Significance
Toll-like receptors (TLRs) that sense microbial or endogenous DNA and RNA have been well implicated in systemic lupus erythematosus (SLE), a multisystem disease characterized by an autoimmune response to nuclear antigens. In mice, both TLR8 and TLR9 control TLR7-mediated lupus, but it is unknown if they have an additive effect in controlling TLR7. We demonstrate that double TLR8/9-deficient mice have increased abnormalities characteristic of SLE and that both TLR8 and TLR9 keep under control TLR7-mediated lupus, but they act on different cell types. TLR8 controls TLR7 function on dendritic cells, and TLR9 restrains TLR7 response on B cells. These TLR interactions have to be taken into account when novel therapeutic approaches are developed that target the blocking of TLRs.
Keywords: knockout mice, innate immunity, endosomal TLRs
Abstract
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with diverse clinical presentations characterized by the presence of autoantibodies to nuclear components. Toll-like receptor (TLR)7, TLR8, and TLR9 sense microbial or endogenous nucleic acids and are implicated in the development of SLE. In mice TLR7-deficiency ameliorates SLE, but TLR8- or TLR9-deficiency exacerbates the disease because of increased TLR7 response. Thus, both TLR8 and TLR9 control TLR7 function, but whether TLR8 and TLR9 act in parallel or in series in the same or different cell types in controlling TLR7-mediated lupus remains unknown. Here, we reveal that double TLR8/9-deficient (TLR8/9−/−) mice on the C57BL/6 background showed increased abnormalities characteristic of SLE, including splenomegaly, autoantibody production, frequencies of marginal zone and B1 B cells, and renal pathology compared with single TLR8−/− or TLR9−/− mice. On the cellular level, TLR8−/− and TLR8/9−/− dendritic cells were hyperesponsive to TLR7 ligand R848, but TLR9−/− cells responded normally. Moreover, B cells from TLR9−/− and TLR8/9−/− mice were hyperesponsive to R848, but TLR8−/− B cells were not. These results reveal that TLR8 and TLR9 have an additive effect on controlling TLR7 function and TLR7-mediated lupus; however, they act on different cell types. TLR8 controls TLR7 function on dendritic cells, and TLR9 restrains TLR7 response on B cells.
Systemic lupus erythematosus (SLE) is a complex chronic autoimmune disease that arises spontaneously and is characterized by production of autoantibodies against self-nucleic acids and associated proteins (1). These autoantibodies bind self-nucleic acids released by dying cells and form immune complexes that accumulate in different parts of the body, leading to inflammation and tissue damage. The kidneys, skin, joints, lungs, serous membranes, as well as, the cardiovascular, nervous and musculoskeletal system become targets of inflammation at onset or during the course of the disease (2). The etiology of SLE is unknown, yet genetics, sex, infectious agents, environmental factors, and certain medications may play a role in the initiation of the disease by causing alterations in lymphoid signaling, antigen presentation, apoptosis, and clearance of immune complexes (3, 4).
Toll-like receptors (TLRs) detect specific microbial components widely expressed by bacteria, fungi, protozoa, and viruses, and initiate signaling pathways critical for induction of immune responses to infection (5). In contrast to the cell surface TLRs that detect bacterial cell wall components and viral particles, nucleic acid-sensing TLRs are localized mainly within endosomal compartments (6). Human endosomal TLRs consist of TLR3, which senses viral double-stranded RNA (dsRNA) (7), TLR7 and TLR8, which recognize viral single-stranded RNA (8–10), and TLR9, which detects bacterial and viral unmethylated CpG-containing DNA motifs (11). Interestingly, these endosomal TLRs are also able to detect self-nucleic acids (12–14). Although the endosomal localization isolate TLR3, TLR7, TLR8, and TLR9 away from self-nucleic acids in the extracellular space, still self-RNA or -DNA can become a potent trigger of cell activation when transported into TLR-containing endosomes, and such recognition can result in sterile inflammation and autoimmunity, including SLE (4, 15, 16). The connection of the endosomal TLRs with SLE originates mainly from mouse models, where TLR7 signaling seems to play a central role. TLR7 gene duplication is the cause for the development of lupus in mice bearing the Y chromosome-linked autoimmune accelerating (Yaa) locus that harbors 17 genes, including TLR7 (17, 18). In TLR7 transgenic mouse lines, a modest increase in TLR7 expression promotes autoreactive lymphocytes with RNA specificities and myeloid cell proliferation, but a substantial increase in TLR7 expression causes fatal acute inflammatory pathology and profound dendritic cell (DC) dysregulation (17). In addition, studies in several lupus-prone mouse strains have revealed that TLR7-deficiency ameliorates disease, but TLR9-deficiency exacerbates it. Interestingly, this controversy can be explained by the enhanced TLR7 activity in the TLR9-deficient lupus mice (19, 20). Although murine TLR8 does not seem so far to be able to sense a ligand (21, 22), we have shown previously that it plays an important biological role in controlling TLR7-mediated lupus. Indeed, TLR8-deficiency in mice (on the C57BL/6 background that is not prone to lupus) leads to lupus development because of increased TLR7 expression and signaling in DCs (23). Thus, tight control and regulation of TLR7 is pivotal for avoiding SLE and inflammatory pathology in mice. Recent studies in humans have also revealed that increased expression of TLR7 is associated with increased risk for SLE (24–26).
Nucleic acid TLRs are expressed in many cell types, including DCs, plasmacytoid DCs (pDCs) and B cells, all of which play a central role in SLE development. TLR7, TLR8, and TLR9 signal through the adaptor molecule myeloid differentiation primary response gene 88 (MyD88), whereas TLR3 signals via the adaptor TRIF (Toll/IL-1 receptor domain-containing adaptor inducing IFN-β) (5). MyD88-deficiency abrogates most attributes of lupus in several lupus-prone mouse strains (19, 27–29). Moreover, deficiency for Unc93B1, a multipass transmembrane protein that controls trafficking of TLRs from the endoplasmic reticulum to endolysosomes and is required for nucleic acid-sensing TLR function (30), also abrogates many clinical parameters of disease in mouse lupus strains, suggesting that endosomal TLRs are critical in this disease (31). Interestingly, TLR9 competes with TLR7 for Unc93B1-dependent trafficking and predominates over TLR7 (32). TLR9 predominance is reversed to TLR7 by a D34A mutation in Unc93B1 and mice that carry this mutation show TLR7-dependent, systemic lethal inflammation (32).
Thus, in mice both TLR8 and TLR9 control TLR7-mediated lupus, but it is unknown if these TLRs act in parallel or in series in the same or different cell types and if they have an additive effect or not in controlling TLR7. To address these issues, we generated double TLR8/TLR9-deficient (TLR8/9−/−) mice and analyzed and compared the lupus phenotype in TLR8−/−, TLR9−/−, and TLR8/9−/− mice. Our data revealed that TLR8/9−/− mice have increased abnormalities characteristic of SLE and that both TLR8 and TLR9 keep TLR7-mediated lupus under control, but they act in different cell types. On DCs TLR7 function is ruled by TLR8, whereas on B cells TLR7 is mastered by TLR9.
Results
Splenomegaly and Reduced Innate B-Cell Populations in TLR8−/− and TLR8/9−/− Mice.
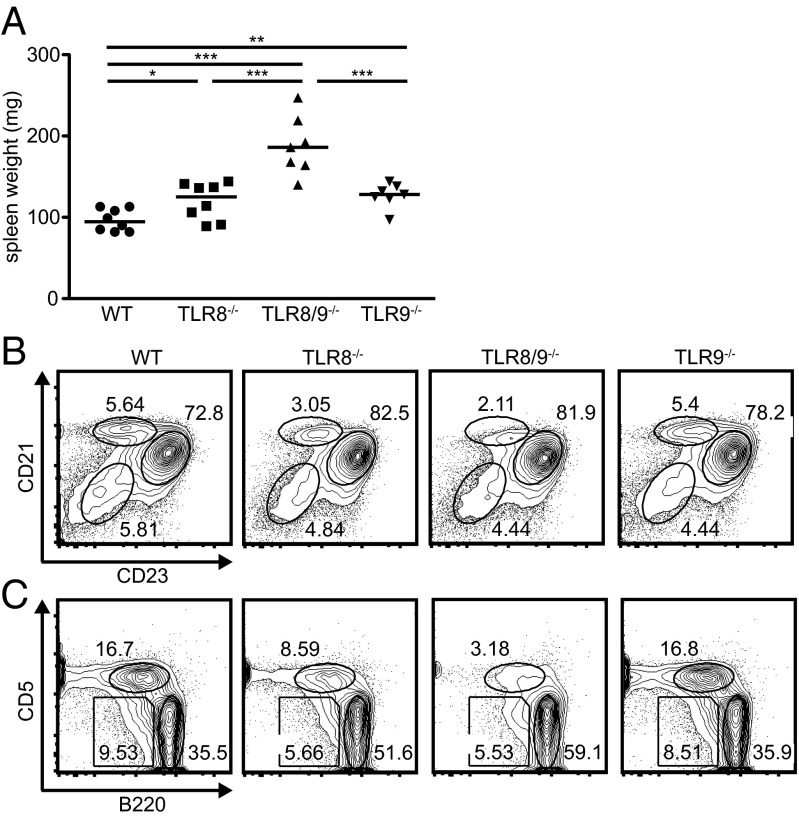
To determine whether TLR8 and TLR9 have an additive effect in the development of lupus autoimmunity on the C57BL/6 background, we generated double TLR8/9−/− mice by intercrossing TLR8−/− to TLR9−/− mice. TLR8/9−/− mice had normal appearance, growth, and fertility, and showed no obvious behavioral abnormalities. Moreover, similar to TLR8−/− and TLR9−/− mice, TLR8/9−/− mice had normal survival rates up to 12 mo of age. We have shown previously that TLR8-deficiency in mice leads to lupus that is accompanied by splenomegaly and a reduction of the marginal zone (MZ) and B1 B cells (23), so first we evaluated these populations. TLR8−/−, TLR8/9−/−, and TLR9−/− mice had splenomegaly compared with age- and sex-matched WT controls, whereas TLR8/9−/− mice showed the most exacerbated phenotype (Fig. 1A and Table S1). Moreover, both TLR8−/− and TLR8/9−/− mice had a defect on the MZ B frequencies (Fig. 1B and Table S2), and an expansion of the CD11c+ and CD19+ populations, whereas TLR9−/− mice looked normal (Table S1). Next, we evaluated the B1 B populations and noticed that B1a (B220loCD5int) and B1b (B220loCD5lo) B cells were reduced in TLR8−/− and TLR8/9−/− mice compared with WT or TLR9−/− mice, and the reduction of B1a B cells was more profound in TLR8/9−/− than in TLR8−/− mice (Fig. 1C and Table S2). Thus, TLR8/9−/− mice have a more severe defect on MZ and B1 B cells than TLR8−/− mice, although these populations appear to be unaffected in TLR9−/− mice.
Fig. 1.
TLR8/9−/− mice show increased splenomegaly and more severe defect on MZ and B1 B cells compared with TLR8−/− or TLR9−/− mice. (A) Spleen weight of 4- to 6-mo-old female WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice. Each point represents one mouse (n = 7–8 mice per genotype) and horizontal bars denote the median. (B) Splenocytes from 12-wk-old female WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice were analyzed by flow cytometry for the expression of CD19, CD21, and CD23. Numbers denote the percentage of MZ (CD21hiCD23lo/-), follicular (CD21intCD23hi), and immature (CD21−CD23−) B cells in the indicated circles. (C) FACS analysis of B220 and CD5 expression on CD19+ gated cells of the peritoneal cavity shows the percentage of B1a (B220loCD5int), B1b (B220loCD5lo), and B2 (B220hiCD5−) B cells. Data in B and C are representative of two to three independent experiments (n = 3–4 per group). *P < 0.05, **P < 0.01, ***P < 0.001.
Double TLR8/9−/− Mice Develop Stronger Lupus Phenotype than TLR8−/− or TLR9−/− Mice.
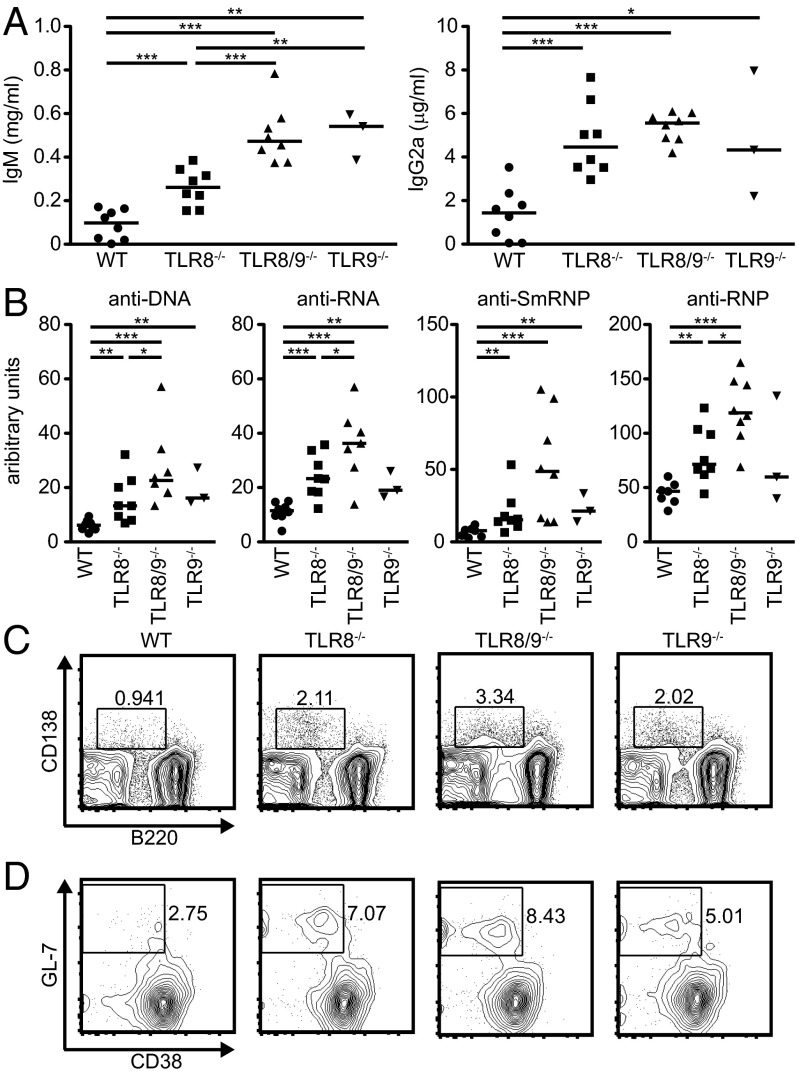
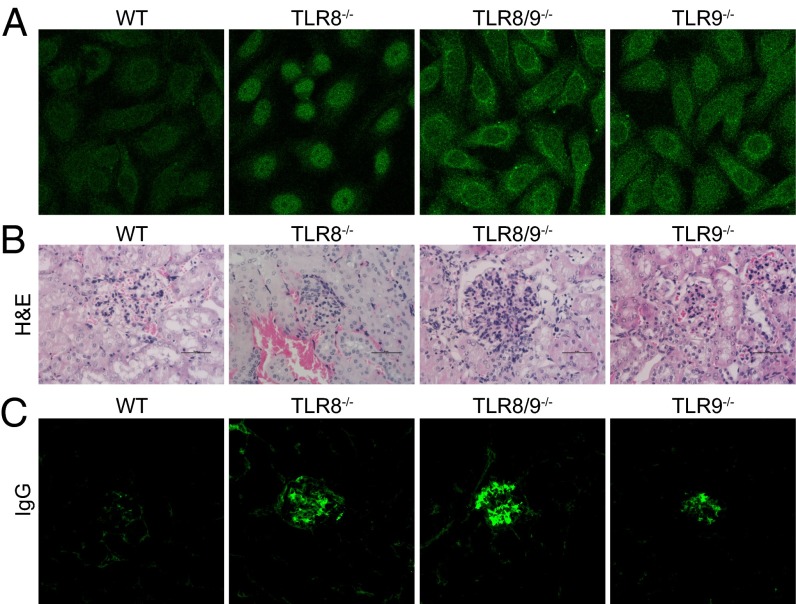
To determine whether the augmented immunological phenotype we observed in TLR8/9−/− mice correlates with increased autoimmunity, serum levels of IgM and IgG2a isotypes were assessed by ELISA. All three TLR-deficient genotypes showed significantly increased IgM and IgG2a levels compared with WT mice, whereas both TLR8/9−/− and TLR9−/− mice also had significantly elevated IgM levels compared with TLR8−/− mice (Fig. 2A). Next, we evaluated the levels of IgG autoantibodies against dsDNA, RNA, Smith ribonucleoprotein (SmRNP) and RNP and found that TLR8−/− and TLR8/9−/− mice had significantly increased titers compared with WT sera (Fig. 2B). Interestingly, TLR8/9−/− mice had also significantly higher levels of anti-dsDNA, -RNA, and -RNP production versus TLR8−/− mice, whereas TLR9−/− mice resembled TLR8−/− mice regarding anti-dsDNA, -RNA, and -SmRNP levels, and showed normal levels of anti-RNP (Fig. 2B). Then, we evaluated the percentages of antibody-producing B cells and found that the B220loCD138+ population was increased in all three TLR-deficient genotypes compared with WT controls, where TLR8/9−/− mice showed the highest increase, followed by TLR8−/− and TLR9−/− mice (Fig. 2C). Moreover, we noticed that there was a twofold increase in the percentage of spontaneous splenic germinal centers in TLR8−/− and TLR8/9−/− mice compared with WT controls (6.9 ± 0.5% and 7.8 ± 0.6%, respectively, vs. 3.0 ± 0.9%), and TLR9−/− mice showed slight increase compared with WT mice (4.5 ± 0.5% vs. 3.0 ± 0.9%) (Fig. 2D). Further analysis of autoantibody production showed that although antinuclear antibody (ANA) patterns varied somehow, TLR8−/− sera appeared mainly nucleolar, and TLR8/9−/− and TLR9−/− sera were both cytoplasmic and nucleolar (Fig. 3A).
Fig. 2.
Increased autoantibodies, plasmablasts, and germinal centers in TLR8/9−/− vs. TLR8−/− or TLR9−/− mice. Sera from 10- to 13-wk-old female WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice were used for the evaluation of (A) serum levels of IgM and IgG2a and (B) dsDNA-, RNA-, smRNP-, and RNP-specific autoantibody production by ELISA. Each point represents value from one mouse and horizontal bars denote the median. Representative flow cytometry plots of (C) B220loCD138+ plasmablasts and (D) B220+GL7+CD38− germinal center B cells in 5 mo old male WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice. Data in C and D are representative of two independent experiments (n= 3–4 per group). *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. 3.
Increased renal pathology in TLR8/9−/− mice compared with TLR8−/− or TLR9−/− mice. (A) ANA staining patterns on Hep2 human epithelial cells for sera derived from 9-wk-old mice (Original Magnification 400×). Kidney sections from 5- to 6-mo-old female WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice were stained with (B) H&E (scale bars, 50 uM) or (C) immunofluorescence anti-IgG (Original Magnification 100×). Data in A and C are representative of two independent experiments (n = 3–4 per group).
To evaluate renal pathology, we used semiquantitative pathological scoring of glomerular and interstitial nephritis. TLR8/9−/− mice had increased glomerular cellularity, glomerular deposits, and interstitial infiltrations compared with TLR8−/− or TLR9−/− mice (Fig. 3B, Fig. S1, and Table S3). In addition, we observed increased IgG and IgM glomerular depositions in TLR8/9−/− kidneys followed by TLR8−/− samples, whereas TLR9−/− mice showed reduced deposition compared with TLR8−/− mice, but stronger than in WT controls (Fig. 3C and Fig. S1B). Collectively, these data indicate that TLR8/9−/− mice show increased autoimmune phenotype compared with TLR8−/− or TLR9−/− mice, suggesting that both TLR8 and TLR9 contribute in lupus development in an additive manner.
Increased Response to R848 and Spontaneous Activation of TLR8−/− and TLR8/9−/− DCs and T Cells.
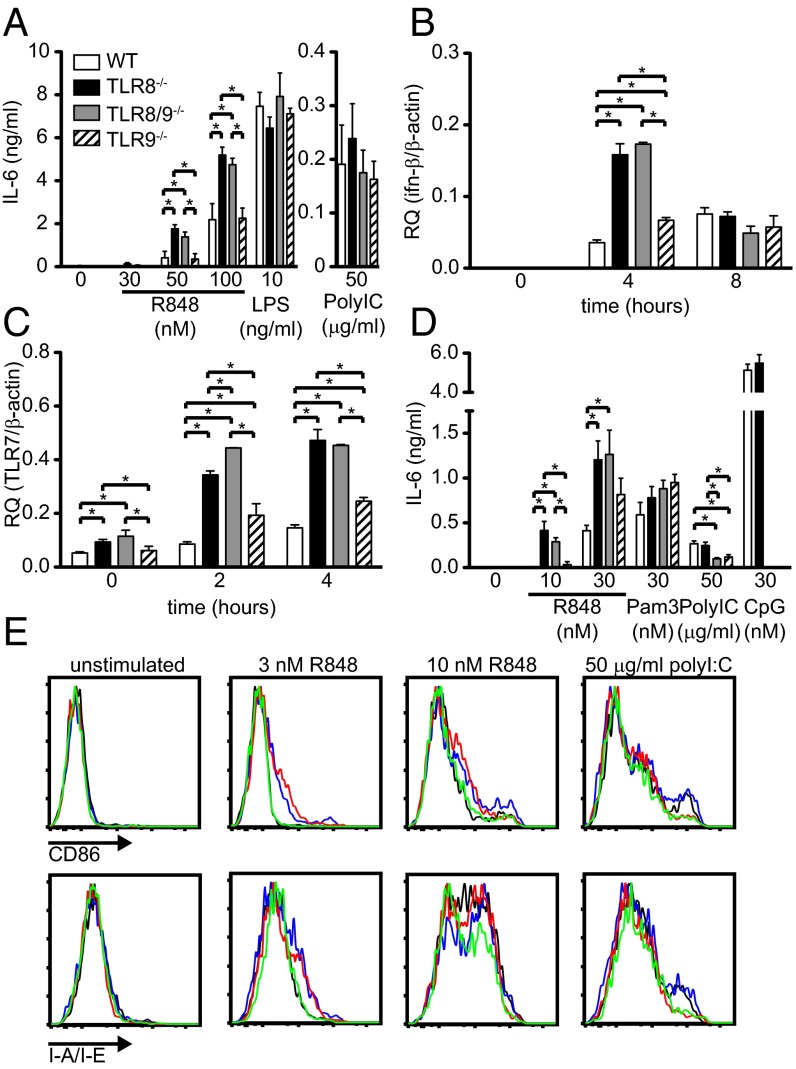
We have shown previously that TLR8-deficiency in mice leads to lupus development because of increased TLR7 expression and signaling in DCs, but not in macrophages (23). Therefore, we evaluated the responses of bone marrow-derived dendritic cells (BM-DCs) to R848 (TLR7 ligand), LPS (TLR4 ligand), and poly I:C (TLR3 ligand). In response to R848, both TLR8−/− and TLR8/9−/− BM-DCs produced similar and significantly higher amounts of IL-6 (Fig. 4A) and TNF (Fig. S2A), compared with WT or TLR9−/− cells, whereas the response to LPS or poly I:C was similar in all four genotypes (Fig. 4A and Fig. S2A). Moreover, R848 stimulation induced IFN-β mRNA expression that was significantly higher in TLR8−/− and TLR8/9−/− BM-DCs vs. WT or TLR9−/− cells at 4 h, and by 8 h the expression was reduced and became similar in all four genotypes (Fig. 4B). The higher cytokine response of TLR8−/− and TLR8/9−/− BM-DCs compared with WT or TLR9−/− cells was accompanied by significantly higher TLR7 mRNA expression in untreated cells that was sustained upon R848 stimulation at 2 or 4 h (Fig. 4C). Bone marrow-derived macrophages from TLR8−/−, TLR8/9−/−, and TLR9−/− mice produced normal levels of IL-6 in response to R848 or poly I:C (Fig. S2B).
Fig. 4.
Enhanced responses of TLR8−/− and TLR8/9−/− DCs and pDCs to TLR7 ligand. (A–C) BM-DCs from WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice were stimulated with R848, LPS, or poly I:C. (A) After 16 h, the concentration of IL-6 in the culture supernatants was assessed by ELISA. (B and C) BM-DCs were left untreated or stimulated with 50 nM R848 for the indicated time points. Total RNA was extracted from the cells and the expression of (B) IFN-β or (C) TLR7 was assessed by quantitative PCR. (D and E) BM-pDCs from WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice were stimulated with R848, Pam3CKS4, poly I:C, or CpG for 16 h. (D) The concentration of IL-6 in culture supernatants was assessed by ELISA. (E) The surface expression of CD86 and MHC class II was analyzed by flow cytometry on B220+CD11cintCD11b− cells from WT (black line), TLR8−/− (blue line), TLR8/9−/− (red line), and TLR9−/− (green line) mice. Data in all panels are representative of two to four independent experiments (n = 3–4 per group). *P < 0.05.
We next tested the responses of BM-pDCs to R848, Pam3CSK4 (TLR2 ligand), and CpG (TLR9 ligand). TLR8−/− and TLR8/9−/− BM-pDCs showed similar but significantly higher IL-6 protein levels to R848 stimulation than WT or TLR9−/− cells, whereas all genotypes responded similarly to the TLR2 agonist (Fig. 4D). As expected, WT and TLR8−/− BM-pDCs showed similar response to CpG, but this response was absent in TLR8/9−/− or TLR9−/− cells (Fig. 4D). Regarding activation, untreated BM-pDCs had similar levels of CD86 or MHC II; however, upon R848 stimulation, TLR8−/− and TLR8/9−/− BM-pDCs showed higher activation than WT or TLR9−/− cells (Fig. 4E). Thus, ex vivo BM-DCs and BM-pDCs from TLR8−/− and TLR8/9−/− mice show higher response to R848 stimulation than their WT or TLR9−/− counterparts.
Next, we evaluated the status of splenic DCs and pDCs. No significant differences were observed regarding the percentages of splenic DCs (CD11chiMHC-II+), pDCs (CD11clowSiglec-H+ or B220+CD11clow), CD11b+-like DCs (CD11chighCD11b+B220−SIRPα+), or CD8α+-like DCs (CD11chighCD11blowB220−SIRPα−CD24+) (33) between the four genotypes (Fig. S2D and Table S1). Nevertheless, we found that TLR8−/− and TLR8/9−/− splenic pDCs showed increased CD86 expression upon R848 stimulation compared with WT or TLR9−/− cells, and CD11b+-like DCs or CD8α+-like DCs showed similar response in all four genotypes (Fig. S2C). Furthermore, we evaluated TNF and IL-12 cytokine production upon R848 stimulation by FACS analysis and found that TLR8−/− and TLR8/9−/− pDCs, as well as TLR8/9−/− CD11b-like DCs, produced significantly higher TNF amounts compared with WT or TLR9−/− cells (Fig. S3 A and B, and Table S4). TLR8−/− and TLR8/9−/− CD11b-like DCs produced also higher amounts of IL-12 compared with WT or TLR9−/− cells (Fig. S3C and Table S4). Nevertheless, the three splenic DC subsets showed similar TNF and IL-12 production in response to poly I:C stimulation in all four mouse genotypes (Fig. S3 and Table S4). Hence, certain splenic TLR8−/− and TLR8/9−/− DCs subsets show higher CD86 expression and ability to produce TNF and IL-12 in response to R848 than WT or TLR9−/− cells.
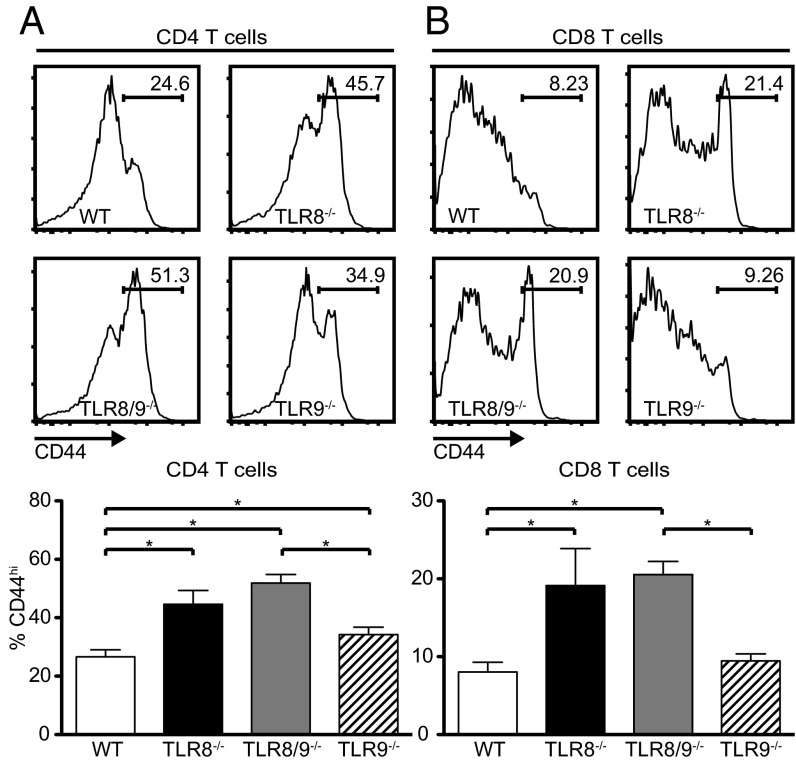
Because DCs play a central role in T-cell activation, we also tested the activation status of splenic T cells and noticed a significantly increased percentage of CD44hi CD4 T cells in TLR8−/−, TLR8/9−/−, and TLR9−/− mice compared with WT controls (Fig. 5A), and of CD44hi CD8 T cells in TLR8−/− and TLR8/9−/− compared with WT or TLR9−/− mice (Fig. 5B). Thus, the percentages of activated/memory CD4 and CD8 T cells are significantly increased in TLR8/9−/− mice compared with WT and TLR9−/− mice, and TLR8−/− mice show significant increased values compared with WT mice.
Fig. 5.
Increased activated memory T cells in TLR8−/− and TLR8/9−/− mice. Flow cytometric histograms (Upper) and graphical analysis (Lower) on splenocytes from 7-mo-old female WT, TLR8−/−, TLR8/9−/−, and TLR9−/− mice, analyzed for the expression of CD3, CD4, CD8, and CD44. CD44 staining profiles of gated (A) CD4+ and (B) CD8+ T cells to identify activated memory (CD44hi) subpopulations. Data are representative of three independent experiments (n = 4 per group). *P < 0.05.
Increased Response to R848 Stimulation of TLR9−/− and TLR8/9−/− B Cells.
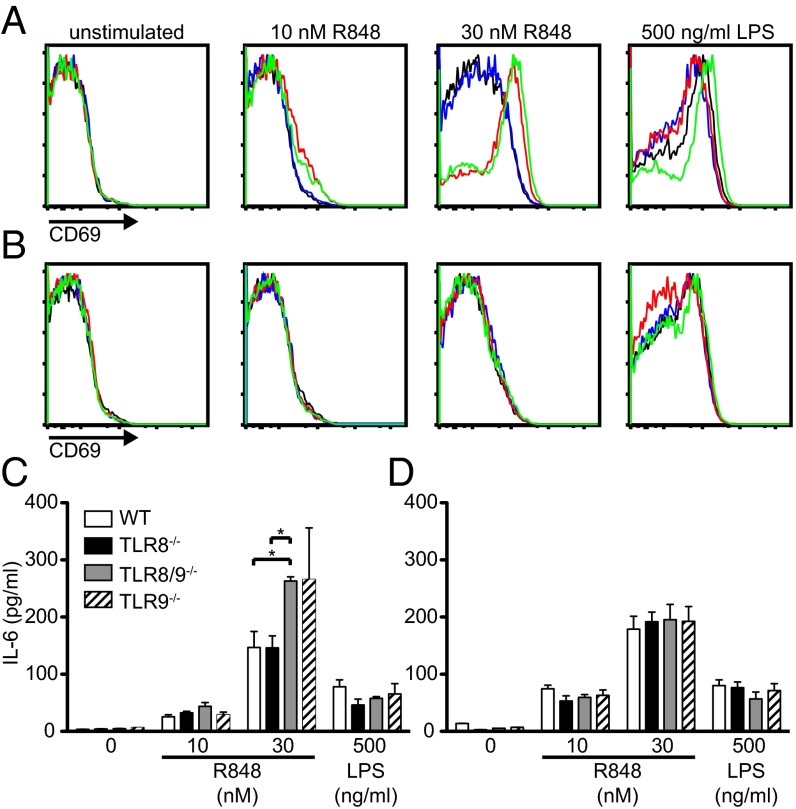
The nucleic acid components of SLE autoantigens have previously been shown to activate B cells through TLR7 and contribute to lupus disease (34). To investigate the contribution of B cells in the lupus phenotype that we observed in TLR8−/−, TLR8/9−/−, and TLR9−/− mice, total splenocytes were stimulated with R848 and the activation status of B cells were assessed by FACS analysis. Upon stimulation with R848, both TLR8/9−/− and TLR9−/− B cells showed a similar and significant up-regulation of CD69 and CD86, compared with TLR8−/− or WT B cells; all four genotypes showed similar response to LPS (Fig. 6A and Fig. S4A). Furthermore, TLR8/9−/− and TLR9−/− splenocytes showed increased production of IL-6 upon R848 stimulation compared with TLR8−/− or WT splenocytes, and all four genotypes responded similarly to LPS stimulation (Fig. 6C). Thus, upon TLR7-stimulation, TLR8/9−/− and TLR9−/− B cells in splenocytes are more responsive than WT or TLR8−/− cells. We wondered if this phenotype was accompanied by increased TLR7 expression, so we tested TLR7 expression in isolated B cells. To our surprise we found that TLR7 mRNA levels were significant higher in TLR8−/− and TLR8/9−/− B cells compared with WT or TLR9−/− cells (Fig. S4B). Moreover, we tested the responses of splenic TLR8−/−, TLR8/9−/−, TLR9−/−, and WT isolated B cells and found that all four genotypes showed similar CD69 up-regulation and IL-6 production upon R848 or LPS stimulation (Fig. 6 B and D). Thus, in splenocyte cultures TLR8/9−/− and TLR9−/− B cells have the capacity to respond higher to R848 stimulation than TLR8−/− or WT B cells, but this capacity is lost in isolated B-cell cultures, suggesting that other factors that are provided from the surrounding splenocytes are necessary. Indeed, it has been shown previously that B-cell responses to the TLR7 ligand depend on type I IFNs (35, 36). Therefore, to test if the lost capacity of isolated TLR9−/− B cells to respond higher to TLR7 ligand than WT cells (as we observed with total splenocytes) could be restored by providing type I IFN, isolated B cells from WT and TLR9−/− mice were stimulated with R848 in the presence of IFN-α. Indeed, IFN-α increased the capacity of B cells to respond to the TLR7 ligand, as assessed by up-regulation of CD69; however, both TLR9−/− and WT isolated B cells showed similar activation (Fig. S4C). Overall, the data demonstrate an increased response of TLR8/9−/− and TLR9−/− B cells to TLR7 ligand, that is lost in isolated B cells and cannot be restored by IFN-α priming.
Fig. 6.
Enhanced response of TLR9−/− and TLR8/9−/− B cells to TLR7 stimulation. Total splenocytes (A and C) or isolated B cells (B and D) from WT (black line), TLR8−/− (blue line), TLR8/9−/− (red line), and TLR9−/− (green line) mice were left untreated or stimulated with R848 or LPS for 16 h. Representative flow cytometry plots of CD69 on (A) CD119+B220+ splenocytes or (B) isolated B cells. IL-6 production on culture supernatants of (C) splenocytes or (D) isolated B cells. Data are representative of two to four independent experiments (n = 3 per group). *P < 0.05.
Discussion
Nucleic acid-sensing TLRs play an important role in SLE not only through their direct signaling upon ligand binding, but also by regulating the expression and function of each other. In the present study we show that in the C57BL/6 genetic background double deficiency for TLR8 and TLR9 leads to spontaneous lupus autoimmunity that is greater than the one that develop single TLR8−/− or TLR9−/− mice. Interestingly, the lupus disease in the TLR8/9−/− mice is caused by increased TLR7 function, where both TLR8 and TLR9 keep under control TLR7 activity, but they act on different cell types. TLR8 controls TLR7 function on DCs, and TLR9 restrains TLR7 response on B cells. Moreover, we are unique in showing that TLR9−/− mice on the C57BL/6 background develop signs of autoimmunity that include splenomegaly, increased serum levels of IgM, and autoantibodies against DNA, RNA, and SmRNP increased frequencies of plasmablasts and renal immunodeposits compared with WT mice.
In vitro studies in HEK293 cells transfected with human TLRs in a pairwise combination have shown that both TLR8 and TLR9 inhibit TLR7, but not vice versa (37). Furthermore, our recent previous studies have revealed that the inhibitory action of TLR8 on TLR7 has important consequences in vivo, because TLR8-deficiency in mice leads to lupus development as a result of increased TLR7 function (23). Studies on C57BL/6 mice that carry the Yaa locus (B6.Yaa mice) revealed that TLR9-deficiency leads to increased TLR7 expression on B cells and BM-pDCs (20). In the same study the authors showed that TLR9-deficiency in C57BL/6 mice congenic for the Nba2 locus (B6.Nba2) in the presence of the Yaa mutation (TLR9−/− B6.Nba2.Yaa mice) is accompanied by increased TLR7-dependent activation of B cells and pDCs, suggesting that TLR9 controls TLR7 function both in B cells and pDCs. However, here we showed that in the C57BL/6 background TLR9-deficiency leads to increased TLR7-dependent activation of B cells, but not of DCs. These discrepancies might be because of the comparison of different autoimmune accelerating loci or might be the outcome of differences in ex vivo culturing conditions. We found that both TLR8−/− and TLR8/9−/− DCs and pDCs show similar but increased response to TLR7 ligand compared with TLR9−/− or WT cells. On the other hand, TLR9−/− and TLR8/9−/− B cells show similar but increased activation and response upon TLR7 stimulation compared with TLR8−/− or WT cells (Fig. 6). Hence, the increased lupus phenotype in TLR8/9−/− mice is the outcome of the additive effect of TLR8 and TLR9 on controlling TLR7 on DCs and B cells, respectively.
Our data suggest that in vivo both TLR8 and TLR9 control TLR7 function, but what is the mechanism by which the absence of TLR8 or TLR9 affect TLR7-mediated lupus-like disease? Unc93B1 associates with and delivers nucleic-acid–sensing TLRs, including TLR7, TLR8, and TLR9, from the endoplasmic reticulum to endolysosomes for ligand recognition; however, the mode of regulation of TLR localization differs for each TLR (30, 38). Elegant studies by the Miyake group have shown that there is a competition of TLR7 and TLR9 for Unc93B1-dependent trafficking, where TLR9 predominates and has higher affinity than TLR7 for Unc93B1 (39). Interestingly, a D34A mutation of Unc93B1 does not affect TLR7 or TLR9 expression, but leads to increased TLR7 trafficking, and mice that carry this D34A mutation develop TLR7-dependent systemic lethal inflammation because of increased response to TLR7 ligand (32). Thus, Unc93B1 controls homeostatic TLR7 activation by balancing TLR9 to TLR7 trafficking. We can hypothesize that TLR8 or TLR9-deficiency decreases the competition of endosomal TLRs for association with Unc93B1 and allows higher availability for TLR7, which results to increased TLR7 trafficking and response that ultimately leads to autoimmunity. In addition, we have shown previously that in the C57BL/6 background TLR8-deficiency leads to TLR7 overexpression in DCs, B cells, macrophages, and MZ B cells (23). However, from all of these cell types only TLR8−/− DCs show increased response to TLR7 stimulation, whereas macrophages, B cells or MZ B cells have a normal response (23). Thus, depending on the cell-type deletion of TLR8 or TLR9 may alter not only TLR7 trafficking, but also TLR7 expression, which dictate TLR7’s ultimate function. Keeping in mind the complexity in expression pattern of the TLRs in a particular cell type and cell compartments, the variation in distribution and response to a given stimuli in different cell types, it is not surprising that TLR8 and TLR9 may control TLR7 function by mechanisms that can vary depending on the cell type.
Cumulative data suggest that from the nucleic-acid–sensing TLRs, TLR7 seems to be the most pathogenic regarding lupus (40); possible explanations could be the increased availability of exogenous or endogenous RNA-containing particles that can be sensed by TLR7 or stronger downstream signaling by TLR7 compared with TLR8 or TLR9. Most of the evidence that endosomal TLRs are implicated in SLE and that tight control and regulation of TLR7 is pivotal for avoiding SLE and inflammatory pathology is coming from studies in various lupus-prone mouse models. However, in our current studies we have used mice on the C57BL/6 background, which is not prone to lupus; we were still able to demonstrate that TLR7 dysregulation because of TLR8- and TLR9-deficiency can lead to autoreactivity and inflammatory pathology. In humans, recent data also incriminate TLR7 as an essential player in SLE pathology. Indeed, two independent studies reported that the SNP rs3853839 (C-G) at the 3′ untranslated region of TLR7 is associated with elevated transcript expression and increased risk for SLE in Eastern Asians (25, 26). Interestingly, further studies revealed that the risk G allele is also linked with increased TLR7 protein levels, and is located within a predicted binding site of microRNA-3148 that is most likely responsible for the observed association with SLE in three populations of non-Asian ancestry (24). Furthermore, increased TLR7 copy number is a risk factor for childhood-onset of SLE in the Mexican population, and correlates significantly with increased TLR7 and IFN-α mRNA levels (41). However, no correlation with SLE has been found regarding the TLR7 SNP rs179008 in Spanish population or variations in the copy number of the TLR7 (42, 43).
In conclusion, TLR8 and TLR9, on the top of their importance in direct signaling upon ligand recognition, also have an essential role in keeping under control TLR7 function to prevent spontaneous triggering of harmful autoreactive and inflammatory responses. Thus, the mechanisms by which TLR8 and TLR9 control TLR7 seem to depend on the given TLR and cell type, and these important TLR interactions have to be taken into account when novel therapeutic approaches are developed that target the blocking of one versus the other endosomal TLR to avoid unwanted consequences.
Materials and Methods
Detailed materials and methods are provided in SI Materials and Methods.
Mice and Serological Analysis.
Double TLR8/9−/− mice were generated by intercrossing TLR8−/− with TLR9−/− mice (11, 23). All mice used in the studies were in the C57BL/6 background. Evaluation of IgM and IgG2a, and IgG autoantibodies on serum samples were performed as described previously (23).
Preparation of Cells and Quantification of Cytokines.
Bone marrow cells from mice were cultured with GM-CSF or M-CSF for the production of BM-DCs or BM-macrophages, respectively, as previously described (23). IL-6 and TNF in culture supernatants were measured by ELISA kits.
RNA Isolation and Quantitative PCR.
Total RNA was isolated with TRIzol reagent or RNA easy kit, reverse-transcribed with Supercsript II reverse transcriptase and quantitiative PCR for TLR7, IFN-β, and β-actin was performed as described previously (23).
Flow Cytometric Analysis.
Cell suspensions were incubated with 24G2 hybridoma supernatant and then stained using immunofluorescence BM-DC antibodies. For intracellular cytokine staining, cells were fixed with Cytofix/Cytoperm and stained using antibodies for TNF and IL12p40/p70. Flow cytometry was conducted using an LSR2 and data were analyzed with FlowJo.
Histology, Immunofluorescence, and ANA Staining.
Kidneys were fixed in formalin, embedded in paraffin, and tissue sections were stained with H&E or periodic acid-Schiff. Renal biopsies were analyzed in a blinded fashion by a pathologist. Immunofluorescence IgG and IgM staining on kidney sections was performed as described previously (23). ANA in mouse sera were tested with Hep2 cells fixed on slides.
Supplementary Material
Acknowledgments
We thank S. Akira for providing TLR9−/− mice; C. Laprie for histological evaluation; L. Chasson (platform RIO/Marseille-Nice Genopole) and A. Zouine for technical help; and the personnel at the animal facility Laboratoire d’Exploration Fonctionelle Service inter-Institut Federatif de Recherche of the Centre d’Immunologie de Marseille Luminy for technical assistance. This work received funding from the Centre National de la Recherche Scientifique and Arthritis Fondation Courtin; a PhD fellowship from Région Provence-Alpes-Côte d’Azur co-financed by Innate Pharma (to A.B.M.); and by SkinDCs Agence Nationale de la Recherche (to S.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314121111/-/DCSupplemental.
References
- 1.Arbuckle MR, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 2.Mak A, Isenberg DA, Lau CS. Global trends, potential mechanisms and early detection of organ damage in SLE. Nat Rev Rheumatol. 2013;9(5):301–310. doi: 10.1038/nrrheum.2012.208. [DOI] [PubMed] [Google Scholar]
- 3.Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann Rheum Dis. 2013;72(Suppl 2):ii56–ii61. doi: 10.1136/annrheumdis-2012-202351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 6.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: Regulation through compartmentalization. Nat Rev Immunol. 2009;9(8):535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 9.Heil F, et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 10.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 12.Avalos AM, Busconi L, Marshak-Rothstein A. Regulation of autoreactive B cell responses to endogenous TLR ligands. Autoimmunity. 2010;43(1):76–83. doi: 10.3109/08916930903374618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green NM, Moody KS, Debatis M, Marshak-Rothstein A. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. J Biol Chem. 2012;287(47):39789–39799. doi: 10.1074/jbc.M112.383000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416(6881):603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 15.Christensen SR, Shlomchik MJ. Regulation of lupus-related autoantibody production and clinical disease by Toll-like receptors. Semin Immunol. 2007;19(1):11–23. doi: 10.1016/j.smim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theofilopoulos AN, et al. Sensors of the innate immune system: Their link to rheumatic diseases. Nat Rev Rheumatol. 2010;6(3):146–156. doi: 10.1038/nrrheum.2009.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deane JA, et al. Control of Toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27(5):801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairhurst AM, et al. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur J Immunol. 2008;38(7):1971–1978. doi: 10.1002/eji.200838138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickerson KM, et al. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol. 2010;184(4):1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santiago-Raber ML, et al. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J Autoimmun. 2010;34(4):339–348. doi: 10.1016/j.jaut.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Alexopoulou L, Desnues B, Demaria O. [Toll-like receptor 8: The awkward TLR] Med Sci (Paris) 2012;28(1):96–102. doi: 10.1051/medsci/2012281023. French. [DOI] [PubMed] [Google Scholar]
- 22.Cervantes JL, Weinerman B, Basole C, Salazar JC. TLR8: The forgotten relative revindicated. Cell Mol Immunol. 2012;9(6):434–438. doi: 10.1038/cmi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demaria O, et al. TLR8 deficiency leads to autoimmunity in mice. J Clin Invest. 2010;120(10):3651–3662. doi: 10.1172/JCI42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng Y, et al. Argentine Collaborative Group; BIOLUPUS and GENLES networks MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 2013;9(2):e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki A, et al. TLR7 single-nucleotide polymorphisms in the 3′ untranslated region and intron 2 independently contribute to systemic lupus erythematosus in Japanese women: A case-control association study. Arthritis Res Ther. 2011;13(2):R41. doi: 10.1186/ar3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen N, et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci USA. 2010;107(36):15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehlers M, Fukuyama H, McGaha TL, Aderem A, Ravetch JV. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. J Exp Med. 2006;203(3):553–561. doi: 10.1084/jem.20052438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groom JR, et al. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J Exp Med. 2007;204(8):1959–1971. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadanaga A, et al. Protection against autoimmune nephritis in MyD88-deficient MRL/lpr mice. Arthritis Rheum. 2007;56(5):1618–1628. doi: 10.1002/art.22571. [DOI] [PubMed] [Google Scholar]
- 30.Lee BL, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. eLife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kono DH, et al. Endosomal TLR signaling is required for anti-nucleic acid and rheumatoid factor autoantibodies in lupus. Proc Natl Acad Sci USA. 2009;106(29):12061–12066. doi: 10.1073/pnas.0905441106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukui R, et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35(1):69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Guilliams M, et al. From skin dendritic cells to a simplified classification of human and mouse dendritic cell subsets. Eur J Immunol. 2010;40(8):2089–2094. doi: 10.1002/eji.201040498. [DOI] [PubMed] [Google Scholar]
- 34.Hwang SH, et al. B cell TLR7 expression drives anti-RNA autoantibody production and exacerbates disease in systemic lupus erythematosus-prone mice. J Immunol. 2012;189(12):5786–5796. doi: 10.4049/jimmunol.1202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekeredjian-Ding IB, et al. Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. J Immunol. 2005;174(7):4043–4050. doi: 10.4049/jimmunol.174.7.4043. [DOI] [PubMed] [Google Scholar]
- 36.Green NM, et al. Murine B cell response to TLR7 ligands depends on an IFN-beta feedback loop. J Immunol. 2009;183(3):1569–1576. doi: 10.4049/jimmunol.0803899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, et al. The functional effects of physical interactions among Toll-like receptors 7, 8, and 9. J Biol Chem. 2006;281(49):37427–37434. doi: 10.1074/jbc.M605311200. [DOI] [PubMed] [Google Scholar]
- 38.Itoh H, et al. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS ONE. 2011;6(12):e28500. doi: 10.1371/journal.pone.0028500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukui R, et al. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med. 2009;206(6):1339–1350. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theofilopoulos AN. TLRs and IFNs: Critical pieces of the autoimmunity puzzle. J Clin Invest. 2012;122(10):3464–3466. doi: 10.1172/JCI63835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Ortiz H, et al. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Ann Rheum Dis. 2010;69(10):1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- 42.Sánchez E, et al. Investigation of TLR5 and TLR7 as candidate genes for susceptibility to systemic lupus erythematosus. Clin Exp Rheumatol. 2009;27(2):267–271. [PubMed] [Google Scholar]
- 43.Kelley J, Johnson MR, Alarcón GS, Kimberly RP, Edberg JC. Variation in the relative copy number of the TLR7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56(10):3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.