common variant studies, will require tens of thousands of cases and careful statistical analysis to achieve adequate power to detect genes underlying disease.
Here, we describe a conceptual framework for thinking about the design of RVAS. Our goal is not to compare specific statistics that have been proposed but rather to extract insight and intuition about how key factors influence RVAS for common disease. For this reason, we focused on a simple situation: a two-class model analyzed with burden tests using frequency cutoffs.
Our main conclusions are as follows.
Disruptive alleles, being a pure class of nulls, provide a key backbone for RVAS. They are powerful and easy to interpret, as the excess in cases directly reflects the effect size.
Missense alleles may potentially play a valuable role as well, by decreasing the required sample size. However, missense alleles are harder to interpret: they are always mixtures of null and neutral alleles, with the proportions depending on the selection coefficient for each gene (which must be inferred), the frequency threshold used (which must be chosen), and the availability and quality of functional predictions (from experimental or computational analysis). Achieving maximal power and accurate inference of effect sizes requires properly accounting for dilution by neutral missense alleles. In practice, the maximal increase in power corresponds to an ∼2.5-fold decrease in sample size for our typical gene with α = 25%. (The improvement is much smaller for genes with protein sequence under weaker constraint.)
The appropriate sample size for RVAS depends on the mutation rate, selection coefficient, and effect size for null alleles in the gene. Based on Fig. 1, a well-powered RVAS might aim for a discovery sample of at least 25,000 cases for 90% power (or ∼15,000 cases for 50% power), together with a substantial replication set. This sample size should allow detection of most genes where null alleles confer at least a 10-fold effect, as well as those with 5- and 3-fold effects provided that s is not too strong (s < 2 × 10−2 and 5 × 10−3, respectively). Fortunately, RVAS can begin by sequencing existing sample collections that have been used for CVAS, many of which contain many tens of thousands of cases. Even larger samples will ultimately be desirable, which should be feasible as genomic sequencing becomes a routine part of medical care.
Three strategies may accelerate progress, by enabling early discoveries. First, isolated populations resulting from recent bottlenecks should make it easier to detect a subset of genes. Examples include Finland, Iceland, Ashkenazi Jews, Amish, Bedouins, and various endogamous groups in India. Studies across multiple such populations could prove valuable. Second, early signals may be provided by the study of gene sets that are likely to be enriched for disease-associated loci. The best sets may consist of genes implicated by CVAS. [Initial efforts to identify rare variants in genes identified by CVAS have met only limited success. However, this is likely due to the small discovery sets used—typically, only a few hundred cases (23).] Whereas CVAS and RVAS are sometimes thought of as alternatives, they are likely to be complementary. Third, de novo mutations can be a valuable tool for detecting genes with large effect sizes. This strategy is likely to be most effective for diseases and genes under extremely strong selection (e.g., severe autism).
Extending RVAS to noncoding regions might seem like an obvious next step, given that CVAS has identified so many common variants affecting common traits in noncoding regions. However, major challenges must be overcome before this could become practical. Without a clear scheme for aggregating alleles in noncoding regions, RVAS would require a 10- to 100-fold larger sample size to detect comparable effect size in noncoding vs. coding regions. Moreover, there is reason to expect that rare variants of large effects may be skewed toward coding regions.
Our simple framework was chosen to maximize insight and intuition. However, it has limitations, including that it assumes (i) a two-class model for allelic effects, (ii) constant selection across history, (iii) absolute frequency cutoffs, (iv) unrelated cases rather than families, (v) discrete traits rather than quantitative traits, and (vi) no gene × environment interaction.
We elaborate on these points in SI Appendix, Section 14. In particular, we show that incorporating hypomorphic alleles would not alter our conclusions significantly. Hypomorphic alleles will typically be common rather than rare owing to weaker selection (s estimated in the range of 10−3–10−4) (13, 29, 59) and rare hypomorphic alleles will increase detection power only modestly, owing to their smaller effect size (SI Appendix, Section 5.1). Both hypomorphic and countervailing alleles will dilute apparent effect sizes. Although we ignore them for initial gene detection in RVAS, it will be important to consider the possibility of such alleles once a gene has been implicated.
Our paper also addresses certain questions in human population genetics. Contrary to some suggestions, the increase in the number of distinct alleles resulting from the human population explosion over the last ∼1,000 generations has not led to an increase in the total frequency of disease or a greater role for younger alleles in disease. The paper also raises a number of further research questions in human genetics, including improved ways to estimating s and α for each gene and analyzing gene sets comprised of genes with varying properties (60).
There is currently little empirical evidence about (i) whether rare variants associated with a disease will reveal many new genes beyond those already implicated by common variants and (ii) whether the contribution of rare variants to the heritability of common disease will be large or small. Most of the loci discovered so far account for only a fraction of a percentage point of the phenotypic variance (with a few exceptions such as LDLR above), but large, systematic studies will be required to assess the combined contribution across loci. The answer may differ across common diseases, with rare variants likely to play a greater role in disorders such as schizophrenia and autism and, perhaps, cancer. Regardless of their overall contribution, rare variants in specific genes can enable clinical insights about function.
Assuming that large-scale sequencing gets underway for many diseases, the coming years should be an exciting period for human genetics as we finally are able to probe the full genetic architecture underlying human disease.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
†We focus on risk to heterozygous carriers. Our calculations implicitly assume that the risk to individuals carrying two null alleles (λC*) is the same as the risk to heterozygous carriers (λC). Although such individuals may well have higher risk, they are much rarer than heterozygous carriers (because fC is small) and thus their impact on RVAS is typically negligible. [The effective relative risk is increased by fC(λC*/λC), which is ≪1 unless λC*/λC is huge; this case would essentially be a monogenic recessive trait.]
‡We focus on the number n of cases needed when the frequency fC in the population is known perfectly based on an (infinitely) large population survey. We make this assumption in the belief that very large datasets will become available in the coming years and that shared population controls can be used across studies. In the meanwhile, one can estimate fD within a study based on the frequency in either unaffected or random individuals. If a study involves cases and unaffecteds in proportion r and 1 − r, one requires approximately n/(1 − r) cases and n/r controls to detect association, where n is the number of cases given in Eq. 1 (SI Appendix, Section 3.3). For a balanced design, this corresponds to 2n cases and 2n controls.
§ , which is approximately
, which is approximately  for small
for small  (0 ≤
(0 ≤ 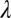 < 1) and
< 1) and  for larger
for larger  (with c in the range 0.7–0.8 for
(with c in the range 0.7–0.8 for  ).
).
¶The expected CAF for new neutral alleles born in a given generation is μC and thus the increase in the expected CAF over k generations cannot exceed kμC (and will be lower because many newborn alleles are lost). It follows that the collection of neutral alleles born since the onset of human population cannot increase the CAF of neutral alleles by more than 5% (= kμC/4μCNeq, where k =1,000 and Neq = 10,000). The actual increase is typically much smaller due to loss of newborn alleles.
||The relevant sampling frame for disease studies involves randomly selecting a chromosome and inspecting it for alleles. Alleles are thus sampled according to their frequency, yielding a frequency-weighted frequency distribution.
**One can also perform an analysis considering only those missense alleles with frequency ≤1% that are predicted to be probably damaging by PolyPhen-2 (that is, RVAS strategy 3). One observes 89 such missense alleles in cases vs. 32 in controls, which corresponds to an apparent excess relative risk λM(1%),PolyPhen2 = 2.5. This result agrees closely with the expectation of λM(1%),PolyPhen2 = 2.3, given the inferred value of s and the frequency threshold of 1%. For this type of analysis (assuming γnull = 80% and γneutral = 20%), the optimal threshold turns out to be T*= 0.15%, which would yield a much higher apparent effect size of λM(T*),PolyPhen2 = 8.2.
††The paper reports that the best nominal P value observed across the genome is 2.7 × 10−8 but does not address whether the value is significant. To correct for scanning the entire genome, one can apply extreme value theory for Orenstein-Uhlenbeck diffusions (SI Appendix, Section 11.1). The probability of such a P value arising by chance somewhere in the genome is ∼70%, and thus the observation is not statistically significant. Genomewide significance at the 5% level corresponds to P ∼ 2 × 10−9 (SI Appendix, Section 11.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322563111/-/DCSupplemental.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109(4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis SE, Chakravarti A, Cohen JC, Hardy J. Mendelian disorders and multifactorial traits: The big divide or one for all? Nat Rev Genet. 2010;11(5):380–384. doi: 10.1038/nrg2793. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492(7429):369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ); International Schizophrenia Consortium (ISC); Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein DB, et al. Sequencing studies in human genetics: Design and interpretation. Nat Rev Genet. 2013;14(7):460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80(4):727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiezun A, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44(6):623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27(7):710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 17.Ahituv N, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80(4):779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnefond A, et al. Meta-Analysis of Glucose and Insulin-Related Traits Consortium (MAGIC) Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44(3):297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 20.Diogo D, et al. Consortium of Rheumatology Researchers of North America; Rheumatoid Arthritis Consortium International. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am J Hum Genet. 2013;92(1):15–27. doi: 10.1016/j.ajhg.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40(5):592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivas MA, et al. National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC); United Kingdom Inflammatory Bowel Disease Genetics Consortium; International Inflammatory Bowel Disease Genetics Consortium. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeo S, et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119(1):70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genet Epidemiol. 2011;35(7):606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stitziel NO, Kiezun A, Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12(9):227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladouceur M, Dastani Z, Aulchenko YS, Greenwood CM, Richards JB. The empirical power of rare variant association methods: Results from sanger sequencing in 1,998 individuals. PLoS Genet. 2012;8(2):e1002496. doi: 10.1371/journal.pgen.1002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Hum Mol Genet. 2005;14(21):3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- 29.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337(6090):100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 32.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173(2):891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177(4):2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jobling M, Hurles M, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples & Disease. New York: Garland Science; 2004. [Google Scholar]
- 35.Kimura M. Diffusion models in population genetics. J Appl Probab. 1964;1(2):177–232. [Google Scholar]
- 36.Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewens WJ. Mathematical Population Genetics: I. Theoretical Introduction. 2nd Ed. New York: Springer; 2004. [Google Scholar]
- 38.Haldane JBS. A mathematical theory of natural and artificial selection. Math Proc Camb Philos Soc. 1927;23(5):607–615. [Google Scholar]
- 39.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 40.Tennessen JA, et al. Broad GO; Seattle GO; NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166(1):351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147(1):32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42(11):969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 45.Haukka J, Suvisaari J, Lönnqvist J. Fertility of patients with schizophrenia, their siblings, and the general population: A cohort study from 1950 to 1959 in Finland. Am J Psychiatry. 2003;160(3):460–463. doi: 10.1176/appi.ajp.160.3.460. [DOI] [PubMed] [Google Scholar]
- 46.Sunyaev SR. Inferring causality and functional significance of human coding DNA variants. Hum Mol Genet. 2012;21(R1):R10–R17. doi: 10.1093/hmg/dds385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest. 1973;52(7):1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levy-Lahad E, et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: Frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet. 1997;60(5):1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 50.Neale BM, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators; Centers for Disease Control and Prevention; Centers for Disease Control and Prevention. Prevalence of autism spectrum disorders—Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61(3):1–19. [PubMed] [Google Scholar]
- 53.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen CT, Wang JC, Cohen BA. The strength of selection on ultraconserved elements in the human genome. Am J Hum Genet. 2007;80(4):692–704. doi: 10.1086/513149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kryukov GV, Schmidt S, Sunyaev S. Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet. 2005;14(15):2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- 58.Morrison AC, et al. Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45(8):899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci USA. 2009;106(10):3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437(7062):1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



