Significance
Methylation of cytosine bases in DNA is an epigenetic modification that influences gene expression. TET (Ten-Eleven Translocation) enzymes regulate DNA methylation status and facilitate DNA demethylation by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and further oxidation products in mammalian genomes. Of the three mammalian TET proteins, Tet1 and Tet2 are the major regulators of 5hmC levels in mouse embryonic stem (ES) cells. We show that Tet1 and Tet2 have distinct roles in mouse ES cells: Tet1 primarily regulates 5hmC levels at gene promoters and transcription start sites, whereas Tet2 mainly regulates 5hmC levels in gene bodies and exon boundaries of highly-expressed genes and exons respectively. Our results suggest a complex interplay between the functions of Tet1 and Tet2 proteins in mESC.
Keywords: DNA methylation, DNA hydroxymethylation, epigenetics, DNA demethylation
Abstract
Dioxygenases of the Ten-Eleven Translocation (TET) family are 5-methylcytosine oxidases that convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and further oxidation products in DNA. We show that Tet1 and Tet2 have distinct roles in regulating 5hmC in mouse embryonic stem cells (mESC). Tet1 depletion diminishes 5hmC levels at transcription start sites (TSS), whereas Tet2 depletion is predominantly associated with decreased 5hmC in gene bodies. Enrichment of 5hmC is observed at the boundaries of exons that are highly expressed, and Tet2 depletion results in substantial loss of 5hmC at these boundaries. In contrast, at promoter/TSS regions, Tet2 depletion results in increased 5hmC, potentially because of the redundant activity of Tet1. Together, the data point to a complex interplay between Tet1 and Tet2 in mESC, and to distinct roles for these two proteins in regulating promoter, exon, and polyadenylation site usage in cells.
The TET proteins TET1, TET2, and TET3 constitute a new family of dioxygenases that use molecular oxygen and the cofactors Fe(II) and 2-oxoglutarate to convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine DNA (together referred to as oxidized methylcytosines or oxi-mC) (1–5). The level of 5hmC is ∼10% the level of 5mC in mouse embryonic stem cells (mESC) (1), whereas 5fC and 5caC are much less abundant (3). Together, Tet1 and Tet2 are responsible for essentially all of the 5hmC present in mESC (6). Tet1 is known to be highly expressed in the blastula (from which ES cells are derived) and in primordial germ cells, whereas Tet2 is expressed in these and several other cell types (7). Loss-of-function mutations in TET2 are associated with the development of myeloid malignancies in humans (7–10). mESC differentiation results in a global decrease of 5hmC, coupled to down-regulation of Tet1 and Tet2 mRNAs and up-regulation of Tet3 mRNA (6).
Increasing evidence suggests that Tet1 and Tet2 have distinct functions in mESC. First, teratomas generated by injection of Tet1-depleted mESC showed increased endoderm, reduced neuroectoderm, and appearance of trophoblastic giant cells, whereas teratomas from Tet2-depleted mESC did not (6, 11). Second, using a cell fusion-based system, Tet1 and Tet2 were shown to have different roles in reprogramming: Tet1 is responsible for imprint erasure in embryonic germ cells, whereas Tet2 is required for 5hmC accumulation at pluripotency loci (such as Oct4) within the somatic genome (12). Third, Tet1 possesses a CXXC domain that likely directs it to DNA containing unmethylated CpG sequences, whereas Tet2 lost its CXXC domain because of a chromosomal inversion event that occurred during evolution (2). The separated region now encodes a unique gene product, IDAX/CXXC4, which also binds unmethylated CpG sequences and continues to be a functional regulator of Tet2 (2, 13). However, IDAX is not expressed in undifferentiated ES cells, suggesting that Tet1 and Tet2 might be directed to different genomic regions in these cells. Fourth, Tet1 was shown to be strongly associated with chromatin in mESC, whereas Tet2 is poorly chromatin-associated (14). However, whether Tet1 and Tet2 regulate 5hmC levels at different genomic regions has not yet been elucidated.
Here, we investigated the roles of Tet1 and Tet2 in mESC. We compared the transcriptional profiles of wild-type, Tet1-depleted, and Tet2-depleted mESC by using RNA sequencing (RNA-seq) and mapped 5hmC genome-wide in the same cells by immunoprecipitation (IP) of bisulfite-treated DNA with antibodies against cytosine-5-methylenesulphonate (CMS) (7, 15, 16). We show that Tet1 and Tet2 regulate 5hmC at different genomic regions in mESC: Tet1 is responsible for 5hmC production at promoter/TSS regions, whereas Tet2 primarily regulates 5hmC levels in gene bodies. RNA-seq analysis showed that Tet1 more frequently altered the expression of entire transcripts, whereas both Tet1 and Tet2 selectively regulated exon expression within transcripts. Tet2 depletion resulted in a significant loss of 5hmC at the boundaries of high-expressed exons, although this loss did not predict whether exon expression was up- or down-regulated. Our findings point to distinct roles for Tet1 and Tet2 in regulating 5hmC deposition in mESC.
Results
Distinct Genome-Wide Distribution of 5hmC in Tet1- Versus Tet2-Depleted mESC.
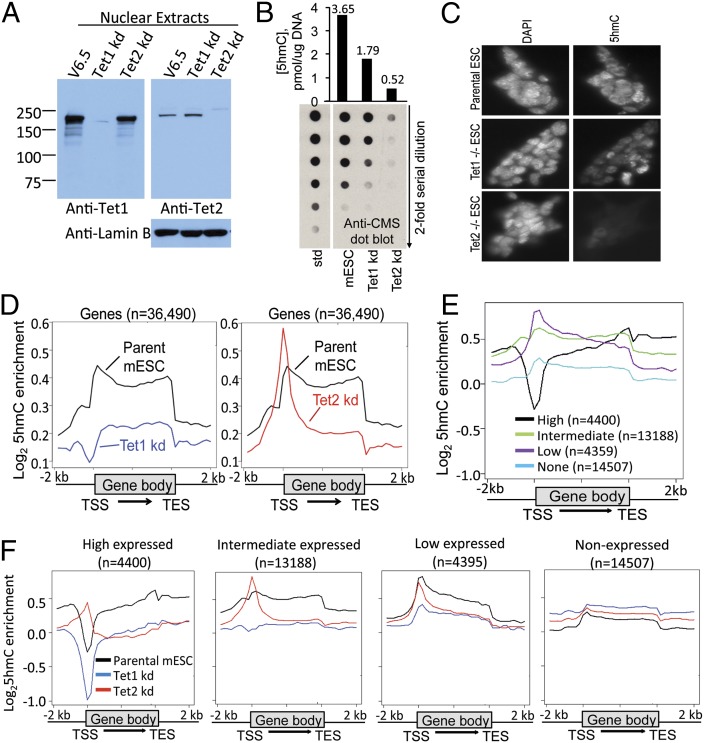
We used shRNAs directed against Tet1 and Tet2 to generate V6.5 mouse embryonic cells (mESC) stably depleted of Tet1 or Tet2 (here termed Tet1 kd and Tet2 kd mESC, respectively). Protein levels of Tet1 and Tet2 were strikingly diminished relative to parental V6.5 mESC; moreover, Tet1-depleted cells showed little or no change in Tet2 mRNA and protein levels and vice versa (Fig. 1A and SI Appendix, Fig. S1 and Table S1). Tet3 mRNA was expressed at much lower levels than Tet1 or Tet2 mRNA, and its expression was unchanged in Tet2 kd mESC and only slightly increased in Tet1 kd mESC (SI Appendix, Table S1). Tet2 depletion resulted in a much greater decrease in genomic 5hmC levels than Tet1 depletion (∼50% vs. ∼15% of control levels respectively, by anti-CMS dot blot; Fig. 1B), as also observed upon transient transfection using Tet1 and Tet2 siRNAs (6) and in mESC generated from Tet1 and Tet2 gene-disrupted mice (17). Immunocytochemistry confirmed that Tet1 kd mESC showed a moderate decrease in staining for 5hmC, whereas Tet2-deficient mESC showed almost no detectable 5hmC (Fig. 1C).
Fig. 1.
Distinct 5hmC distribution in Tet1 kd or Tet2 kd mESC. (A) Western blot showing that v6.5 mESC with stable shRNA-mediated depletion of Tet1 and Tet2 show ∼90% depletion of Tet1 and Tet2 protein levels, with no or little change in protein levels of the other Tet protein. (B) Levels of 5hmC in parental v6.5 ESC and ESC stably depleted of Tet1 or Tet2, quantified by dot blot using anti-CMS antiserum. (Lower) A dot-blot with twofold dilutions of genomic DNA is shown. (Upper) Quantification based on a standard curve of twofold dilutions of a 5hmC-containing oligonucleotide. (C) Immunocytochemical staining of 5hmC in parental, Tet1 or Tet2 deleted mESC. Left, DAPI; Right, 5hmC. Consistent with the dot-blot assay, 5hmC staining is weaker in Tet2 KO mESC than that in Tet1 KO mESC. (D) Distribution of averaged 5hmC enrichment at all genes in parental, Tet1 kd, or Tet2 kd mESC. (E) Distribution of averaged 5hmC enrichment in parental mESC ranked by gene expression. (F) Distribution of average 5hmC enrichment at high, intermediate, low and non-expressed genes in parental (black), Tet1 kd (blue) and Tet2 kd (red) mESCs. For a heatmap representation of the data in Fig. 1 E and F, see SI Appendix, Fig. S2.
We mapped the genomic distribution of 5hmC in parental V6.5, Tet1 kd, and Tet2 kd mESC by IP of bisulfite-treated DNA with antibodies against CMS (7, 15, 16). Compared with parental mESC, Tet1 kd mESC displayed decreased 5hmC both at transcription start sites (TSS) and in the gene body; in Tet2 kd cells, the decrease in 5hmC was largely restricted to the gene body, with the remaining 5hmC disproportionately present at the TSS (Fig. 1D). The effects of Tet1 and Tet2 depletion showed a striking dependence on gene expression levels (Fig. 1 E and F and SI Appendix, Fig. S2). As reported (18, 19), the most highly expressed (top 10%) genes in ES cells have low 5hmC at their promoter/TSS regions but high 5hmC in their gene bodies (Fig. 1E and SI Appendix, Fig. S2A); in this category of genes, Tet1 depletion resulted in even further loss of 5hmC at the promoter and in the gene body, whereas Tet2 depletion resulted in increased 5hmC at the promoter/TSS with a major loss in the gene body (Fig. 1F, Far Left and Center Left and SI Appendix, Fig. S2B). Genes expressed at low levels (bottom 10%) had a small peak of 5hmC at their promoter/TSS regions and low 5hmC in their gene bodies, both of which were diminished by Tet1 depletion, but again Tet2 kd mESC showed diminished 5hmC only in the gene body of this category of genes (Fig. 1F, Center Right). Nonexpressed genes had barely any 5hmC (Fig. 1F, Far Right, and SI Appendix, Fig. S2A).
To further examine 5hmC at promoters, we used the classification of high, intermediate, and low CpG promoters—high-CpG promoters (HCP), which generally show low DNA methylation (20). On average, HCP—which correspond to CpG island promoters of highly expressed genes—lost 5hmC in Tet1 kd cells but gained 5hmC in Tet2 kd cells (SI Appendix, Fig. S2C); this effect was not observed for intermediate and low CpG promoters.
Together, these data show clearly that Tet1 and Tet2 control the deposition of 5hmC at distinct genomic regions in mESC, albeit with some overlap: Gene body 5hmC is positively correlated with gene transcription in parental mESC and is largely deposited by Tet2, whereas promoter 5hmC is negatively correlated with gene transcription and is largely deposited by Tet1.
Gain and Loss of 5hmC in Tet2 kd mESC.
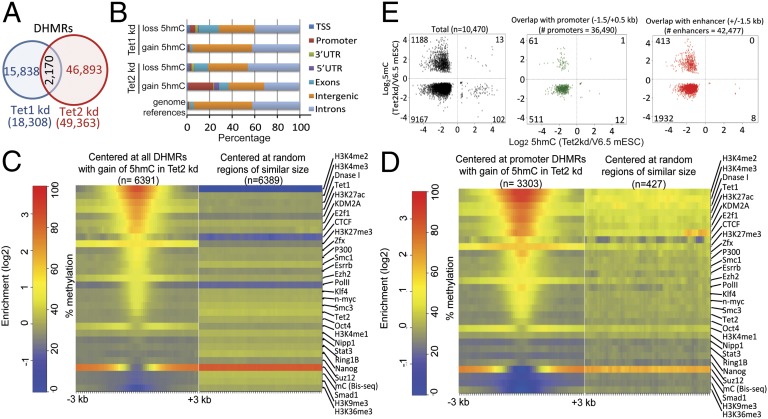
To further investigate the differences in 5hmC distribution between Tet1 and Tet2 kd mESC, we took a genome-wide approach, as opposed to the gene-centric approach of Fig. 1. Comparing 5hmC levels in adjacent (nonoverlapping) 300-bp genomic windows, we found that Tet1 kd and Tet2 kd mESC significantly lost 5hmC at 8,965 and 60,023 windows, respectively (SI Appendix, Fig. S3A), consistent with the greater overall loss of 5hmC upon Tet2 versus Tet1 depletion (Fig. 1 B and C). Unexpectedly, a substantial fraction of 300-bp windows showed an apparent significant gain rather than loss of 5hmC (SI Appendix, Fig. S3A), although the amount of 5hmC in these windows was lower than for windows that lost 5hmC (SI Appendix, Fig. S3B). We defined differentially hydroxymethylated genomic regions (DHMRs) in Tet1 or Tet2 kd mESC compared with parental mESC by merging adjacent windows showing the same direction of change of 5hmC. There was only a small overlap of DHMRs in Tet1 kd versus Tet2 kd mESC (Fig. 2A), again showing that Tet1 and Tet2 control 5hmC levels at different regions of the genome. The majority of the overlap (90%) was at regions with loss of 5hmC in both cell types (SI Appendix, Fig. S3C). There was no evidence for increased 5hmC enrichment after Tet2 knockdown in regions where the bulk of 5hmC was deposited by Tet1 (SI Appendix, Fig. S3D), arguing against the possibility that the global loss of 5hmC in Tet2 kd mESC results in apparent read enrichment at regions whose 5hmC levels are in fact unchanged. In fact, detailed examination identified several promoter regions with no 5hmC in parental V6.5 mESC, which nevertheless showed strong gain of 5hmC in Tet2 kd mESC (SI Appendix, Fig. S4).
Fig. 2.
Distribution of DHMRs in Tet1 kd and Tet2 kd mESC. (A) Venn diagram showing the overlap of DHMRs in Tet1 kd compared with Tet2 kd mESC. (B) Genomic distribution of DHMRs in Tet1 kd or Tet2 kd mESC. DHMRs with gain of 5hmC in Tet2 kd specifically show enrichment at promoter-TSS regions. An alternative representation is shown in SI Appendix, Fig. S5A. (C and D) Heat map of histone modification and transcription factor enrichment at all DHMRs with gain of 5hmC (C), or DHMRs with gain of 5hmC at promoter regions (D), in Tet2kd mESC. (E) Relation of differentially methylated (y axis) and hydroxymethylated (x axis) 300-bp windows (assessed by MeDIP and CMS-IP, respectively) in Tet2 kd mESC. Only 300-bp windows with significant changes in both 5hmC and 5mC are shown. In all cases (Left, all windows; Center, windows overlapping with promoters; Right, windows overlapping with enhancers defined by the presence of H3K4me1 but the absence of H3K4me3), the majority of windows show loss of both 5hmC and 5mC.
Notably, DHMRs that gained 5hmC in Tet2 kd cells were strongly enriched for promoters/ transcription start sites and 5′ untranslated regions (UTRs) of the transcribed regions of genes (Fig. 2B and SI Appendix, Fig. S5A; for detailed information, see SI Appendix, Table S2). Since the only Tet proteins expressed at high levels in mESC are Tet1 and Tet2 (ref. 6; SI Appendix, Table S1), and because Tet1 controls 5hmC at promoters (Fig. 1 and SI Appendix, Fig. S2B), it seemed plausible that Tet1 was responsible for 5hmC enrichment at this subset of promoters in Tet2 kd mESC. Indeed, DHMRs that showed gain of 5hmC in Tet2 kd mESC were enriched for Tet1 binding sites (21), which tend to be present at regions of DNase I hypersensitivity that bear histone marks associated with promoters and active enhancers (H3K4me2, H3K4me3 and H3K27Ac) (22) (Fig. 2 C and D). DHMRs that lost 5hmC upon Tet1 depletion also possessed these histone marks as expected, albeit to a lesser degree (SI Appendix, Fig. S5B, Lower Left); however the marks were not present, or were barely present, at DHMRs that gained 5hmC upon Tet1 depletion or lost 5hmC upon Tet2 depletion (SI Appendix, Fig. S5B, Upper Left and Lower Right). These features were even more obvious in DHMRs specific for either Tet1 or Tet2 (SI Appendix, Fig. S5C, Upper Right and Lower Left), but were much less pronounced at DHMRs not localized at promoters (SI Appendix, Fig. S5D). A plausible explanation is that in the absence of Tet2, Tet1 occupies Tet2 target sites in promoters and redundantly regulates 5hmC in mESC.
Relation Between 5hmC and DNA Methylation.
The oxidized methylcytosines produced by TET proteins are likely to be players in several pathways of DNA demethylation (1, 4, 5), thus loss of TET proteins is generally expected to result in increased DNA methylation. To test this hypothesis, we performed MeDIP experiments to assess 5mC distribution in parental and Tet2 kd mESC. Of a total of 10,470 300-bp windows with significant changes in both 5hmC and 5mC, the vast majority (9,167; ∼88%) showed loss of both 5hmC and 5mC and only a minority (1,188; 11%) showed the “expected” loss of 5hmC and gain of 5mC (Fig. 2E, Left). The identical pattern was observed when considering the small subset of 585 windows with significant changes in both 5hmC and 5mC that overlapped with promoters (Fig. 2E, Center): 511 of these (∼87%) showed losses of both 5hmC and 5mC.
Two previous studies reported that 5hmC is enriched at enhancers (23, 24). We analyzed DHMRs at 42,477 enhancers marked by the presence of H3K4me1 but the absence of H3K4me3 (25). Of these enhancers, 2353 showed significant changes in both 5hmC and 5mC; again, the majority of these (1,932; 82%) also showed decreased 5mC in Tet2-depleted cells (Fig. 2E, Right).
Overall, loss of function of a single TET protein does not necessarily lead to loss of 5hmC and a corresponding gain of 5mC, as often assumed from the postulated roles of TET proteins and oxidized methylcytosines in DNA demethylation pathways (5). Nevertheless, it was possible to find clear examples of regions showing this pattern (SI Appendix, Fig. S6A), as well as the more frequent pattern of loss of both 5hmC and 5mC (SI Appendix, Fig. S6B).
Transcriptional Changes in Tet1- or Tet2-Depleted mESC.
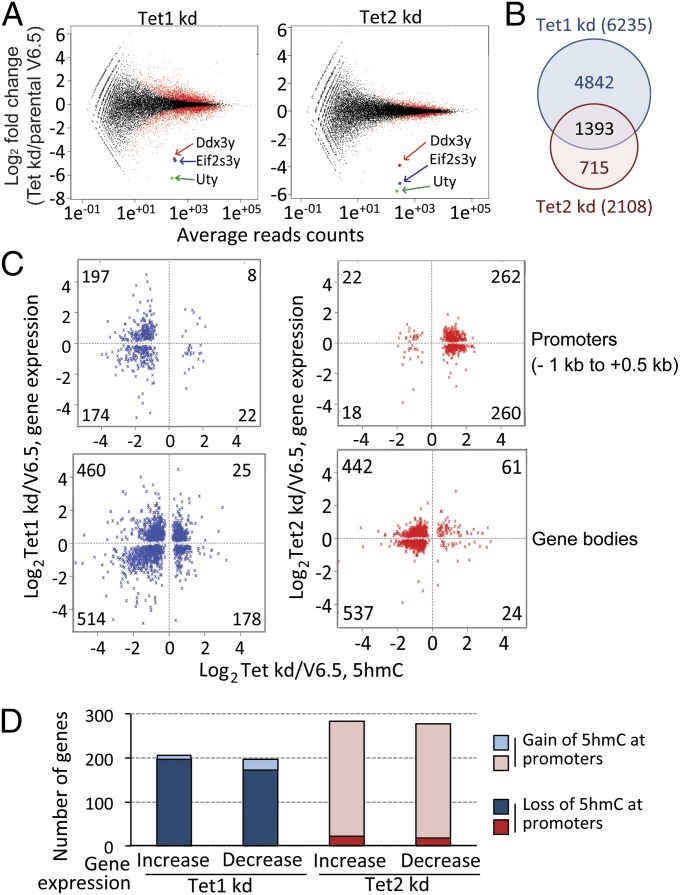
To investigate the relation between 5hmC and gene expression, we performed RNA-seq of polyA+ RNA from Tet1 kd and Tet2 kd mESC. Tet1 or Tet2 depletion altered the expression of 6,235 and 2,108 genes, respectively, with an equivalent number of genes being up- or down-regulated in each case (Fig. 3 A and B and SI Appendix, Table S1). Of these genes, 1393 showed overlapping regulation (Fig. 3B and SI Appendix, Fig. S7); these genes constitute 66.0% of Tet2-regulated genes compared with only 22.3% of Tet1-regulated genes, indicating that the majority of Tet2-regulated genes can also be regulated by Tet1. Notably, three genes located on the Y chromosome—Eif2s3y, Ddx3y, and Uty—showed a considerable decrease in expression in both Tet1 kd and Tet2 kd mESC (Fig. 3A and SI Appendix, Fig. S8A). a result validated by RT-PCR (SI Appendix, Fig. S8B) and supported by a previous microarray analysis of gene expression in Tet1 kd mESC (21). The decreased expression of these genes was not due to loss of the entire Y chromosome, as judged by genomic PCR for two Y chromosome genes, Zfy1 and Sry (SI Appendix, Fig. S8C).
Fig. 3.
Differentially expressed genes and 5hmC distribution in Tet1 kd or Tet2 kd mESC compared with parental mESC. (A) Genes differentially expressed in Tet1 kd and Tet2 kd compared with parental mESC were identified using DESeq. The numbers of genes significantly up- or down-regulated in Tet1 and Tet2 kd mESC are indicated. Three genes (Uty, Ddx3y, Eif2s3y) located on the Y chromosome were highly down-regulated in both Tet1 and Tet2 kd mESC. (B) Venn diagram showing the overlap of differentially expressed genes in Tet1 kd and Tet2 kd mESC (also see SI Appendix, Fig. S7). (C) Correlation of significant change of gene expression (y axis) with promoter (−1 kb to +0.5 kb relative to the TSS; Upper) and gene body (Lower) 5hmC (x axis) in Tet1 kd (blue) and Tet2 kd (red) mESC. The figure shows all genes for which RNA-seq and promoter or gene body CMS-IP data are available. (D) Graphical representation of C.
To relate these gene expression data to 5hmC levels, we analyzed the 5hmC data of Fig. 1 at the level of individual promoters. Consistent with the average profiles shown in Fig. 1D, detailed views of individual promoter regions (−1 kb to +0.5 kb) confirmed that Tet1 kd tended to lose 5hmC, whereas Tet2 kd mESC tended to gain 5hmC at promoters relative to parental mESC (SI Appendix, Fig. S9). This pattern was even stronger when we examined only the subset of genes that showed significant differential expression relative to parental mESC as well as a significant change of 5hmC at their promoter/TSS regions (Fig. 3 C and D and SI Appendix, Table S1), confirming that Tet1 maintains, whereas Tet2 limits, 5hmC levels at gene promoters. For this minority of genes (401 and 562, respectively) that showed significant changes in gene expression and 5hmC levels at the promoter in Tet1 or Tet2 kd mESC, there was no correlation of 5hmC changes with changes in gene expression, because loss of promoter 5hmC in Tet1 kd mESC and gain of promoter 5hmC in Tet2 kd mESC could both be associated with either increased or decreased expression of the corresponding genes (Fig. 3 C, Upper and D). The same was true for 5hmC at gene bodies: Although more genes lost 5hmC in gene bodies in both Tet1 and Tet2 kd mESC, there was no correlation of loss or gain of gene body 5hmC with the direction of changes in gene expression. SI Appendix, Fig. S10 shows individual examples of genes that lose promoter 5hmC in Tet1 kd mESC and gain promoter 5hmC in Tet2 kd mESC, but nevertheless can show either up- or down-regulation of gene expression.
5hmC at Exon Boundaries in Tet1- or Tet2-Depleted mESC.
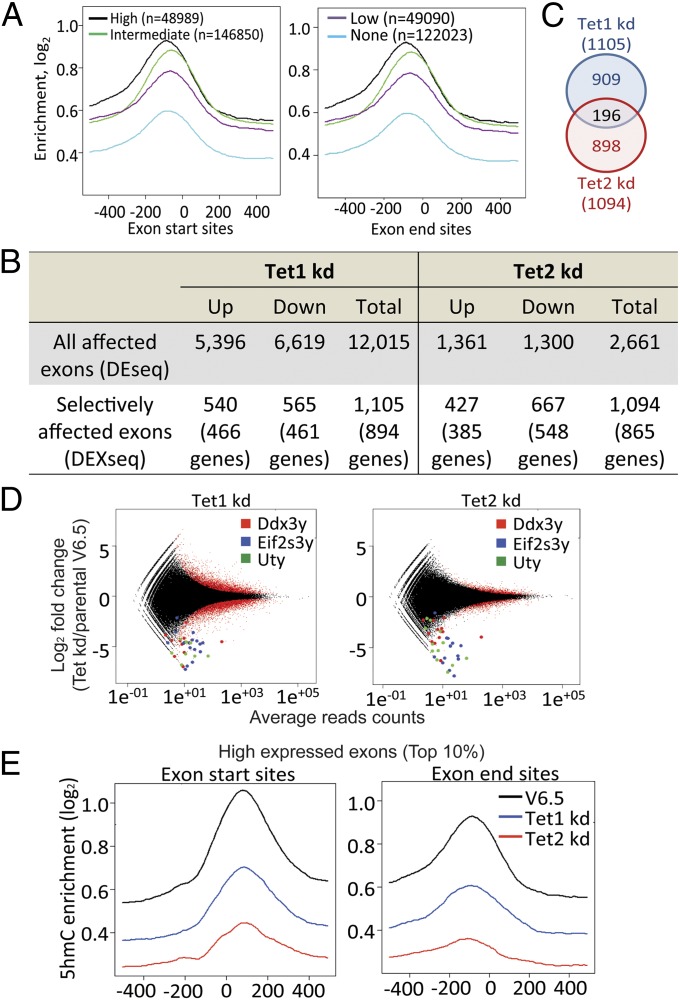
Notably, our RNA-seq analysis showed a clear enrichment of 5hmC near exon start and end sites, particularly prominent in the 10% of most highly expressed exons (Fig. 4A). Reanalyzing our RNA-seq data by DESeq. (26) and DEXSeq. (27) (Fig. 4 B–D), we found that Tet2 kd mESC showed altered expression of a much smaller number of exons relative to parental mESC than did Tet1 kd mESC (2,661 versus 12,015 exons, respectively; Fig. 4B), consistent with the smaller overall change in gene expression observed in Tet2 kd mESC relative to Tet1 kd mESC (Fig. 3A). However, Tet2 kd ESC showed selective exclusion or inclusion of exons within a transcript far more frequently (1,094/2,661; 41.1%) than did Tet1 kd mESC (1,105/12,015; 9.2%) (Fig. 4B), indicating that Tet1 more frequently alters the expression of entire transcripts whereas both Tet1 and Tet2 can selectively regulate exon expression within transcripts. Of the selectively regulated exons, 45% and 37% corresponded to annotated alternatively used exons in Tet1 kd and Tet2 kd mESC, respectively (SI Appendix, Fig. S11 and Table S6) (28). The total number of differentially affected exons was similar between Tet1 and Tet2 kd mESC (1,105 and 1,094, respectively), but only 196 of these exons showed overlapping regulation (Fig. 4C and SI Appendix, Table S6). Thus, exons selectively regulated by Tet1 within larger transcripts do not overlap substantially with exons selectively regulated by Tet2.
Fig. 4.
Differential exon use and 5hmC distribution at exons in parental, Tet1 kd, and Tet2 kd mESC. (A) Enrichment of 5hmC at exon start and exon end sites for exons with high (top 10%, black), intermediate (10–90%, green), low (bottom 10%, purple), or no (blue) expression in parental mESC. (B) Numbers of differentially expressed exons in Tet1 kd and Tet2 kd. All affected exons, exons located within differentially expressed genes; selectively affected exons, exons that are differentially expressed relative to the entire transcript. (C) Venn diagram showing the overlap of selectively affected exons in Tet1kd or Tet2kd mESC. (D) Differentially expressed exons were analyzed by DEseq. Exons located within the three most significantly down-regulated genes on the Y chromosome (Uty, Ddx3y, and Eif2s3y) are highlighted. (E) Distribution of averaged 5hmC enrichment at exon start (Left) and end (Right) sites for the top 10% of high-expressed exons in parental, Tet1 kd and Tet2 kd mESC.
Querying only the 10% most highly expressed exons, we found that these exons show the highest levels of 5hmC at their boundaries, and both Tet1 and Tet2 depletion led to decreased 5hmC levels on average, but Tet2 depletion had a more striking effect (Fig. 4E). For exons with intermediate levels of expression, Tet1 and Tet2 depletion led to equivalent losses of 5hmC at exon boundaries (SI Appendix, Fig. S12), whereas for exons with the lowest levels of expression (bottom 10%), the effect of Tet1 depletion was greater than that of Tet2 depletion (SI Appendix, Fig. S12). Thus, Tet2 maintains 5hmC at exon boundaries of highly expressed genes, whereas Tet1 seems to be responsible for the lower level of 5hmC observed at exon boundaries of poorly expressed genes. Reanalysis of the data at the level of individual exons confirmed the tendency for a more pronounced loss of 5hmC in Tet2 kd mESC compared with Tet1 kd mESC near exon start and end sites (SI Appendix, Fig. S13 A–E). However, a change of 5hmC at exon boundaries did not necessarily predict a change in exon expression, because both up- and down-regulated exons showed loss of 5hmC (see examples for Tet2 kd mESC in SI Appendix, Fig. S14).
Discussion
Our studies establish that Tet1 and Tet2 have distinct functions in mESC. Despite some overlap, Tet1 is the major regulator of 5hmC levels at promoter/TSS regions, whereas a primary function of Tet2 is to regulate 5hmC in gene bodies, especially those of high-expressed genes (Fig. 1 and SI Appendix, Fig. S2). This dichotomy may reflect the fact that Tet1 possesses a CXXC domain, whereas Tet2 does not (1, 2, 13, 19); CXXC domains generally bind unmethylated CpGs (5, 13), explaining the preferential localization of Tet1 at CpG islands and high CpG promoters that largely lack methylation (29, 30). Our data suggest that Tet2 may be responsible for the gene body 5hmC observed in diverse cell types and tissues, including mouse and human ESCs, mouse liver, mouse brain, and human melanomas (15, 18, 19, 21, 24, 31–38), as well as the enrichment of 5hmC at intron-exon boundaries in brain and primordial germ cells (39, 40). The ability of Tet2 to control 5hmC deposition at gene bodies may reflect its reported association with the SET1/COMPASS complex (41), which travels with RNA polymerase II (42). Previous studies showed that gene body methylation is also positively correlated with transcription levels (43–46), but these studies were performed using bisulfite-based methods, which cannot distinguish between 5mC and 5hmC (47).
Notably, loss of Tet2 resulted in an apparent increase of 5hmC at promoter/TSS (Figs. 1 and 2). One possibility is that in the absence of Tet2, Tet1 occupies a subset of Tet2-binding sites at promoter/TSS regions and is enzymatically more active than Tet2 at these regions. Notably, however, the vast majority (>97%) of changes in 5mC were observed at promoters and enhancers that lost 5hmC; of these promoters and enhancers, the majority (>88% of promoters, >82% of enhancers) also lost 5mC (Fig. 2E). We previously observed a similar result—a decrease in DNA methylation at the majority of tested CpG sites—in bone marrow samples from patients with myeloid malignancies and low 5hmC, compared with healthy control individuals and patients with normal high levels of 5hmC (7). As postulated here for Tet1 in mESC, Tet2-binding sites may be occupied in the bone marrow cells by a redundantly acting Tet enzyme, in this case most likely Tet3. Indeed, redundant actions of Tet proteins may explain the confusing findings of no relation, or only a modest relation, between loss of function of Tet proteins and 5mC/5hmC levels at their binding sites in the genome (reviewed in ref. 5).
Consistent with previous reports (18), our study shows that in resting mESC, promoter and gene-body 5hmC are correlated with low and high gene expression, respectively (Fig. 1 and SI Appendix, Fig. S2). Approximately equal numbers of genes and exons were up- and down-regulated in both Tet1 and Tet2 kd mESC (Figs. 3A and 4B), a finding seemingly at odds with our findings that (i) Tet1 and Tet2 are primarily responsible for promoter and gene-body 5hmC and (ii) that promoter 5hmC correlates with low gene expression, whereas gene body 5hmC correlates with high gene expression. Because Tet1 depletion decreases promoter 5hmC, which is associated with low gene expression, Tet1 depletion might be expected to result primarily in decreased gene expression; similarly, because Tet2 depletion increases promoter 5hmC and simultaneously decreases gene body 5hmC, Tet2 depletion might be expected to result primarily in increased gene expression. However, many of the observed changes in gene expression are likely to be indirect. For instance, if Tet2 depletion resulted in decreased expression of a widely acting transcriptional repressor, it would indirectly cause up-regulation of a large number of target genes.
A major finding of this study is that the changes in gene/exon expression observed in Tet1 and Tet2 kd mESC showed no clearcut correlation with changes in 5hmC levels at promoter/exon boundaries (Fig. 3C and SI Appendix, Fig. S9 and S12). Possible reasons include (i) the compensating/redundant and (ii) indirect effects of Tet1 and Tet2 kd as discussed above; (iii) the fact that our measurements were all made in resting mESC, whereas the effects of Tet depletion may be most striking during or after differentiation; (iv) possible effects of Tet1 and Tet2 depletion that do not depend on their catalytic functions (14, 41, 48). Analysis of individual genes that show up- or down-regulated expression together with loss or gain of promoter 5hmC (SI Appendix, Table S2) will likely be most informative in this regard.
Overall, our study distinguishes the effects of Tet1 and Tet2 in regulating 5hmC, gene expression, and exon use in mESC. Our primary finding—that Tet1 and Tet2 differ in terms of the genomic regions at which they preferentially deposit 5hmC—point to distinct roles for Tet1 and Tet2 in regulating 5hmC deposition and exon use in mESC. The biological roles of oxidized methylcytosines are still not clearly understood, and a detailed analysis of the differential properties of Tet1 and Tet2 will increase our understanding of how TET proteins and oxi-mC affect gene expression and the use of alternative promoters, exons, and polyadenylation sites in mESC as well as differentiated cells.
Experimental Procedures
CMS-IP.
CMS-IP was performed as described. Briefly, DNA fragments were ligated with methylated adaptors and treated with sodium bisulfite (Invitrogen). The denatured DNA fragments were incubated with 1 μL of our in-house anti-CMS antiserum in 1× IP buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl, and 0.05% Triton X-100) for 2 h at 4 °C, and precipitated with protein G beads. Precipitated DNA was eluted with proteinase K, purified with phenol chloroform, amplified, and enriched by four to six cycles of PCR using Pfu TurboCx hotstart DNA polymerase (Stratagene). DNA sequencing was carried out using Illumina Genome Analyzer 2 and HiSeq sequencing systems.
RNA-seq.
Total RNA was isolated with an RNeasy kit (Qiagen). Four hundred micrograms of total RNA were isolated and selected twice with Ambion MicroPoly(A)Purist Kit. cDNA conversion and library preparation were performed by using SOLiD RNA-seq library prep kit. RNA-seq was performed on SOLiD 4 instrument. Each sample contains three biological replicates.
Supplementary Material
Acknowledgments
We thank Drs. Harri Lähdesmäki and Tarmo Äijö (Aalto University School of Science) for suggestions on data analysis and Dr. Bing Ren (University of California at San Diego) and Suhua Feng (University of California, Los Angeles) for assistance with next generation sequencing. Y.H. is supported by a postdoctoral Fellow Award from the Leukemia and Lymphoma Society, L.C. by a Feodor Lynen postdoctoral Research Fellowship from the Alexander von Humboldt Foundation, W.A.P. by an National Science Foundation predoctoral graduate research fellowship and a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research, and J.K. by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. S.E.J. is an Investigator of the Howard Hughes Medical Institute. This study was supported by National Institutes of Health Grants R01 HL114093, U19 AI100275 (to A.R. and B.P.), R01 GM60398 (to S.E.J.), R01 AI44432, HD065812, and CA151535, California Institute of Regenerative Medicine Grant RM1-01729, and Leukemia and Lymphoma Society Translational Research Program Grant 6187-12 (to A.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this study have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE50198 and GSE50200).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322921111/-/DCSupplemental.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 9.Tefferi A, Lim KH, Levine R. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;361(11):1117. doi: 10.1056/NEJMc091348. author reply 1117–1118. [DOI] [PubMed] [Google Scholar]
- 10.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccolo FM, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell. 2013;49(6):1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko M, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497(7447):122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49(4):645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Pastor WA, Zepeda-Martínez JA, Rao A. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat Protoc. 2012;7(10):1897–1908. doi: 10.1038/nprot.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 23.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13(1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10(15):2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 32.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szulwach KE, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7(6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150(6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3(2):291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 39.Khare T, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19(10):1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32(5):645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shilatifard A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315(5815):1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi H, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8(1):e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493(7433):561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.