Significance
This study reports on the beneficial effects of forcing high expression of the transcription factor activating transcription factor 3 (ATF3) in ALS. ALS is a noncurable adult-onset disease that attacks motor neurons, resulting in paralysis and death. ATF3 overexpression in motor neurons in an ALS mouse model modifies gene expression and drives the neurons into a prosurvival and proregenerative state, increasing motor neuron survival and maintaining axonal connection with muscle by promoting axonal sprouting. ATF3 overexpression results in markedly improved muscle strength and function and delayed disease onset but only slightly increased lifespan. Molecular mechanisms that promote axonal sprouting could substantially improve quality of life in ALS, although additional approaches will be required to overcome progressive motor neuron deterioration.
Abstract
ALS is a fatal neurodegenerative disease characterized by a progressive loss of motor neurons and atrophy of distal axon terminals in muscle, resulting in loss of motor function. Motor end plates denervated by axonal retraction of dying motor neurons are partially reinnervated by remaining viable motor neurons; however, this axonal sprouting is insufficient to compensate for motor neuron loss. Activating transcription factor 3 (ATF3) promotes neuronal survival and axonal growth. Here, we reveal that forced expression of ATF3 in motor neurons of transgenic SOD1G93A ALS mice delays neuromuscular junction denervation by inducing axonal sprouting and enhancing motor neuron viability. Maintenance of neuromuscular junction innervation during the course of the disease in ATF3/SOD1G93A mice is associated with a substantial delay in muscle atrophy and improved motor performance. Although disease onset and mortality are delayed, disease duration is not affected. This study shows that adaptive axonal growth-promoting mechanisms can substantially improve motor function in ALS and importantly, that augmenting viability of the motor neuron soma and maintaining functional neuromuscular junction connections are both essential elements in therapy for motor neuron disease in the SOD1G93A mice. Accordingly, effective protection of optimal motor neuron function requires restitution of multiple dysregulated cellular pathways.
Amyotrophic lateral sclerosis (ALS) is an adult-onset degenerative disorder of motor neurons that manifests as progressive paralysis and results in inevitable death, typically within 5 y (1–3). Most cases of ALS are sporadic, with no apparent familial history, whereas 10% of cases are inherited (2, 4, 5). Identifying the underlying dysregulated gene pathways and pathological processes in ALS has been a major research focus over the last decade. Mice that harbor a transgene-expressing human cytosolic superoxide dismutase with an ALS-associated mutation (designated hSOD1G93A) develop motor neuron disease that closely mimics the phenotype of both familial and sporadic ALS (6, 7).
Motor neuron cell death, accompanied by astrogliosis and microglial activation, is the characteristic pathological feature in ALS (8, 9). Additional pathological features include neuromuscular junction (NMJ) denervation, axonal degeneration, and abortive collateral sprouting (10, 11). The combination of progressive motor neuron cell death and the retraction of axons from NMJs together with an impaired ability of surviving intact axons to generate compensatory axonal collateral sprouts results in progressive irreversible muscle atrophy (2, 12, 13).
Activating transcription factor 3 (ATF3), a basic leucine zipper transcription factor, is induced under a variety of stress conditions (14, 15). ATF3 transcriptional targets vary in different cell types and conditions, resulting in diverse influences on cellular phenomena, such as cell survival, proliferation, and death (15, 16). In neurons (17), induction of ATF3 expression prevents cell death and promotes neurite formation and elongation, leading to enhanced nerve regeneration through transcriptional induction of survival and growth-associated genes (18–26). Intact adult motor neurons do not normally express ATF3. However, after sciatic nerve injury, lumbar motor neurons induce and retain ATF3 expression (27). ATF3 has not been detected in end stage spinal cord samples of ALS mouse models or postmortem samples of ALS patients (28–32). In the SOD1G93A ALS mouse and rat models, ATF3 expression was detected in only a small proportion of spinal motor neurons (33–36). However, it remains unclear whether ATF3 induction in the motor neurons drives prosurvival or even possibly, detrimental proapoptotic mechanisms (33, 36). Because constitutive ATF3 expression in an ATF3 transgenic mouse model has no adverse phenotype (19) and ATF3 has neuroprotective and regeneration-promoting actions in diverse neurons, we examined the effect of constitutive ATF3 expression on disease phenotype in the ALS mouse model. Here, we report that transgenic expression of ATF3 in adult motor neurons of SOD1G93A mice altered the transcriptome to promote motor neuron survival and maintain axonal integrity and NMJ innervation. ATF3 expression in SOD1G93A mice resulted in a delay in disease deterioration that manifested in delayed muscle atrophy, improved muscle performance, and a small increase in lifespan.
Results
ATF3 Promotes Motor Neuron Survival in ATF3/SOD1G93A Mice.
To address the effect of forced ATF3 expression on motor neuron survival in the SOD1 mutant mice, ATF3 transgenic mice (19) were crossed with hSOD1G93A transgenic mice that harbor the mutant gene at high copies (6), and their progeny was analyzed. These ATF3 transgenic mice express the ATF3 transgene under control of the thy1.2 promoter (19), which drives transgene expression specifically in postnatal neurons (37). To exclude transgene expression variability, two independent ATF3 transgenic founder lines that express ATF3 abundantly in most spinal motor neurons were crossed with SOD1G93A mice and generated identical results.
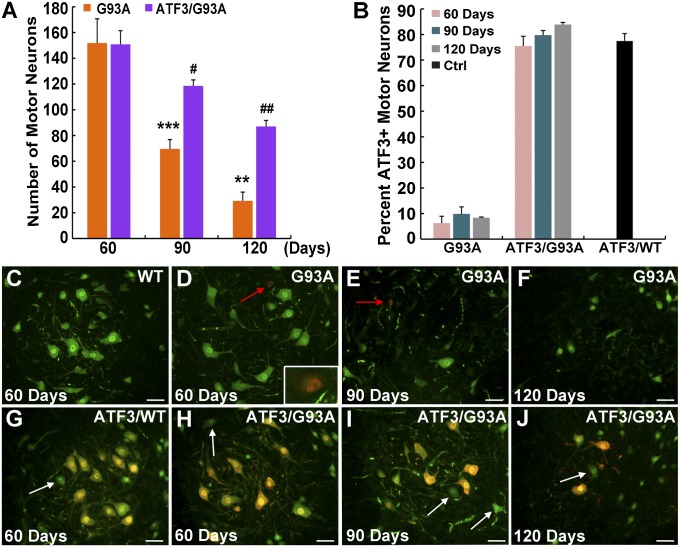
Progressive motor neuron cell death is a hallmark of ALS. Motor neuron survival was quantified at 60 d of age (a presymptomatic stage), 90 d of age (when disease onset is apparent), and 120 d of age [by which time the mice show clear muscle weakness but are not at end stage (130–140 d of age)]. Large lumbar motor neurons with a cell body area of ≥450 μm2 were quantified using the neuronal marker NeuN. Thus, only the large α-motor neurons most vulnerable to cell death in ALS (2, 38, 39) were quantified, and the γ-type motor neurons that are resistant to the disease were excluded. In SOD1G93A littermates at the presymptomatic stage, no motor neuron cell death was detected, but by the time of disease onset, only 45.8% of motor neurons had survived, with 19.3% of motor neurons surviving at 120 d of age (Fig. 1 A and D–F). In contrast, motor neuron survival in the ATF3/SOD1G93A double transgenic mice was substantially higher; 78.6% and 57.6% of the motor neurons survived at 90 and 120 d, respectively (Fig. 1 A and H–J).
Fig. 1.
Forced ATF3 expression promotes motor neuron survival. (A) Lumbar ventral horn motor neurons immunostained for the neuronal marker NeuN with an area ≥ 450 μm2 were quantified in the SOD1G93A compared with ATF3/SOD1G93A mice at 60, 90, and 120 d. A significant difference in the number of surviving motor neurons was identified. Data are presented as mean ± SEM with n = 5 mice per group per time point. **P < 0.01; ***P < 0.001 by ANOVA with Bonferroni postanalysis. A significant difference in the number of motor neurons in ATF3/SOD1G93A at 90 and 120 d relative to 60 d was also detected. #P < 0.05; ##P < 0.001 by ANOVA with Bonferroni postanalysis. (B) A significant difference was found in the percentage of ATF3-positive motor neurons in ATF3/SOD1G93A compared with SOD1G93A mice at all time points (P < 0.0001 by ANOVA). No difference in the proportion of ATF3-positive motor neurons was detected within each group over time (P > 0.05 by ANOVA with Bonferroni postanalysis). ATF3 expression in control WT and ATF3/WT mice at age 120 d is presented. (C–J) Representative merged images of motor neurons double stained for NeuN (green) and ATF3 (red) (nonmerged images are presented in Fig. S1). (Scale bar: 50 μm.) (D and E) Some endogenous ATF3 induction is detected in the SOD1G93A mice (red arrows and Inset). (G–J) Motor neurons that do not express ATF3 in the ATF3/WT and ATF3/SOD1G93A mice are marked by a white arrow.
Less than 10% of large motor neurons in SOD1G93A mice induced endogenous ATF3 expression, which was detected by double immunostaining for ATF3 and NeuN (Fig. 1 B–F). As anticipated, in the presence of the ATF3 transgene, 79.71 ± 2.14% and 77.53 ± 2.8% of the motor neurons were ATF3-positive in ATF3/SOD1G93A and ATF3/WT mice, respectively (Fig. 1 B and G–J). The fraction of non-ATF3–expressing motor neurons (∼20%) in ATF3/WT transgenic mice and in ATF3/SOD1G93A mice was similar. This fraction in ATF3/SOD1G93A mice remained unaltered over time (Fig. 1 B and H–J). A cell-autonomous mechanism of ATF3 action would be expected to enhance the survival of only ATF3-positive neurons and thus, lead to a decrease in the ATF3-negative fraction over time. However, the lack of change in this fraction suggests that the beneficial influence of ATF3 may also be mediated by non-cell-autonomous actions that protect neighboring motor neurons that do not express ATF3 (Fig. 1 B and H–J). For example, a non-cell-autonomous action could be achieved if ATF3 induces the expression of secreted factors that then exert the protective action on the non-ATF3–expressing cells or because of reduced neuroinflammation caused by the increased neuronal survival.
ATF3 Delays the Loss of Axonal Integrity in ATF3/SOD1G93A Mice.
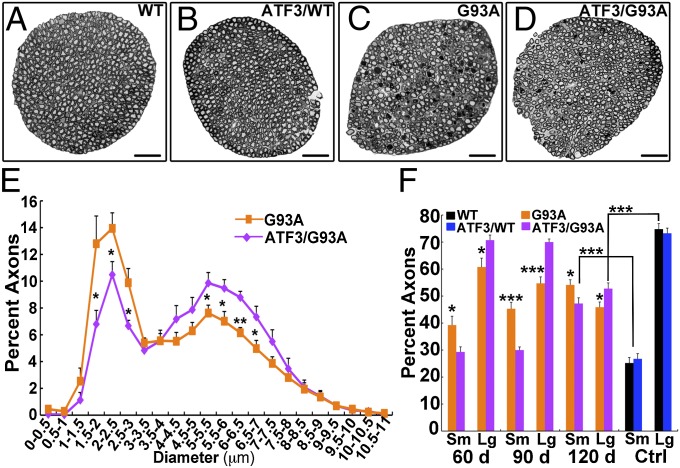
Analysis of lumbar L5 ventral roots (Fig. 2) revealed axonal damage early on at the presymptomatic stage (60 d) in SOD1G93A mice. Large-caliber α-axons with a diameter >3.5 μm degenerate in ALS. This degeneration is observed in the ventral root as a decline in diameter size. A continuous decline in large axons with a corresponding increase in the number of small axons was observed over time in SOD1G93A mice (Fig. 2). In contrast, significant axonal damage in the ventral roots was detected only at 120 d in ATF3/SOD1G93A mice (Fig. 2F). The most extreme differences were detected at 90 d, at which time ATF3/SOD1G93A axons remained intact; by contrast, in SOD1G93A mice at this time point, there was an increase of 51% in small-caliber axons because of the rapid degeneration of SOD1G93A large-caliber axons (Fig. 2 C–F).
Fig. 2.
Axonal integrity is maintained in the ATF3/SOD1G93A mice. (A–D) Representative cross-section images of L5 ventral roots isolated from WT, ATF/WT, SOD1G93A, and ATF3/SOD1G93A mice at 90 d of age. (Scale bar: 50 μm.) (E and F) Quantification of axonal size per ventral root. Note that there is a reduction in the percentage of large-caliber axons and a concurrent increase in the percentage of small-caliber axons in the SOD1G93A mice compared with the ATF3/SOD1G93A mice (n = 6–9 mice per group per time point). *P < 0.05; **P < 0.001; ***P < 0.0001 by ANOVA with Bonferroni postanalysis. The total mean ± SEM values of axons counted per ventral root in SOD1G93A mice at 60, 90, and 120 d were 904 ± 24.56, 677.7 ± 23.08, and 586.54 ± 28.26, respectively. The total mean ± SEM values of axons counted per ventral root in ATF3/SOD1G93A mice at 60, 90, and 120 d were 781.83 ± 30.81, 803 ± 31.85, and 653.81 ± 9.27, respectively. In WT and ATF3/WT, the numbers of axons were 911.6 ± 14.99 and 980.5 ± 58.67, respectively. (E) Distribution of axonal diameter size in SOD1G93A compared with ATF3/SOD1G93A mice at 90 d of age is presented. (F) Percent of small-diameter axons (Sm; 0–3.5 μm) compared with large-diameter axons (Lg; 3.5–19 μm) per ventral root. Control (Ctrl) WT and ATF3/WT mice were 120 d of age.
ATF3 Delays NMJ Denervation and Muscle Atrophy.
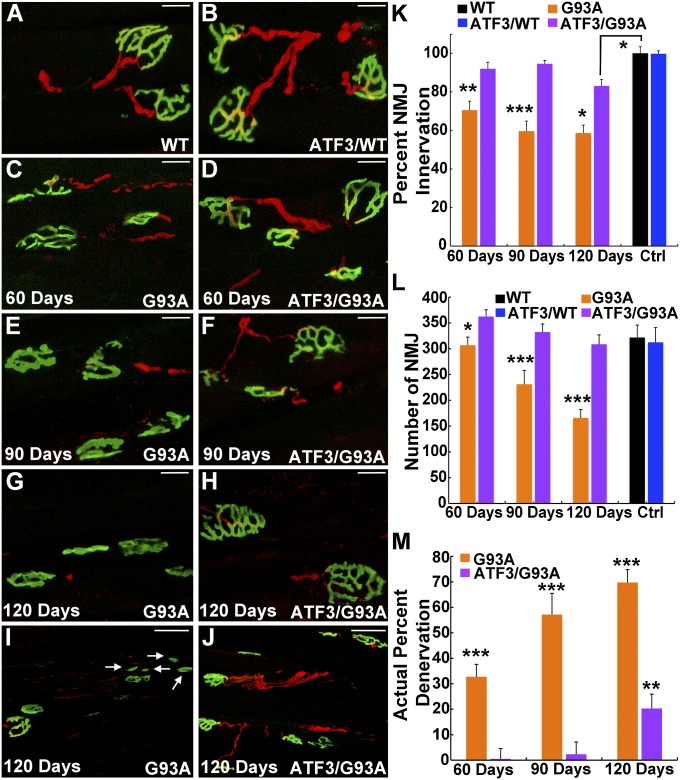
Evaluation of NMJs in the gastrocnemius muscle of SOD1G93A mice revealed denervation as early as 60 d of age, with only 70.5 ± 4.6% of the NMJs remaining innervated at this time (Fig. 3 C and K). Denervation of the NMJs accelerated in SOD1G93A mice as the disease progressed, and it was accompanied by a collapse of the NMJ structures, which were revealed by a reduction in the number of pretzel-shaped NMJs (Fig. 3L). NMJ collapse was apparent at 90 d in SOD1G93A mice and rapidly accelerated, with only 56.6% of NMJs retaining their normal morphology in the gastrocnemius muscle at 120 d (Fig. 3 L and I).
Fig. 3.
Innervation of the NMJ is maintained in ATF3/SOD1G93A mice. (A–M) Immunostaining with α-bungarotoxin (green) to label the NMJs in the gastrocnemius muscle and antineurofilament (red) to mark innervating axons reveals severe NMJ denervation and NMJ collapse in SOD1G93A mice compared with ATF3/SOD1G93A mice that worsens as the disease progresses. Only NMJs with a pretzel-shaped morphology were quantified. Nonmerged images are presented in Fig. S2. (Scale bar: A–H, 20 μm; I and J, 50 μm.) (I) Arrows show collapsed NMJs. (K–M) n = 9 mice per group per time point. (K) This graph represents the percent of innervated NMJs out of the total number of intact pretzel-shaped NMJs within each group. *P < 0.01; **P < 0.001; ***P < 0.0001 by ANOVA with Bonferroni postanalysis. (L) There is a decline in the number of pretzel-shaped NMJs in the SOD1G93A mice over time. *P < 0.05; ***P < 0.0001 by ANOVA with Bonferroni postanalysis. No difference in the number of NMJs in ATF3/SOD1G93A over time and compared with WT was observed (P > 0.05). (M) Actual percent denervation represents the percent of denervated NMJs in each group relative to the total number of NMJs in WT mice at 90 d of age. **P < 0.001; ***P < 0.0001 by ANOVA with Bonferroni postanalysis.
The rate of NMJ denervation extrapolated from Fig. 3K does not include the acceleration in NMJ collapse. Evaluation of the actual percent of NMJ denervation, which takes into account the loss of NMJs, reached 69.7 ± 5.2% at 120 d in SOD1G93A mice (Fig. 3M). In contrast, denervated NMJs were detected only at 120 d of age in ATF3/SOD1G93A mice; even then, 83.1 ± 3.4% of the NMJs remained innervated, and there was no apparent NMJ collapse (Fig. 3 H, J, K, and M). No difference in the extent of NMJ innervation between WT and ATF3/WT littermates was detected, indicating that ATF3 in WT mice did not induce hyperinnervation under normal conditions (Fig. 3 A, B, and K).
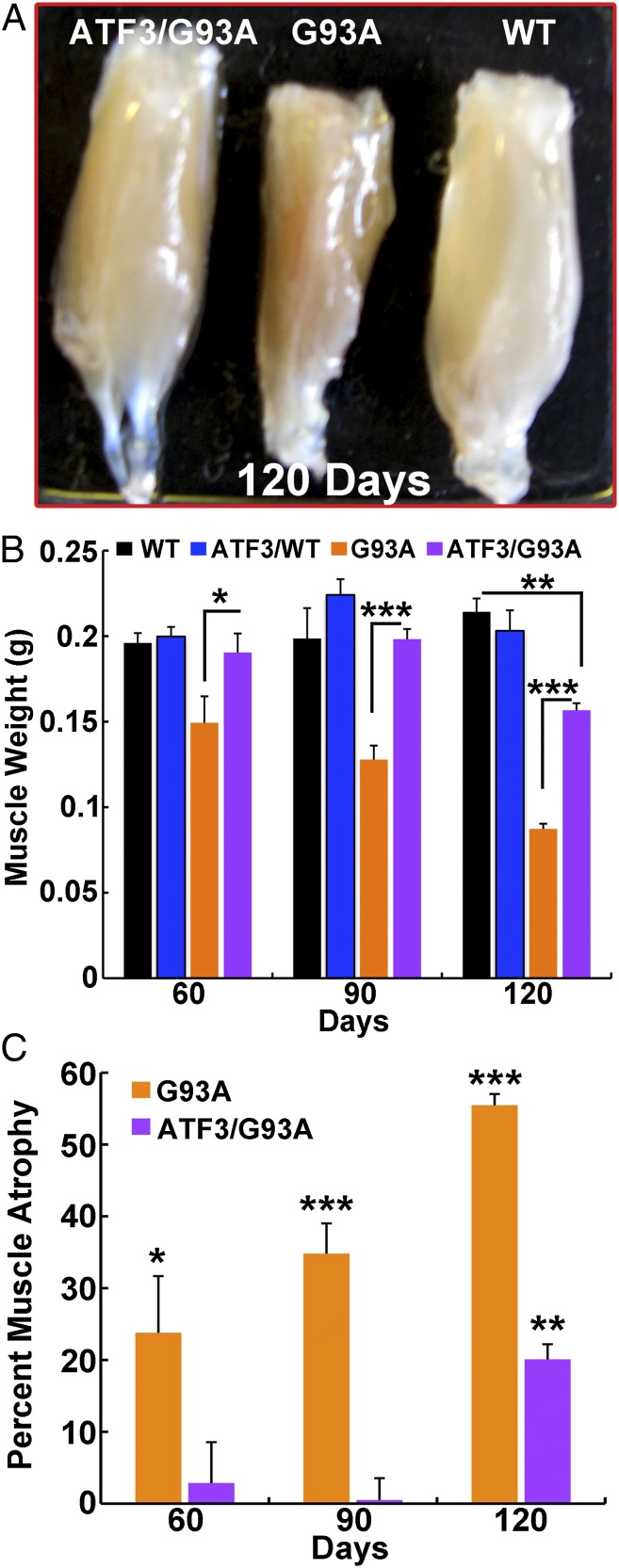
Measurements of gastrocnemius muscle weight in the SOD1G93A mice revealed muscle wasting (24% weight loss) at the presymptomatic stage (60 d), correlating with the early NMJ denervation detected at this time (Figs. 3 and 4). The decline in muscle mass in SOD1G93A mice advanced over time; weight reduction in the gastrocnemius muscle was 35% and 55.5% at 90 and 120 d, respectively, compared with WT mice (Fig. 4). In contrast, ATF3/SOD1G93A mice preserved muscle mass and only showed muscle wasting at 120 d of age (20%) (Fig. 4).
Fig. 4.
ATF3 expression in SOD1G93A mice delays muscle atrophy. (A) Representative image of the gastrocnemius muscle showing delayed muscle atrophy in ATF3/SOD1G93A compared with SOD1G93A mice at 120 d of age. (B and C) Muscle wasting was calculated by measurement of the gastrocnemius muscle weight in male mice littermates (the same trend was found in female mice). Data are presented in grams as mean ± SEM (n = 4–6 per group). *P < 0.01; **P < 0.001; ***P < 0.0001 by ANOVA with Bonferroni postanalysis. (C) Percent muscle atrophy was calculated relative to WT mice.
As the disease progressed, measurement of end plate innervation in SOD1G93A mice showed that motor axons retracted from the NMJs concomitantly with the development of muscle atrophy, which preceded motor neuron cell body death (Fig. 5 A and B). Such axonal dying back phenomenon is well-characterized in both human ALS and mouse disease models (12). However, although some motor neuron cell death was already apparent in ATF3/SOD1G93A mice at 90 d of age, NMJs denervation was not detected in these mice until 120 d (Fig. 5 A and C). This discrepancy suggests that, as NMJs are dennervated by motor neuron loss, they become reinnervated by sprouts extended from the intact axons of surviving motor neurons, contributing to the delay in muscle atrophy (Fig. 5B). Extension of collateral and terminal axonal sprouts is detected in ALS disease, but this sprouting process is usually unable to compensate for loss of axonal connections with muscle (11). In SOD1G93A mice, quantification of terminal sprouts in the gastrocnemius muscle detected no sprouting at 60 d. The amount of terminal sprouting at 90 and 120 d was similar, with no increase over time (Fig. 5D). In ATF3/SOD1G93A mice, terminal sprouting was also first detected at 90 d and similar to the sprouting in SOD1G93A mice. However, at 120 d, the degree of terminal sprouting was significantly higher in ATF3/SOD1G93A mice than SOD1G93A mice (Fig. 5 D–G). These data suggest that ATF3 promotes the capacity to form and maintain terminal axonal sprouts in SOD1 mutant mice to maintain NMJ innervation and prevent muscle atrophy.
Fig. 5.
Changes in architecture of the motor unit induced by SOD1G93A are attenuated by ATF3 transgene coexpression. (A) Comparison of progressive changes in percentages of remaining motor neurons (black diamonds), large axons (blue squares), innervated NMJs (extrapolated from the actual denervation data in Fig. 3M) (green triangles), and weight (red circles) in (Left) SOD1G93A and (Right) ATF3/SOD1G93A mice. In SOD1G93A mice, all parameters rapidly decline over time. By contrast, in ATF3/SOD1G93A mice, all four parameters are indistinguishable from WT at 60 d, and only numbers of large motor neurons declined by 90 d. At 120 d, all parameters are improved in ATF3/SOD1G93A mice compared with SOD1G93A counterparts. Arrow highlights the 90-d time point at which ATF3/SOD1G93A mice clearly show a decline in numbers of surviving motor neurons without a decline of innervated NMJs or other parameters. (B and C) A schematic model of the disease course illustrates the dying back phenomenon in SOD1G93A mice and induction of collateral sprouts in ATF3/SOD1G93A mice as a mechanism to maintain maximal NMJ innervation. Intact motor neurons and axons are in purple/blue. Diseased motor neurons are indicated with white specks, and degenerating axons are indicated with white and black. New collateral axonal sprouts are yellow; intact muscle is red, and NMJs are green. (D) Terminal sprouts were determined when the nerve extended beyond the acetylcholine receptor (AChR) clusters in the innervated NMJ at any direction. Sprouting is presented as the percent of NMJs with sprouts relative to the total number of innervated NMJs in each group. Increased sprouting is detected at 120 d in ATF3/G93A compared with G93A mice. *P < 0.01 by ANOVA with Bonferroni postanalysis (n = 4–5 mice per time point). (E–G) Immunostaining with α-bungarotoxin (green) to label NMJs in the gastrocnemius muscle and antineurofilament (red) to mark innervating axons reveals sprouting at the NMJ in ATF3/G93A mice. The terminal sprout is indicated with an arrow.
ATF3 Delays Motor Deficits and Disease Onset in ATF3/SOD1G93A Mice.
We next tested whether the beneficial effects of ATF3 on motor neuron survival and axonal connectivity with muscle identified in this study had an impact on muscle performance and disease progression. ATF3/SOD1G93A double transgenic mice displayed increased muscle strength compared with SOD1G93A littermates as determined by the hind limb grip strength assay (Fig. 6A). The improved motor performance in ATF3/SOD1G93A mice directly reflected an impact of the ATF3 transgene on the disease; the ATF3 transgene did not enhance muscle strength in ATF3/WT transgenic littermates compared with WTs.
Fig. 6.

ATF3 neuronal expression prolongs survival and delays motor deficits in the SOD1 mutant mice. (A) Hind limb grip strength analysis for WT, ATF3/WT, SOD1G93A, and ATF3/SOD1G93A mice. Data are mean ± SEM. *P < 0.05; ***P < 0.001 by repeated measures ANOVA with Bonferroni postanalysis (n = 30 mice per group). (B and C) ATF3/SOD1G93A mice display delayed onset of disease and prolonged survival (P = 0.0026 and P = 0.0013, respectively, by Kaplan–Meier log rank test for survival; n = 29 mice per group).
Onset of the disease in ATF3/SOD1G93A mice was delayed by an average of 11.3 d. Age at onset was 91.72 ± 2.44 d in SOD1G93A mice compared with 103.1 ± 2.2 in ATF3/SOD1G93A double transgenic mice (P < 0.01 by Kaplan–Meier log rank test) (Fig. 6B). Similarly, mean survival in ATF3/SOD1G93A mice was prolonged by 7.7 d. The average ages at death were 137.2 ± 1.4 and 144.9 ± 2.1 d in the SOD1G93A and ATF3/SOD1G93A mice, respectively (P < 0.01 by Kaplan–Meier log rank test for survival) (Fig. 6C). However, no difference was identified in disease duration between the groups (P > 0.05). Although ATF3 expression was sufficient to modestly delay the onset of the disease and consequently, prolong survival, disease duration was not extended. Nonetheless, as the disease progressed and SOD1G93A mice reached end stage, they were weak and largely immobile, whereas ATF3/SOD1G93A mice were in much better health. They were grooming and exploring their cages (Movies S1 and S2), illustrating the marked beneficial effects of ATF3 on muscle performance as the disease progressed.
Discussion
This study shows that forced expression of the transcription factor ATF3 in SOD1G93A ALS mice slows the loss of motor neurons, delays terminal axonal atrophy, and augments axonal sprouting by driving the motor neurons into a prosurvival and regenerative state. In the ALS mouse model, forced ATF3 expression modifies the cell transcriptome (Figs. S3, S4, and S5) to support compensatory axonal sprouting and neuronal survival. At 90 d, the ATF3/SOD1G93A mice showed no loss of NMJ innervation, despite some motor neuron loss, suggesting that denervated NMJs in ATF3/SOD1G93A mice are rapidly reinnervated by terminal and collateral sprouts extending from neighboring intact motor neurons that are in a proregenerative state. Moreover, at this age, the ATF3/SOD1G93A mice showed little or no loss of muscle mass and showed improved muscle strength relative to SOD1G93A mice, such that the overall health of the animals was remarkably improved. However, although disease onset and death were slightly delayed, disease duration was not extended, indicating that induction of an intrinsic growth state in SOD1 mutant motor neurons by ATF3 was not sufficient to halt disease progression. Although ATF3-dependent compensation improved motor neuron viability, axonal dynamics, and motor performance, it did not arrest the ongoing motor neuron loss sufficiently to prevent death.
It is well-established that terminal axonal degeneration with NMJ denervation is an early component of motor neuron disease in ALS (12, 40, 41) (Fig. 5). Whereas intact motor neurons undergo distal axonal sprouting, this remodeling is insufficient to innervate neighboring denervated end plates and compensate for motor neuron loss, leading to progressive paralysis and death (11). Regardless of whether these deficits in axonal innervation or capacity to sprout arise as a consequence of events in the soma, such as excitotoxicity, endoplasmic reticulum stress, or mitochondrial pathology, or distally at peripheral nerve endings, it is evident from this study that the functional reserve provided by surviving motor neurons is enhanced by mechanisms that increase their axonal integrity, growth, and sprouting and thereby, maintain maximal connectivity with muscle.
The functional importance of the response of the axon to motor neuron pathology is illustrated by experimental paradigms that prevent death of the motor neuron soma. Three different experimental manipulations can delay motor neuron soma death in SOD1 mutant ALS mice: overexpression of the antiapoptotic protein Bcl-2, KO of the proapoptotic Bax protein, and double KO of BAX and BAC (42–44). However, even in the BAX/BAC double KO mice (42), in which a substantial delay in motor neurons cell death is observed, axonal integrity is not preserved to the same degree, and the motor axon terminals continue to gradually die back from the NMJ. Despite the substantial preservation of motor neuron soma achieved in these mouse model experiments, motor neuron survival alone does not prevent axons from degenerating.
The strong propensity for axonal degeneration in SOD1 mutant ALS mice is shown in an additional model. In Wlds mice, a unique fusion protein of ubiquitin conjugation factor E4B and nicotinomide mononucleotide adenylyltransferase 1 dramatically delays Wallerian degeneration after nerve injury and protects against axonal degeneration in several neuropathies and neurodegenerative models. However, when it is bred into the SOD1G93A mice, the Wlds gene fails to block axonal degeneration (45, 46). The molecular events controlled by Wlds expression that blunt axonal degeneration do not suffice to slow axonal degeneration triggered by mutant SOD1 protein. From this perspective, it is striking that, by contrast, the transcription factor ATF3 substantially augments axonal protection and sprouting, at least through ∼90–100 d.
Mechanisms that act to protect only one compartment, either the motor neuron soma or the axons, do not seem sufficient to preserve a functional motor unit in ALS (42–46). The importance of both maintaining axonal connectivity with the muscle and protecting the motor neurons from cell death for preservation of a functional motor unit is clearly illustrated in this study. The modest degree of muscle denervation and atrophy at 120 d in ATF3/SOD1G93A mice is comparable with the denervation detected in SOD1G93A mice 2 mo earlier (at 60 d of age). However, muscle performance is not maintained to the same extent. This difference implies that newly collaterally innervated NMJs, although sufficient to prevent muscle atrophy, do not maintain full muscle strength as the disease progresses, which may reflect the limited function of collateral sprouts relative to normal inputs. The preservation of large α-motor axon integrity found in this study is crucial for maintaining muscle mass and function, but the fact that axonal damage and progression of motor neuron cell death are observed in ATF3/SOD1G93A mice at 120 d suggests that deleterious processes continue to impair motor neuron function over time and secondarily, may compromise the axonal maintenance, dynamic axonal sprouting, and remodeling that are promoted by ATF3. Future studies, including longitudinal observations on motor neuron survival, axonal remodeling, and NMJ function in ATF3/SOD1G93A and ATF3/SOD1G93A mice with knockdown of the BAX/BAC proapoptotic genes, may help tease out why the beneficial influence of ATF3 on disease onset and lifespan is so modest relative to the substantial effect that it has on reducing neuronal loss, axonal damage, and denervation of the NMJ. These studies may also help identify key disease-inducing pathways that are resistant to ATF3-mediated transcriptional changes. However, the early benefits of ATF3 on overall function are substantial and if they could be translated into patients, might dramatically improve quality of life. Additional interventions would be required to slow significantly the inexorable ATF3-resistant cell death.
Transcription factors control the expression of many genes, and therefore, they serve as good candidates to govern the synchronization and coordination between the many different gene pathways required to achieve regeneration and neuronal survival. ATF3, a basic leucine zipper transcription factor (14), regulates gene expression either directly by binding the DNA as a homodimer or heterodimer or indirectly by sequestering activators and repressors, and thus, its transcriptional targets depend on the specific cellular milieu. Neuronal expression of ATF3 induced favorable transcriptional changes in the ventral horn of ATF3/SOD1G93A mice (Figs. S3, S4, and S5), including reduced induction of neuroinflammatory and apoptotic gene pathways, improved nerve transmission, and enhanced expression of gene pathways involved in cytoskeleton organization, axon guidance, and neurogenesis. These findings suggest that ATF3 modifies the intrinsic growth state of the motor neurons to enable axonal sprouting and remodeling in the ATF3/SOD1G93A mice (Figs. S4 and S5). Interestingly, the set of genes that is uniquely up-regulated in ATF3/SOD1G93A clusters in pathways that support axonal myelination and nerve transmission (Fig. S5), supporting a role for ATF3 in maintaining axonal integrity and function and thereby, retaining muscle mass and function.
This study (i) supports the view that neuroprotection to improve motor neuron viability will be required to slow disease progression and (ii) shows, in addition, that, to counteract motor neuron loss, it may also be critical to induce regenerative mechanisms that reduce terminal axonal degeneration and enhance compensatory axonal sprouting and functional reinnervation of denervated NMJs. Because the cell survival and regenerative mechanisms that ATF3 induces are not sufficient to completely repair motor neurons in ALS, complementary protective mechanisms that target the multiple dysregulated cellular pathways will be required for full restitution, further reflecting the complexity of the disease.
Materials and Methods
ATF3 transgenic mouse lines (19) that constitutively express ATF3 in neurons, including spinal motor neurons, were crossed with high-copy number SOD1G93A transgenic mice (6) (stock number 002726; Jackson Laboratories). Protocols for the behavioral examinations are described in SI Materials and Methods. Muscle, motor neuron, ventral root, and NMJ analyses, including staining protocols and antibodies used, and additional methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Igor Bagayev at the Confocal Core Facility at Massachusetts General Hospital for assistance. We also thank the Molecular Genetics Core Facility at Children's Hospital of Boston for providing microarray support, which was supported by National Institutes of Health Grants P50NS40828 and P30HD18655. This study was supported by the ALS Therapy Alliance (R.S.), Israel ALS Research Association (IsrALS) (R.S.), National Institutes of Health Grant NS038253 (to C.J.W.), and the Miriam and Sheldon G. Adelson Medical Research Foundation (C.J.W.). R.H.B. was supported by the Angel Fund, the ALS Therapy Alliance, Project ALS, P2ALS, the Pierre L. De Bourgknecht ALS Research Fund, and National Institute of Neurological Disorders and Stroke Awards 1RC2NS070342-01, 1RC1NS068391-01, R01NS050557-05, U01NS05225-03, and R01NS065847-01A1.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314826111/-/DCSupplemental.
References
- 1.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7(9):710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell JD, Borasio GD. Amyotrophic lateral sclerosis. Lancet. 2007;369(9578):2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]
- 3.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(Suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 4.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136(6):1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 6.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 7.Bruijn LI, Cudkowicz M. Therapeutic targets for amyotrophic lateral sclerosis: Current treatments and prospects for more effective therapies. Expert Rev Neurother. 2006;6(3):417–428. doi: 10.1586/14737175.6.3.417. [DOI] [PubMed] [Google Scholar]
- 8.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187(6):761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadimitriou D, et al. Inflammation in ALS and SMA: Sorting out the good from the evil. Neurobiol Dis. 2010;37(3):493–502. doi: 10.1016/j.nbd.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer LR, Glass JD. Axonal degeneration in motor neuron disease. Neurodegener Dis. 2007;4(6):431–442. doi: 10.1159/000107704. [DOI] [PubMed] [Google Scholar]
- 11.Gordon T, Hegedus J, Tam SL. Adaptive and maladaptive motor axonal sprouting in aging and motoneuron disease. Neurol Res. 2004;26(2):174–185. doi: 10.1179/016164104225013806. [DOI] [PubMed] [Google Scholar]
- 12.Fischer LR, et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 14.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene. 2001;273(1):1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 15.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7(4-6):321–335. [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 2009;87(11):1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt D, Raivich G, Anderson PN. Activating transcription factor 3 and the nervous system. Front Mol Neurosci. 2012;5:7. doi: 10.3389/fnmol.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seijffers R, Allchorne AJ, Woolf CJ. The transcription factor ATF-3 promotes neurite outgrowth. Mol Cell Neurosci. 2006;32(1-2):143–154. doi: 10.1016/j.mcn.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27(30):7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis JS, Dragunow M, During MJ. Over expression of ATF-3 protects rat hippocampal neurons from in vivo injection of kainic acid. Brain Res Mol Brain Res. 2004;124(2):199–203. doi: 10.1016/j.molbrainres.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 21.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci. 2003;23(12):5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, et al. ATF3 plays a protective role against toxicity by N-terminal fragment of mutant huntingtin in stable PC12 cell line. Brain Res. 2009;1286:221–229. doi: 10.1016/j.brainres.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson AG, et al. ATF3 enhances c-Jun-mediated neurite sprouting. Brain Res Mol Brain Res. 2003;120(1):38–45. doi: 10.1016/j.molbrainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Lindwall C, Dahlin L, Lundborg G, Kanje M. Inhibition of c-Jun phosphorylation reduces axonal outgrowth of adult rat nodose ganglia and dorsal root ganglia sensory neurons. Mol Cell Neurosci. 2004;27(3):267–279. doi: 10.1016/j.mcn.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang SJ, et al. A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J Neurosci. 2011;31(13):4978–4990. doi: 10.1523/JNEUROSCI.2672-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, et al. Increased inflammation and brain injury after transient focal cerebral ischemia in activating transcription factor 3 knockout mice. Neuroscience. 2012;220:100–108. doi: 10.1016/j.neuroscience.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Tsujino H, et al. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15(2):170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 28.Offen D, et al. Spinal cord mRNA profile in patients with ALS: Comparison with transgenic mice expressing the human SOD-1 mutant. J Mol Neurosci. 2009;38(2):85–93. doi: 10.1007/s12031-007-9004-z. [DOI] [PubMed] [Google Scholar]
- 29.Wang XS, Simmons Z, Liu W, Boyer PJ, Connor JR. Differential expression of genes in amyotrophic lateral sclerosis revealed by profiling the post mortem cortex. Amyotroph Lateral Scler. 2006;7(4):201–210. doi: 10.1080/17482960600947689. [DOI] [PubMed] [Google Scholar]
- 30.Dangond F, et al. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol Genomics. 2004;16(2):229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- 31.Jiang YM, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57(2):236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 32.Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 2007;8:26. doi: 10.1186/1471-2164-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlug AS, et al. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur J Neurosci. 2005;22(8):1881–1894. doi: 10.1111/j.1460-9568.2005.04389.x. [DOI] [PubMed] [Google Scholar]
- 34.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12(5):627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 35.Lobsiger CS, Boillée S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci USA. 2007;104(18):7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malaspina A, et al. Activation transcription factor-3 activation and the development of spinal cord degeneration in a rat model of amyotrophic lateral sclerosis. Neuroscience. 2010;169(2):812–827. doi: 10.1016/j.neuroscience.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 37.Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J Neurosci Methods. 1997;71(1):3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]
- 38.Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 2. Etiopathogenesis. Ann Neurol. 1985;18(4):419–431. doi: 10.1002/ana.410180402. [DOI] [PubMed] [Google Scholar]
- 39.Bruijn LI, Cleveland DW. Mechanisms of selective motor neuron death in ALS: Insights from transgenic mouse models of motor neuron disease. Neuropathol Appl Neurobiol. 1996;22(5):373–387. doi: 10.1111/j.1365-2990.1996.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 40.Hegedus J, Putman CT, Gordon T. Time course of preferential motor unit loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neurobiol Dis. 2007;28(2):154–164. doi: 10.1016/j.nbd.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Durand J, Amendola J, Bories C, Lamotte d’Incamps B. Early abnormalities in transgenic mouse models of amyotrophic lateral sclerosis. J Physiol Paris. 2006;99(2-3):211–220. doi: 10.1016/j.jphysparis.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Reyes NA, et al. Blocking the mitochondrial apoptotic pathway preserves motor neuron viability and function in a mouse model of amyotrophic lateral sclerosis. J Clin Invest. 2010;120(10):3673–3679. doi: 10.1172/JCI42986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould TW, et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26(34):8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostic V, Jackson-Lewis V, de Bilbao F, Dubois-Dauphin M, Przedborski S. Bcl-2: Prolonging life in a transgenic mouse model of familial amyotrophic lateral sclerosis. Science. 1997;277(5325):559–562. doi: 10.1126/science.277.5325.559. [DOI] [PubMed] [Google Scholar]
- 45.Vande Velde C, Garcia ML, Yin X, Trapp BD, Cleveland DW. The neuroprotective factor Wlds does not attenuate mutant SOD1-mediated motor neuron disease. Neuromolecular Med. 2004;5(3):193–203. doi: 10.1385/NMM:5:3:193. [DOI] [PubMed] [Google Scholar]
- 46.Fischer LR, et al. The WldS gene modestly prolongs survival in the SOD1G93A fALS mouse. Neurobiol Dis. 2005;19(1–2):293–300. doi: 10.1016/j.nbd.2005.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.