Significance
Theory predicts that chronic pathogens with vertical or familial transmission should become less virulent over time because of coevolution. Although transmitted in this way, Helicobacter pylori is the major causative agent of gastric cancer. In two distinct Colombian populations with similar levels of H. pylori infection but different incidences of gastric cancer, we examined human and pathogen ancestry in matched samples to assess whether their genomic variation affects the severity of premalignant lesions. Interaction between human Amerindian ancestry and H. pylori African ancestry accounted for the geographic disparity in clinical presentation. We conclude that coevolutionary relationships are important determinants of gastric disease risk and that the historical colonization of the Americas continues to influence health in modern American populations.
Keywords: admixture, histopathology, inflammation, Latin America
Abstract
Helicobacter pylori is the principal cause of gastric cancer, the second leading cause of cancer mortality worldwide. However, H. pylori prevalence generally does not predict cancer incidence. To determine whether coevolution between host and pathogen influences disease risk, we examined the association between the severity of gastric lesions and patterns of genomic variation in matched human and H. pylori samples. Patients were recruited from two geographically distinct Colombian populations with significantly different incidences of gastric cancer, but virtually identical prevalence of H. pylori infection. All H. pylori isolates contained the genetic signatures of multiple ancestries, with an ancestral African cluster predominating in a low-risk, coastal population and a European cluster in a high-risk, mountain population. The human ancestry of the biopsied individuals also varied with geography, with mostly African ancestry in the coastal region (58%), and mostly Amerindian ancestry in the mountain region (67%). The interaction between the host and pathogen ancestries completely accounted for the difference in the severity of gastric lesions in the two regions of Colombia. In particular, African H. pylori ancestry was relatively benign in humans of African ancestry but was deleterious in individuals with substantial Amerindian ancestry. Thus, coevolution likely modulated disease risk, and the disruption of coevolved human and H. pylori genomes can explain the high incidence of gastric disease in the mountain population.
The bacterium Helicobacter pylori colonizes the gastric mucosa of approximately half the world’s population. Although all infected individuals develop gastric inflammation, only a small fraction (<1%) develop gastric adenocarcinoma, which accounts for 10% of total cancer-related mortality worldwide (1, 2). The prevalence of H. pylori infection in a population generally does not predict the incidence of serious clinical sequelae, suggesting that host and pathogen genetic variation, as well as dietary and other environmental factors, play an important role (3–6). However, these factors analyzed in isolation have failed to provide adequate explanations for the variability in infection outcomes.
Since accompanying Homo sapiens out of Africa, H. pylori has evolved in concert with geographically defined human populations (7). Both evolutionary theory and empirical comparisons predict that chronic pathogens with vertical or familial transmission, such as H. pylori, should become less virulent over time (8–10). Indeed, most H. pylori infections are well tolerated by humans, leading only to low-grade inflammation in most adult carriers, while possibly conferring protection against asthma and some esophageal disorders (11–13). However, H. pylori also can be transmitted horizontally, especially under poverty-associated conditions common in the developing world (14). In such situations, infection with multiple strains may be more common than in developed countries (15). Because H. pylori is known to be highly recombinogenic (16), infection with multiple strains can lead to horizontal gene transfer of segments that have not coevolved with their hosts, disrupting the selection for reduced virulence (17–19). Thus, in regions where humans are highly admixed, such as in South America, the human–H. pylori relationship may be less likely to reflect a coadapted, reduced-virulence complex.
In many parts of South and Central America, the incidence of gastric cancer is higher in mountainous areas than in coastal regions (20). To explore whether this phenomenon can be explained by host and microbial genetic variation, we took advantage of a natural laboratory consisting of two Colombian communities that have virtually identical prevalence of H. pylori infection (∼90%) but extremely different incidence rates of gastric cancer. The reported gastric cancer incidence rate in the town of Tuquerres in the Andes Mountains (∼150 per 100,000) is ∼25 times higher) than that in the town of Tumaco only 200 km away on the coast (∼6 per 100,000) (21). In addition, people from the mountains exhibit a higher incidence of precancerous gastric lesions (22).
Admixture analyses of H. pylori have been used to describe the evolutionary history of the bacteria in relation to human migrations (23–25). In the present study, we extend these analyses to elucidate the role that admixture in both H. pylori and humans has played in gastric disease (i.e., histopathology scores) by explicitly assessing whether variation in the ancestries of both host and pathogen and their coevolution affect disease trajectories and risk.
Results
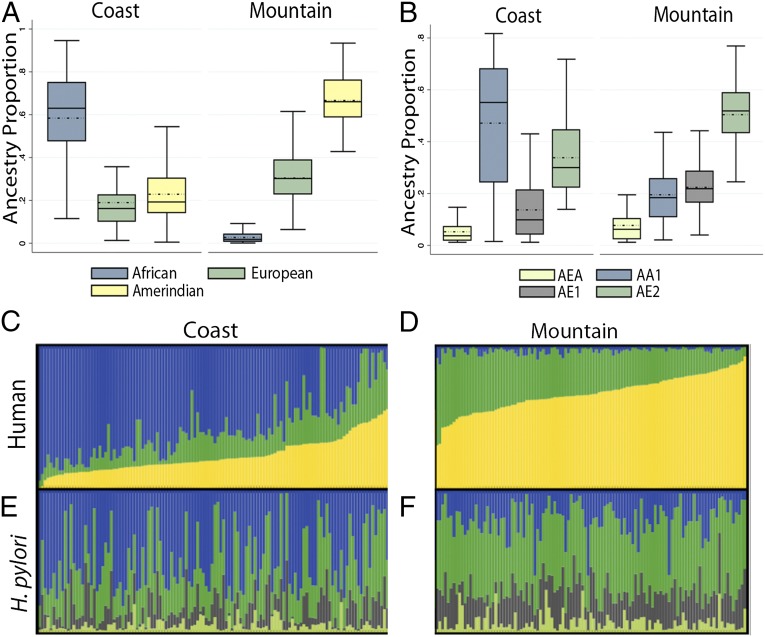
Examining host genetics in the mountain and coastal communities, we assessed whether the populations differed in ethnic composition (see Table S1 for demographic information on participants). Admixture analysis of high-density human genotypic data revealed a major African ancestral cluster in the coastal population (mean proportion of 0.58 ± 0.23; n = 122), with significant Amerindian (0.23 ± 0.13) and European (0.19 ± 0.13) ancestry (Fig. 1 A and C). In contrast, the mountain population was mostly Amerindian (0.67 ± 0.12; n = 120), with significant European (0.31 ± 0.11) and a negligible proportion of African ancestry (0.03 ± 0.02, Fig. 1 A and D). Amerindian and European ancestry estimates had an approximately normal distribution in the mountain cohort (Shapiro–Wilk test, P > 0.37), whereas all coastal distributions were skewed (P < 0.001 for all tests). The lack of normality in the admixture proportions of coastal individuals is consistent with recent migrations and/or nonrandom mating. Principal components analysis (PCA) also was used to assess human ancestry and showed a pattern similar to the admixture analyses (Fig. S1).
Fig. 1.
Human and H. pylori ancestry. (A) Human ancestry distributions of Colombian study participants from the coastal region of Tumaco (n = 122) and the mountain region of Tuquerres (n = 120). Black lines indicate median ancestry proportions; dotted lines indicate means. Box limits mark the 25th and 75th percentiles. (B) H. pylori ancestry distributions from the coastal region of Tumaco (n = 156) and the mountain region of Tuquerres (n = 132). Mean estimates were 0.47 ± 0.23 AA1, 0.34 ± 0.14 AE2, 0.14 ± 0.12 AE1, and 0.05 ± 0.04 AEA on the coast and 0.20 ± 0.12 AA1, 0.50 ± 0.11 AE2, 0.22 ± 0.09 AE1, and 0.08 ± 0.07 AEA in the mountain region. Black lines indicate median ancestry proportions; dotted lines indicate means. In the H. pylori ancestries: AEA, Ancestral East Asia; AA1, Ancestral Africa1; AE1, Ancestral Europe1; and AE2, Ancestral Europe2. Box limits mark the 25th and 75th percentiles. (C–F) Admixture proportions of human (C and D) and H. pylori (E and F) ancestry (n = 233). Each human host and his or her corresponding H. pylori isolate are represented by a vertical bar spanning both panels, with admixture proportions denoted by color. (C) Humans from the coastal region. (D) Humans from the mountain region. (E) H. pylori from the coastal region. (F) H. pylori from the mountain region. For the human admixture, blue = African, green = European, and Amerindian = yellow. For the H. pylori admixture, blue = AA1, green = AE2, gray = AE1, and lime green = AEA.
We found that the Colombian H. pylori strains are derived from four ancestral populations: Africa1 (AA1), European (AE1 and AE2), and East Asia (AEA) (Fig. 1 B, E, and F). Although most isolates contained substantial fractions of both African and European ancestry, AA1 predominated on the coast, and AE2 predominated in the mountain region (see the legend of Fig. 1B for proportions). Consistent with replacement of ancestral Amerindian/East Asian H. pylori, we observed that the major ancestral cluster in the mountains is AE2, the cluster most common in southern Europe, not AEA, which would be expected in an Asia-derived human population. This observation indicates a recent transfer of H. pylori strains from Europeans to Amerindians postcolonization.
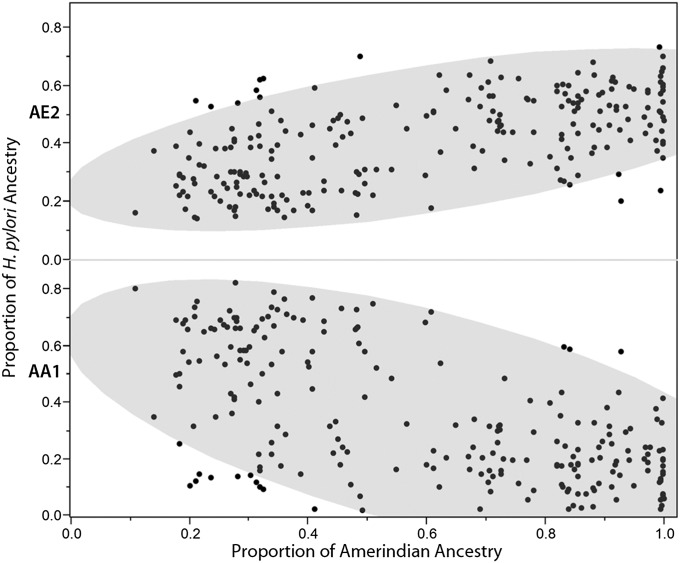
Both human and H. pylori ancestry data were available for 233 isolates (Fig. 1 C–F), and these isolates were tested for correlations between the human and H. pylori ancestries. We found that all pairwise Pearson correlations between proportions of host ancestry (African, European, or Amerindian) and AA1, AE1, and AE2 H. pylori ancestry were significant at the 0.0005 level, whereas those correlations with AEA were significant at the 0.041 level (Table 1). Spearman’s ρ values yielded similar results (Table S2). The proportion of host African ancestry correlated positively with AA1 (r = 0.62; P < 0.0001) and negatively with AE2 (r = −0.59; P < 0.0001), consistent with vertical or familial transmission and coevolution. Host Amerindian ancestry correlated negatively with AA1 ancestry (r = −0.60), and positively with AE2 ancestry (r = 0.58; P < 0.0001 for both) (Fig. 2).
Table 1.
Correlations between human and H. pylori ancestry (n = 233)
| Human | H. pylori | r | P value | 95% CI |
| African | AEA | –0.21 | 0.0015 | –0.33 to –0.08 |
| European | 0.13 | 0.0406 | 0.01 to 0.26 | |
| Amerindian | 0.19 | 0.0035 | 0.06 to 0.31 | |
| African | AA1 | 0.62 | <0.0001 | 0.53 to 0.69 |
| European | –0.34 | <0.0001 | –0.45 to –0.23 | |
| Amerindian | –0.60 | <0.0001 | –0.68 to –0.51 | |
| African | AE1 | –0.39 | <0.0001 | –0.49 to –0.28 |
| European | 0.22 | 0.0005 | 0.10 to 0.34 | |
| Amerindian | 0.38 | <0.0001 | 0.26 to 0.48 | |
| African | AE2 | –0.59 | <0.0001 | –0.67 to –0.50 |
| European | 0.32 | <0.0001 | 0.20 to 0.43 | |
| Amerindian | 0.58 | <0.0001 | 0.48 to 0.66 |
H. pylori ancestry: AEA, Ancestral East Asia; AA1, Ancestral Africa1; AE1, Ancestral Europe1; AE2, Ancestral Europe2. Includes host and H. pylori ancestries for participants with both available, in both mountain and coastal populations.
Fig. 2.
Relationship between H. pylori ancestry and human Amerindian ancestry. The proportion of European H. pylori ancestry (AE2) correlates positively and African H. pylori ancestry (AA1) correlates negatively with host Amerindian ancestry. The 90% density ellipses are demarcated by the shaded area in each plot. The wider minor axis of the lower ellipse indicates that AA1 is dispersed more broadly than AE2 in Amerinds. n = 233.
We next evaluated the effect of human ancestry on gastric histopathology, scored on a continuous scale of increasing severity from 2 (gastritis) to 6 (cancer), according to Correa’s Cascade model for progression toward the intestinal subtype of gastric cancer (26, 27). For this analysis, we used generalized mixed linear models with data for participants 40 y of age and older (n = 121; Materials and Methods and Table S3). Within this older group (40–65 y), age was not significantly associated with disease severity (P = 0.58). Both Amerindian and European host ancestries were associated with more severe lesions (β = 1.16, P = 0.003; and β = 2.06, P = 0.002, respectively, where β can be interpreted as the expected increase in histopathology score as ancestry proportion increases from 0 to 1). In contrast, African host ancestry conferred protection (β = −1.11, P < 0.001). Area of residence also was associated with histopathology, with the mountain population exhibiting more advanced lesions (P = 0.007), but this association became nonsignificant when host genetic ancestry was considered, indicating that host ancestry, not location or altitude, was driving the discrepancy in disease severity. Consistent with this concept, the proportion of Amerindian host ancestry was associated with more severe lesions in the coastal population alone (β = 2.49, P = 0.017). Univariate ordinal regression on the discrete histopathology scores provided similar results, with severity of lesions increasing with European and Amerindian host ancestry and decreasing with African host ancestry (P values of 0.002, 0.002, and <0.001, respectively).
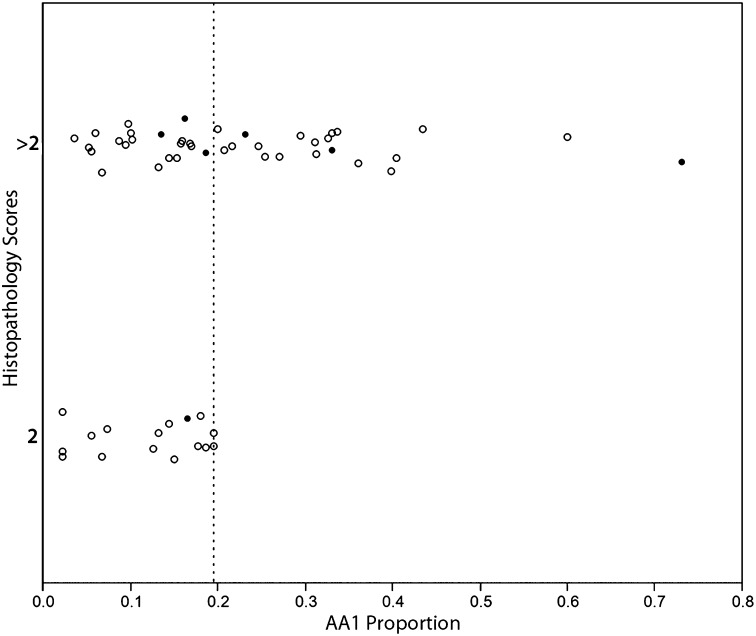
None of the four H. pylori ancestral clusters was associated with histopathology in univariate analyses of the entire cohort. However, in an analysis of the mountain cohort, the AA1 and AE2 clusters displayed highly significant but opposite effects. AA1 was associated with worse outcomes, and AE2 was associated with more favorable outcomes (β = 3.38, P = 0.002 and −3.42, P = 0.012, respectively). Ordinal regression on discrete histopathology scores confirmed these results (corresponding P values of 0.009 and 0.023 for AA1 and AE2, respectively). Because the mountain population had minimal host African ancestry, we asked whether AA1 increased severity of gastric lesions in all subjects with low host African ancestry, regardless of area of residence. Such a scenario could signify disrupted coadaptation. When we considered the 56 individuals with less than 17.6% African ancestry (the lowest decile of African host ancestry on the coast), we found that every person who carried H. pylori with >19.8% AA1 ancestry (n = 20) had lesions more severe than gastritis alone, irrespective of location (P < 0.001; Fig. 3).
Fig. 3.
All subjects with low proportions of host African ancestry (<17.6%, the lowest decile on the coast) and carrying an H. pylori strain with >19.8% AA1 ancestry (dotted line) had advanced lesions (histopathology scores >2), regardless of place of residence, indicating the importance of the interaction of host Amerindian ancestry and H. pylori AA1. Closed circles represent coastal residents; open circles represent mountain residents. n = 56.
Because AA1 was predictive of more serious disease in the subjects with low African ancestry, we tested whether there was an interaction between human and H. pylori ancestry that affected histopathology. Using a generalized multivariate mixed linear model, we found a significant interaction between host Amerindian ancestry and H. pylori AA1 ancestry (β = 5.08, P = 0.010). The lack of an effect of AA1 in the univariate model of the entire cohort thus was an artifact of its opposite effects in people of African vs. Amerindian ancestries. With the interaction term included in the model, human Amerindian ancestry and area of residence were no longer significant as independent factors (P = 0.62 and P = 0.89, respectively). H. pylori strains also were assessed for the presence or absence of cytotoxin-associated gene A (cagA), which encodes a secreted virulence factor with oncoprotein-like activities (Table S1) (28, 29). The presence of cagA was a significant predictor of premalignant gastric pathology independent of all other variables in the model (β = 0.98; P = 0.001), but the interaction between human and H. pylori ancestry that affected histopathology was not impacted by presence or absence of cagA.
Discussion
Both the H. pylori and human genomes had high levels of admixture, but the ancestral proportions of each differed significantly by region. The mountain population had only a small amount of human African ancestry (∼3%) but a large proportion of AA1 H. pylori ancestry (∼20%). The largest proportion of H. pylori ancestry in the mountains was European. Some of the AA1 ancestry in the mountains may have originated from strains once carried by slaves of West African origin. However, given the large European (AE1 and AE2) components in the mountain strains, it is probable that most of the AA1 is derived from strains acquired from the Spanish following their 15th century conquest of the Americas. Falush et al. documented a mean of ∼17% AA1 in 37 Spanish strains (23).
In Latin America, some strains from isolated indigenous populations are uniquely Amerindian, with affinities to East Asian H. pylori subtypes (30, 31). However, in mestizo communities where Amerindian populations have mixed with introduced European populations, there is evidence that Amerindian strains have been replaced by European strains with a competitive advantage, representing an exception to the pattern of vertical/familial transmission (32, 33). Our results are consistent with a general replacement of Amerindian H. pylori with European ancestral strains (as shown by low AEA, high AE2) and significant correlations between AE2 and human Amerindian ancestry. In contrast, we showed in the coastal population a high correlation of human African ancestry with AA1, consistent with vertical/familial transmission.
Because our current approach used matched samples and examined admixture, we were able to assess how coevolution may have affected gastric disease. We previously characterized the ancestry of H. pylori strains in these communities, using phylogenetic reconstruction without analysis of admixture (34). However, disregarding admixture in the H. pylori isolates represents an oversimplification, as would denoting human residents of these admixed communities as only African or only Amerindian. Conventional strain typing would have missed the AA1 ancestry in H. pylori from the mountains that, in our analyses, was critical evidence in detecting an interaction. Remarkably, the interaction effect between Amerindian host ancestry and AA1 was about five times larger than the effect of cagA (β = 5.08 vs. 0.98). We included cagA status in our analysis, because it is a known H. pylori virulence factor associated with a higher risk of severe gastritis and both premalignant and malignant lesions (28, 29, 35); cagA previously has been shown to differ in prevalence in the two locales, with higher prevalence in the mountains (22). We were able to replicate this difference and also demonstrate that cagA alone could not account for the difference in risk of premalignant lesions between populations, whereas the interaction between host ancestry and H. pylori ancestry did account for these differences, whether cagA was included in the model or not. The presence of a cagA gene, however, does not necessarily provide information about CagA functionality, which is dependent on many factors that were not assessed in the present study (36, 37). In contrast to cagA, which was a risk factor for premalignant histopathology independent of all other variables, geographic location and human ancestry did not independently predict disease outcome in a generalized multivariate mixed linear model with the interaction included.
We conclude that both human and H. pylori genetics influence susceptibility and pathogenicity but depend strongly on context. The effect of human ancestry on histopathology depends on the ancestry of the infecting H. pylori strain, and vice versa. Our data satisfy conditions essential to support a coevolutionary model (38) in which a phenotypic outcome must be the result of an interspecific genotype-by-genotype interaction, with the genotypes reflecting adaptive changes rather than random processes or genetic drift. H. pylori variation is known to mirror human migration patterns (23–25, 39), but this association alone does not demonstrate coevolution. We have shown that interactions between human and H. pylori ancestral clusters can confound the effects of each on prediction of histopathology scores; this result is evidence not only of parallel evolution but also of a functional relationship between the two genomes. Because our analyses demonstrated that area of residence could be ruled out as a factor when genetic ancestry was included in the model, the key variables influencing disease risk are likely to be genomic patterns of variation in both host and pathogen, interacting to predict clinical phenotype.
Our findings indicate that information about H. pylori admixture, particularly in the context of human ancestry, can have significant clinical implications, as has been anticipated by Falush et al. (23). For example, based on our current data, a patient at the 95th percentile of Amerindian ancestry (82.6%) has a vastly different risk profile depending on the percentage of AA1 in the infecting H. pylori strain; 73.1% AA1 (95th percentile) predicts a histopathology score of 4.8 [95% confidence interval (CI), 3.5–6.1], whereas 5.6% AA1 (fifth percentile) predicts a histopathology score of 3.1 (95% CI 2.6–3.6). In contrast, a patient at the fifth percentile of Amerindian ancestry (10.5%) harboring H. pylori with 73.1% AA1 will present with a histopathology score of only 2.4 (95% CI 2.0–2.8), according to our model. These results may be relevant to the “African Enigma,” i.e., the relatively low incidence of gastric cancer in Africa despite the high prevalence of H. pylori infection (3, 22, 40). Host and pathogen genomic variation are more predictive of histopathology when analyzed together than either one is alone and can be used to inform prevention efforts to eradicate H. pylori in those at greatest risk. In summary, coevolutionary relationships are important determinants of gastric disease, and the disruption of these relationships and subsequent adverse health outcomes may reflect a continued legacy of European colonization.
Materials and Methods
Study Participants.
Men and women were recruited from two locations in the State of Nariño, Colombia: from Tumaco, on the Pacific coast (n = 151, 72 males) and Tuquerres, in the Andes Mountains (n = 141, 66 males). Ages ranged from 18 to 65 y (mean 40.7 ± 11.8). Inclusion criteria were presentation with dyspeptic symptoms meriting upper gastrointestinal tract endoscopy. Patients treated with proton pump inhibitors, H2-receptor antagonists, or antimicrobials in the month before the endoscopy were excluded from this study. Also excluded were patients with chronic conditions such as diabetes, heart disease, or a prior gastrectomy. Informed consent was obtained from all participants. The ethics committees of the participating hospitals and the Universidad del Valle in Cali, Colombia and the Institutional Review Board of Vanderbilt University approved all study protocols.
Histopathology.
Gastric mucosa biopsy samples were taken from all participants from the gastric antrum (greater curvature, within 3 cm of the pylorus), incisura angularis (lesser curvature), and corpus (middle anterior wall) for histopathology. One additional antral biopsy from each participant was frozen in glycerol/thioglycolate for subsequent culturing of H. pylori organisms. All endoscopies were performed by a single experienced gastroenterologist. Frozen biopsies were shipped on dry ice to Vanderbilt University in Nashville, TN and stored at −80 °C.
One biopsy from each of the three gastric sites was fixed in buffered formalin and embedded in paraffin for histopathology. After staining with H&E, samples from individuals 40 y or older (77 from Tumaco; 49 from Tuquerres) were evaluated independently by two pathologists blinded to information about sample provenance. Diagnostic categories were based on the updated Sydney system for gastritis (41) and the Padova International Classification for Dysplasia (42). Cases with discordant diagnoses were reviewed until a consensus was reached. Ordinal values of 1–6 were assigned as follows: 1 = normal, 2 = nonatrophic gastritis, 3 = multifocal atrophic gastritis without intestinal metaplasia (MAG), 4 = intestinal metaplasia (IM), 5 = dysplasia, and 6 = carcinoma. Morphological differences within these categorical diagnoses also have been shown to be associated with gastric cancer risk (43–45). Therefore, a more detailed scoring system was implemented to take such differences into account. Within the category of MAG, the presence of indefinite (borderline) atrophy raised the score to 3.25, mild atrophy to 3.5, moderate atrophy to 3.75, and severe atrophy to 4.0 (46). The IM scores were augmented in two ways. First, the extent of the biopsy section exhibiting IM was assessed as <30%, between 30% and 60%, or >60% and was given a corresponding value of 0.2, 0.4, or 0.6, respectively. The second adjustment was based on the IM type and was ascertained via special staining with periodic acid Schiff/Alcian blue and high iron diamine/Alcian blue, as previously described, when needed (46). Complete IM, mixed predominant complete IM, mixed predominant incomplete IM, and incomplete IM were assigned values of 0.1, 0.2, 0.3, and 0.4, respectively. The two sets of measures were added to create a final IM score ranging from 4.3 to 5.0. Scores for dysplasia were refined by assigning a score of 5.25 for the category indefinite for dysplasia, 5.5 for low-grade dysplasia, and 5.75 for high-grade dysplasia. The detailed histopathology scoring system was used for statistical analyses, except where otherwise noted.
H. pylori Culture, Genotyping, and Multilocus Sequence Typing.
H. pylori were cultured from antral biopsies as previously described (34). Bacterial pellets grown from single colonies of H. pylori (one per subject, except for 13 Tumaco residents from whom two distinct strains per person were identified) were digested with proteinase K, and DNA was isolated by phenol/chloroform extraction and ethanol precipitation. Each strain was subjected to multilocus sequence typing (MLST) analysis (see below) and tested for the presence of cagA as previously described (47).
Characterizing H. pylori Ancestry.
For 288 strains, fragments of seven unlinked housekeeping genes (atpA, efp, ureI, ppa, mutY, trpC, and yphC), ranging from 398 to 627 bp per gene for a total of 3,406 bp, were PCR-amplified and sequenced on both strands for all samples, as previously described (34). On the basis of differences in allele frequency in these core genes, previous studies (23, 24) have used the Bayesian algorithm STRUCTURE (48, 49) to assign H. pylori isolates from multiple geographic sites to one of seven distinct populations (hpAfrica1, hpAfrica2, hpEurope, hpEAsia, hpNEAfrica, hpAsia2, or hpSahul). A set of 712 sequences used in these previous studies was retrieved from an MLST database (http://pubmlst.org/helicobacter), encompassing all seven global populations, including known subsets of hpAfrica1 (hspWAfrica and hspSAfrica) and hpEAsia (hspEAsia, hspMaori, and hspAmerind). After alignment with the Colombian sequences (n = 288), preliminary runs of STRUCTURE (version 2.3) demonstrated that the two highly isolated groups, hpAfrica2 and hpSahul, showed no evidence of ancestral overlap with the Colombian strains. Consequently hpAfrica2 and hpSahul sequences were dropped from the analysis, yielding a final dataset of 1,477 polymorphic sites derived from the 638 remaining MLST database sequences, 156 sequences from Tumaco, and 132 from Tuquerres.
The Admixture model of STRUCTURE assigns proportions of ancestry to each individual sample across K inferred ancestral clusters. The Linkage option of this model takes the chromosomal position of polymorphisms into account, incorporating into ancestry estimates the linkage disequilibrium that is expected after population admixture. Previous studies have used this model to characterize six ancestral clusters in globally distributed H. pylori isolates: AA1, AEA, AE1, AE2, and also two clusters, AA2 from hpAfrica2 samples and an isolated cluster from hpSahul samples, which are rare or nonexistent in Colombian samples (ref. 23 and Fig. S2). Our analyses with K = 4 maximized the model probability and generated the highest consistency of clustering by assigning fractions to AA1, AEA, AE1, or AE2 in all samples (Fig. S3). Graphical displays of population structure were created using DISTRUCT (50).
Characterizing Human Ancestry of Participants.
Human DNA from 242 study participants (122 from Tumaco and 120 from Tuquerres) was extracted from blood samples using Puregene kits (Qiagen). DNA was genotyped using the Immunochip (51), a platform that contains 196,524 SNPs from most of the genes known to be involved in immune disorders at the time of its design. To enrich for SNPs in linkage equilibrium, SNPs in linkage disequilibrium (LD) were removed using PLINK (r2 threshold >0.1) (52). Preliminary analyses with STRUCTURE (Admixture model) revealed that the model probability was maximized at K = 3, with the distribution of the three inferred ancestral groups matching the expected proportions of European, African, and Amerindian ancestry at both locations (53, 54). To validate these results and to refine ancestry estimates, the following reference populations were merged with existing data, using the PLINK toolset: all nonrelated individuals from the European (CEU) and African (YRI) HapMap populations (Phase 2, release 23) taken from files hosted on the PLINK website (55); the Iberian Spanish (IBS) population from the 1000 Genomes Project (56); and the H952 subset of the Karitania, Surui, and Colombian populations in the Human Genome Diversity Project (57, 58). The final dataset contained 5,947 SNPs and 514 individuals. The Admixture model of STRUCTURE (assuming correlated allele frequencies) was run 10 times (50,000 iterations after a burn-in of 50,000 iterations) under default settings and was supervised by reference population information. CLUMPP was used to collate replicate runs and calculate means of individual ancestry (59).
A complementary PCA of human ancestry was performed using the R program SNPRelate (60). Before PCA, the merged set of genotype data from the Colombian and YRI, CEU, and East Asian (CHB + JPT) founder populations was reduced to 30,735 SNPs using an LD threshold of 0.5 in the R environment using SNPRelate.
Statistical Analysis.
Descriptive statistics of ancestry were calculated for 242 human subjects and 288 Colombian H. pylori strains (from 275 subjects). Of these, 233 overlapped and were used to compute pairwise Pearson correlations between human ancestry (African, European, Amerindian) and H. pylori ancestry (AA1, AEA, AE1, AE2). Spearman ρ also was calculated to validate the Pearson correlations, because Shapiro–Wilk tests revealed lack of normality among some ancestries. Regression analyses assessing the dependence of histopathology on human and H. pylori ancestry are based on data for individuals age 40 y and over (n = 121). Because five of the individuals (all from the coast) contributed two strains each to the analysis, mixed linear models were implemented. Ordinal regression, a nonparametric approach, was used on discrete histopathology scores to determine if the likelihood of categorical increases in scores varied significantly with ancestry proportions, with the assumption of proportional odds tested using Brant’s test. A stepwise multivariate mixed linear model was built to assess whether human and H. pylori ancestry, their interaction, and cagA status could account for the known association between histopathology and area of residence. Residuals were tested for normality using the Shapiro–Wilk test. STATA 11 (StataCorp LP) was used for all analyses.
Supplementary Material
Acknowledgments
We thank Dr. Holli Dilks, Cara Sutcliffe, Lindsay Williamson, Nila Gillani, and Jamie L. Roberson of the VANTAGE Genotyping Core of the Center for Human Genome Resources of Vanderbilt University for helpful advice and assistance and Drs. William Bush and Peter Straub for help in accessing the 1000 Genomes data. This study was supported by Grant UL1 RR024975-01 from the National Center for Research Resources (now at the National Center for Advancing Translational Sciences); National Institutes of Health Grants 2 UL2 TR000445, P01 CA28842 (to P.C.), R01 CA77955 (to R.M.P.), P01 CA116087 (to R.M.P.), P30 DK058404 (to R.M.P.), R01 DK053620 (to K.T.W.), R01 DK058587 (to R.M.P.), P20 GM103534 (to N.K. and S.M.W.), R01 AI039657 (to T.L.C.), R01 AI068009 (to T.L.C.), and T32 AI007474 (to C.L.S.); and Grants 1I01BX001453 (to K.T.W.) and 01BX000627 (to T.L.C.) from the Office of Medical Research in the Department of Veterans Affairs. Additional support was provided by the Vanderbilt-Ingram Cancer Center, the Wendy Dio family, and the T.J. Martell Foundation (P.C. and R.M.P.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318093111/-/DCSupplemental.
References
- 1.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Holcombe C. Helicobacter pylori: The African enigma. Gut. 1992;33(4):429–431. doi: 10.1136/gut.33.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiba T, Seno H, Marusawa H, Wakatsuki Y, Okazaki K. Host factors are important in determining clinical outcomes of Helicobacter pylori infection. J Gastroenterol. 2006;41(1):1–9. doi: 10.1007/s00535-005-1743-4. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Perez GI, Rothenbacher D, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9(Suppl 1):1–6. doi: 10.1111/j.1083-4389.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- 6.Ghoshal UC, Chaturvedi R, Correa P. The enigma of Helicobacter pylori infection and gastric cancer. Indian J Gastroenterol. 2010;29(3):95–100. doi: 10.1007/s12664-010-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moodley Y, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull JJ, Molineux IJ, Rice WR. Selection of benevolence in a host-parasite system. Evolution. 1991;45(4):875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x. [DOI] [PubMed] [Google Scholar]
- 9.Agnew P, Koella JC. Virulence, parasite mode of transmission, and host fluctuating asymmetry. Proc Biol Sci. 1997;264(1378):9–15. doi: 10.1098/rspb.1997.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messenger SL, Molineux IJ, Bull JJ. Virulence evolution in a virus obeys a trade-off. Proc Biol Sci. 1999;266(1417):397–404. doi: 10.1098/rspb.1999.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser MJ, Chen Y, Reibman J. Does Helicobacter pylori protect against asthma and allergy? Gut. 2008;57(5):561–567. doi: 10.1136/gut.2007.133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll IM, Khan AA, Ahmed N. Revisiting the pestilence of Helicobacter pylori: Insights into geographical genomics and pathogen evolution. Infect Genet Evol. 2004;4(2):81–90. doi: 10.1016/j.meegid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Vaezi MF, et al. CagA-positive strains of Helicobacter pylori may protect against Barrett’s esophagus. Am J Gastroenterol. 2000;95(9):2206–2211. doi: 10.1111/j.1572-0241.2000.02305.x. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz S, et al. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 2008;4(10):e1000180. doi: 10.1371/journal.ppat.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghose C, Perez-Perez GI, van Doorn LJ, Domínguez-Bello MG, Blaser MJ. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J Clin Microbiol. 2005;43(6):2635–2641. doi: 10.1128/JCM.43.6.2635-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suerbaum S, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95(21):12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvani A. Epidemiology meets evolutionary ecology. Trends Ecol Evol. 2003;18(3):132–139. [Google Scholar]
- 18.Herre EA. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science. 1993;259(5100):1442–1445. doi: 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- 19.Stewart AD, Logsdon JM, Jr, Kelley SE. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution. 2005;59(4):730–739. [PubMed] [Google Scholar]
- 20.Torres J, et al. Gastric cancer incidence and mortality is associated with altitude in the mountainous regions of Pacific Latin America. Cancer Causes Control. 2013;24(2):249–256. doi: 10.1007/s10552-012-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correa P, et al. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst. 1976;57(5):1027–1035. doi: 10.1093/jnci/57.5.1027. [DOI] [PubMed] [Google Scholar]
- 22.Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: Do they explain the African enigma? Am J Gastroenterol. 2002;97(11):2839–2842. doi: 10.1111/j.1572-0241.2002.07031.x. [DOI] [PubMed] [Google Scholar]
- 23.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 24.Moodley Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323(5913):527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth T, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc Natl Acad Sci USA. 2004;101(14):4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48(13):3554–3560. [PubMed] [Google Scholar]
- 27.Correa P. Human Gastric Carcinogenesis: A multistep and multifactorial process–First American Cancer Society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52(24):6735–6740. [PubMed] [Google Scholar]
- 28.Tummuru MKR, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: Evidence of linkage to cytotoxin production. Infect Immun. 1993;61(5):1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Covacci A, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90(12):5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camorlinga-Ponce M, et al. Helicobacter pylori genotyping from American indigenous groups shows novel Amerindian vacA and cagA alleles and Asian, African and European admixture. PLoS ONE. 2011;6(11):e27212. doi: 10.1371/journal.pone.0027212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kersulyte D, et al. Helicobacter pylori from Peruvian amerindians: Traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS ONE. 2010;5(11):e15076. doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kersulyte D, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182(11):3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domínguez-Bello MG, et al. Amerindian Helicobacter pylori strains go extinct, as european strains expand their host range. PLoS ONE. 2008;3(10):e3307. doi: 10.1371/journal.pone.0003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Sablet T, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60(9):1189–1195. doi: 10.1136/gut.2010.234468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noto JM, Peek RM., Jr The Helicobacter pylori cag pathogenicity Island. Methods Mol Biol. 2012;921:41–50. doi: 10.1007/978-1-62703-005-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olbermann P, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6(8):e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102(1):36–43. doi: 10.1111/j.1349-7006.2010.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32(4):569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 39.Latifi-Navid S, et al. Ethnic and geographic differentiation of Helicobacter pylori within Iran. PLoS ONE. 2010;5(3):e9645. doi: 10.1371/journal.pone.0009645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell DI, et al. The African enigma: Low prevalence of gastric atrophy, high prevalence of chronic inflammation in West African adults and children. Helicobacter. 2001;6(4):263–267. doi: 10.1046/j.1083-4389.2001.00047.x. [DOI] [PubMed] [Google Scholar]
- 41.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20(10):1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Rugge M, et al. Gastric dysplasia: The Padova international classification. Am J Surg Pathol. 2000;24(2):167–176. doi: 10.1097/00000478-200002000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Ohata H, et al. Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. Int J Cancer. 2004;109(1):138–143. doi: 10.1002/ijc.11680. [DOI] [PubMed] [Google Scholar]
- 44.Filipe MI, et al. Intestinal metaplasia types and the risk of gastric cancer: A cohort study in Slovenia. Int J Cancer. 1994;57(3):324–329. doi: 10.1002/ijc.2910570306. [DOI] [PubMed] [Google Scholar]
- 45.González CA, et al. Gastric cancer occurrence in preneoplastic lesions: A long-term follow-up in a high-risk area in Spain. Int J Cancer. 2010;127(11):2654–2660. doi: 10.1002/ijc.25273. [DOI] [PubMed] [Google Scholar]
- 46.Mera R, et al. Long term follow up of patients treated for Helicobacter pylori infection. Gut. 2005;54(11):1536–1540. doi: 10.1136/gut.2005.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sicinschi LA, et al. Non-invasive genotyping of Helicobacter pylori cagA, vacA, and hopQ from asymptomatic children. Helicobacter. 2012;17(2):96–106. doi: 10.1111/j.1523-5378.2011.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg NA. DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 51.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3(11):e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4(3):e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70(Pt 6):841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 58.Cavalli-Sforza LL. The Human Genome Diversity Project: Past, present and future. Nat Rev Genet. 2005;6(4):333–340. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- 59.Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 60.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28(24):3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.