Significance
Transthyretin, a normal circulating plasma protein, is inherently amyloidogenic. It forms abnormal, insoluble, extracellular amyloid fibrils in the elderly, sometimes causing structural and functional damage leading to disease, senile amyloidosis. More than 100 different point mutations in the transthyretin gene cause earlier adult-onset, autosomal-dominant, fatal, hereditary amyloidosis. The transthyretin variant Ser52Pro is responsible for the most aggressive known clinical phenotype. Here we identify the crucial pathogenic role of specific proteolytic cleavage at residue 48 in triggering fibril formation by this variant. Genuine amyloid fibril formation in vitro is much more extensive than previously reported for wild-type transthyretin or any other transthyretin variant. Characterization of the fibrillogenic effect of this cleavage powerfully informs drug design and targeting for transthyretin amyloidosis.
Keywords: misfolding, protein aggregation
Abstract
The Ser52Pro variant of transthyretin (TTR) produces aggressive, highly penetrant, autosomal-dominant systemic amyloidosis in persons heterozygous for the causative mutation. Together with a minor quantity of full-length wild-type and variant TTR, the main component of the ex vivo fibrils was the residue 49-127 fragment of the TTR variant, the portion of the TTR sequence that previously has been reported to be the principal constituent of type A, cardiac amyloid fibrils formed from wild-type TTR and other TTR variants [Bergstrom J, et al. (2005) J Pathol 206(2):224–232]. This specific truncation of Ser52Pro TTR was generated readily in vitro by limited proteolysis. In physiological conditions and under agitation the residue 49-127 proteolytic fragment rapidly and completely self-aggregates into typical amyloid fibrils. The remarkable susceptibility to such cleavage is likely caused by localized destabilization of the β-turn linking strands C and D caused by loss of the wild-type hydrogen-bonding network between the side chains of residues Ser52, Glu54, Ser50, and a water molecule, as revealed by the high-resolution crystallographic structure of Ser52Pro TTR. We thus provide a structural basis for the recently hypothesized, crucial pathogenic role of proteolytic cleavage in TTR amyloid fibrillogenesis. Binding of the natural ligands thyroxine or retinol-binding protein (RBP) by Ser52Pro variant TTR stabilizes the native tetrameric assembly, but neither protected the variant from proteolysis. However, binding of RBP, but not thyroxine, inhibited subsequent fibrillogenesis.
Amyloidosis is caused by extracellular accumulation of abnormal protein fibrils in various tissues throughout the body (1). Hereditary systemic amyloidosis is a variably penetrant autosomal dominant condition caused by mutations encoding usually single-residue substitutions in certain globular proteins, including transthyretin (TTR), apolipoprotein-AI, fibrinogen A α-chain (2), gelsolin, lysozyme, cystatin C, and β2-microglobulin (3). Individuals heterozygous for the causative mutated genes produce variant proteins that are less stable than their wild-type counterparts and consequently have a propensity to misfold and then aggregate in the characteristic amyloid fibril conformation and deposit in the tissues. TTR variants, encoded by more than 100 different amyloidogenic mutations (http://amyloidosismutations.com/attr.html), are by far the most common cause of hereditary amyloidosis, although there are only ∼10,000 cases worldwide.
Elucidation of the mechanism of amyloidogenic conversion of TTR has been challenging and hitherto has not been revealed by any obvious feature of the native protein structure despite very high-resolution crystallography of the wild-type and several amyloidogenic variants (4). The native tetrameric assemblies of all amyloidogenic TTR variants studied so far are less stable in vitro than wild-type TTR, whereas the Thr119Met variant is more stable and is known to protect carriers of amyloidogenic mutations from developing TTR amyloidosis (5). Therefore it is widely accepted that tetramer dissociation and partial denaturation of released monomers is the crucial and rate-limiting prerequisite for the formation of TTR amyloid fibrils (2). However, there is no direct supportive evidence for this mechanism in vivo, and a key role for posttranslational modifications of TTR, including partial proteolysis has been debated extensively (6).
In addition to full-length TTR monomers, Lundgren’s group identified the residue 49-127 fragment, generated by proteolytic cleavage of the peptide bond Lys48–Thr49 (7), as the main fragment present in ex vivo TTR amyloid fibrils. The protease responsible has not yet been identified, but the highly specific cleavage suggests that it could be a trypsin-like serine protease. Meanwhile Westermark and collaborators have elegantly characterized the constituents of TTR amyloid fibrils extracted from cardiac amyloid deposits and adipose tissue, identifying two clearly distinct categories (8, 9). Type A fibrils contain a high proportion of truncated species that are not found in type B fibrils, which are composed almost entirely of full-length TTR (8). The presence of cleaved fragments apparently is not influenced by the nature or the position of the amino acid substitution (8). Rather, it correlates with the abundance of the cardiac amyloid deposits (10), with increased patient age, and with late-onset disease, being most common in persons with senile systemic amyloidosis caused by wild-type, not variant, TTR. The possible pathogenic role of proteolytic cleavage is suggested further by the clinical course in persons with hereditary TTR amyloidosis who undergo liver transplantation to replace their variant TTR production with wild-type TTR. Individuals with type A fibrils in their cardiac deposits have a notably poorer outcome, often with rapidly progressive cardiac involvement (11).
Here we report a large kindred in which affected individuals carrying the mutation encoding Ser52Pro variant TTR developed unusually early-onset, aggressive, and fatal systemic amyloidosis. Rapidly progressive amyloid polyneuropathy, soon followed by symptomatic cardiac amyloidosis, developed in the third decade, typically leading to death within 5 y. We now have fully characterized the native structure of the variant and the composition of the ex vivo amyloid fibrils, which contain abundant truncated TTR cleaved at the C terminus of residue 48. This specific cleavage was remarkably reproduced by limited proteolysis of the isolated recombinant variant TTR in vitro, and we have established that, once released from the native tetrameric protein, the polypeptide 49-127 rapidly self-aggregates into genuine amyloid fibrils.
Results
Hereditary Amyloidosis Phenotype.
The proband (Fig. S1A, III-2) presented at age 30 y with autonomic dysfunction, peripheral neuropathy, euthyroid multinodular goiter, and sicca syndrome. A needle biopsy of the thyroid contained extensive TTR amyloid deposits, and the causative mutation encoding the Ser52Pro TTR variant was detected by DNA sequencing. 123I-serum amyloid P component (SAP) scintigraphy (12) demonstrated substantial visceral amyloidosis affecting the spleen, liver, kidneys, and adrenal glands. Echocardiography and subsequent cardiac angiography demonstrated an infiltrative restrictive cardiomyopathy with well-preserved ventricular function, and there were abundant TTR amyloid deposits in an endomyocardial biopsy. He underwent liver transplantation in an attempt to reduce abundance of the amyloidogenic variant, but amyloidosis nevertheless progressed rapidly, and he died 5 mo later with cardiac failure and progressive neuropathy. The proband's mother (II-1) died at age 34 y from a progressive neuromuscular disorder, and extensive systemic amyloidosis was discovered at post mortem. The clinical picture in subjects II-3 and II-5 was consistent with neuropathic systemic amyloidosis, but no histological confirmation was available. The maternal uncle of the propositus (II-3) was diagnosed as paraplegic before his death at age 42 y in renal failure. Subject II-5 developed thyromegaly from about age 20 y and died at age 30 y from a neuromuscular disease with a presumptive diagnosis of multiple sclerosis. SAP scintigraphy in the proband's brother (III-3) at age 28 y showed extensive amyloid in the spleen, kidneys, and adrenal glands, similar to the propositus. He developed amyloid polyneuropathy 1 y later and died with progressive amyloidosis, including severe cardiac involvement, 3 mo after liver transplantation. The third brother does not carry the TTR mutation and remains asymptomatic. Two cousins died before age 40 y with a similar pattern of disease but including clinically significant leptomeningeal TTR amyloid deposits. One of these subjects underwent liver transplantation when he had only the very earliest symptoms of neuropathy and no definite cardiac amyloid deposition, but he died 8 y later having developed profound neuropathy, severe cardiac amyloidosis, and dementia. The clinical phenotype in this kindred is among the most severe ever described in TTR-related amyloidosis.
DNA Sequencing.
The proband, his affected brother, and the two affected cousins were heterozygotes for a single-point mutation in exon 3 of the TTR gene, encoding substitution of a proline (codon CCT) for serine (codon TCT) at residue 52 of the mature TTR protein (Fig. S1B). The father, unaffected brother, and unaffected cousins were homozygous for the wild-type sequence.
Amyloid Fibril Protein Typing.
Immunohistochemical staining of the cardiac biopsy from the propositus identified TTR as the amyloid fibril protein (Fig. S1C), and the deposits also stained for SAP, the universal nonfibrillar component of all human amyloid.
Analysis of ex Vivo Fibrils.
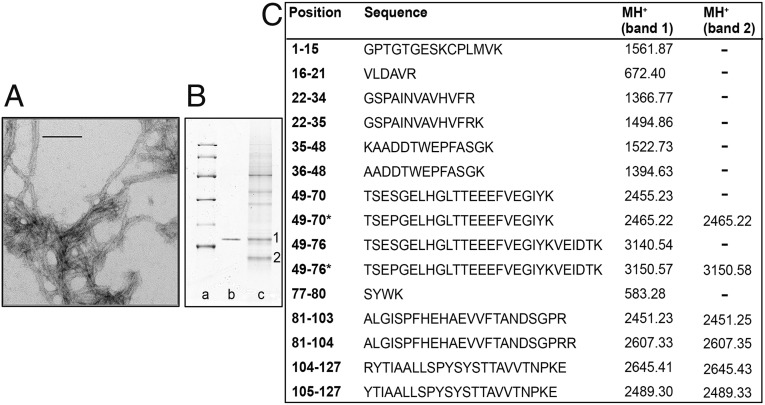
Ex vivo amyloid fibrils were isolated from the spleen of the propositus removed during liver transplantation and were separated by SDS/PAGE (Fig. 1 A and B) for further proteomic analysis. The mass mapping analysis of the tryptic peptides from bands 1 and 2 in the gel are shown in Fig. 1C. The MALDI-MS spectrum of band 1 showed the presence of several mass signals corresponding to TTR tryptic peptides leading to high protein-sequence coverage. In particular, the mass signal at m/z 3,150.57 was assigned to the tryptic peptide 49-76 carrying the Ser-to-Pro amino acid substitution. A small signal at m/z 3,140.54 corresponding to the wild-type peptide was detected also. LC-MS/MS analysis of tryptic peptides from band 1 showed that both the wild-type and Ser52Pro full-length TTR were present with two mass signals at m/z 819.8 and 823.0 corresponding to the triply charged wild-type and variant peptides 49-70, respectively. This assignment was confirmed by the MS/MS spectra of the two precursor ions. In contrast, MS analysis of band 2 showed only a single protein species, corresponding to the C-terminal fragment of the variant TTR generated by cleavage at residue Lys48. Five cycles of N-terminal Edman sequencing of band 2 yielded Thr-Ser-Glu-Pro-Gly, i.e., residues 49–53 of variant TTR, unequivocally confirming the presence of proline at only position 52 (Fig. S2).
Fig. 1.
Ex vivo amyloid fibrils. (A) Transmission electron microscopy of amyloid fibrils extracted from spleen. (Direct magnification: 230,000×; scale bar, 100 nm.) (B) SDS 15% PAGE under reducing conditions. Lane a: marker proteins (14.4, 20.1, 30.0, 45.0, 66.0, and 97.0 kDa, respectively); lane b: 0.5 μg recombinant Ser52Pro TTR; lane c: 60 μg ex vivo amyloid fibrils from spleen. 1 and 2 indicate bands subjected to mass mapping analysis. (C) Tryptic peptides obtained by digestion of SDS/PAGE bands were analyzed by MALDI-MS and nano-LC MS-MS. MH+ monoisotopic values are reported for each peptide; asterisks indicate the presence of the Ser52Pro substitution. Cysteine is carbamidomethylated.
Authenticity and Stability of Recombinant TTR Proteins.
The subunits of recombinant wild-type and Ser52Pro variant TTR proteins had molecular masses of 13,892.2 ± 0.9 Da and 13,902.1 ± 1.5 Da, respectively, corresponding to the expected theoretical mass values of 13,892.6 Da and 13,902.7 Da, including the additional N-terminal methionine residue, Met0. Correct assembly of the subunits into native homotetrameric wild-type and variant TTR was confirmed by both size-exclusion chromatography and native mass spectrometry, and these intact proteins bound the natural ligand, thyroxine, normally, showing the same IC50 with mds84 and Tafamidis (13, 14) as native wild-type TTR isolated from normal human serum (Table S1). However, the guanidine thiocyanate (Gdn-SCN)–mediated transition from folded tetramer to unfolded monomers (Fig. S3) (5) showed that the native tetrameric Ser52Pro variant TTR was destabilized significantly, by 2.3 kcal/mol, compared with the wild-type protein, with a midpoint concentration of Gdn-SCN (Cm) shifting from mean (SD) of 1.03 (0.02) M for the wild type to 0.8 (0.06) M for the variant (n = 3) (Table S2).
Limited Proteolysis of Recombinant Wild-Type and Variant TTR.
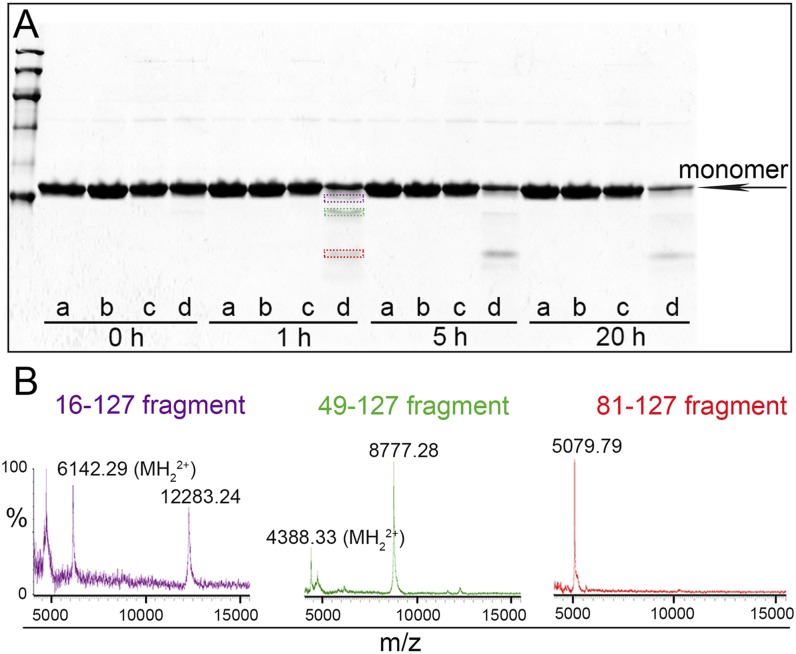
Proteolysis at a very low enzyme:substrate ratio is a powerful and highly discriminating approach for comparing the most flexible and solvent-exposed sites and thus monitoring focal stability in the surface topology of native globular proteins. It revealed a surprisingly dramatic difference between wild-type and Ser52Pro variant TTR, the former being completely resistant to cleavage even after long incubation or increased protease concentration, whereas the latter was proteolyzed rapidly. Fig. 2A shows the electrophoretic pattern of the product of trypsin digestion. After 1 h of incubation the variant TTR released three main polypeptides which we identified by MALDI-MS analysis of the corresponding electroeluted bands as peptides 16-127 (purple in Fig. 2), 49-127 (green in Fig. 2), and 81-127 (red in Fig. 2), respectively. In the same lane of the gel the band of the undigested monomer of Ser52Pro TTR was also present (Fig. 2A).
Fig. 2.
Limited trypsin proteolysis of wild-type and Ser52Pro TTR. (A) SDS (15%) PAGE under reducing conditions of wild-type and Ser52Pro TTR after incubation at 37 °C with trypsin at an enzyme:substrate ratio of 1:200 for the times shown. Wild-type TTR alone (a) or with trypsin (b), Ser52Pro TTR alone (c) or with trypsin (d). The three main polypeptides released from Ser52Pro TTR after 1 h of digestion are highlighted with purple, green, and red boxes. (B) MALDI-MS spectra of the fragments electroeluted from the bands shown in A and acquired in linear mode.
Consistent with the protease resistance of wild-type TTR, and in sharp contrast to the exquisite sensitivity of the Ser52Pro variant, none of the other recombinant amyloidogenic TTR variant proteins which we tested (Val30Met, Leu55Pro, and Val122Ile) was cleaved at all under the conditions used (Fig. S4). In this experiment we monitored the proteolytic cleavage in the early phase of incubation up to 2 h, thus showing that the peptide 49-127 (green box in Fig. S4) is formed already at 3 min of trypsin exposure. The peptide 81-127 (red box in Fig. S4) is most likely a later product derived from a secondary proteolytic cleavage, as are other minor unidentified lower molecular weight fragments.
TTR Cleavage and Fibril Formation.
Like wild-type TTR and all other amyloidogenic TTR variants previously tested, the full-length Ser52Pro variant did not form amyloid fibrils in vitro when stirred in solution at physiological pH and ionic strength (Fig. 3A). In contrast, when trypsin was added to the native Ser52Pro TTR solution in PBS at 37 °C while the solution was stirred at 900 rpm, thioflavin T (ThT) fluorescence increased very rapidly (Fig. 3A). Fibrillogenesis of this variant under physiological conditions was strictly dependent on both proteolytic cleavage and stirring, and the main component of this fibrillar and insoluble material was the same residue 49-127 fragment (Fig. 3B, lane c) identified in the natural fibrils (Fig. 3B, lane d). The fibrils formed by residue 49-127 variant TTR peptide in vitro showed the pathognomonic amyloid green birefringence under polarized light after staining with Congo red. Transmission electron microscopy showed that the aggregated TTR was almost entirely fibrillar, with minimal amorphous material (Fig. 3C), indicating much more efficient fibrillar conversion than previously reported for in vitro TTR aggregation (15). Atomic force microscopy (Fig. 3D) confirmed the fibrillar ultrastructure. The diffraction pattern obtained from partially aligned Ser52Pro TTR variant amyloid fibrils showed diffraction signals at 4.7 Å and 9.5 Å (Fig. 3E), consistent with the cross-β structure previously reported for many different types of amyloid fibrils, including Val30Met TTR (16). Although the fibrils were difficult to orient, probably because of their tangled nature (Fig. 3C), the diffraction pattern shows increased intensity on the vertical axis compared with horizontal axes supporting the cross-β arrangement whereby β-strands are hydrogen bonded along the length of the fibril with a regular 4.7-Å distance between them. The side chains are accommodated between the β-sheets that are associated with a distance of ∼9.5 Å. In addition to the cross-β signals, a strong reflection at 18 Å was observed on the equator of the pattern; this signal may arise from the association of protofilaments within the fibrils.
Fig. 3.
Fibrillogenesis of Ser52Pro TTR triggered by release of the residue 49-127 fragment. (A) Normalized ThT fluorescence emission from wild-type TTR (black) and Ser52Pro TTR (red) fibrillogenesis in the presence (solid lines) and absence (dotted lines) of trypsin 5 ng/μL. (B) SDS 15% PAGE under reducing conditions. Lane a: marker proteins (14.4, 20.1, 30.0, 45.0, 66.0, and 97.0 kDa); lane b: recombinant Ser52Pro TTR; lane c: in vitro Ser52Pro fibrils; lane d: ex vivo amyloid fibrils from spleen. (C) Negatively stained transmission electron micrograph of Ser52Pro TTR amyloid fibrils generated in the presence of trypsin and shaking. (Direct magnification: 175,000×; scale bar, 100 nm.) (D) Tapping mode atomic force microscopy image (height data) of Ser52Pro TTR fibrils obtained as in A. Scan size: 2.0 μm; Z range: 10 nm. (E) The X-ray fiber diffraction pattern collected from partially aligned amyloid fibrils formed by Ser52Pro TTR (λ = 1.5417 Å; sample-to-detector distance = 100 nm; exposure time = 30 s) shows a reflection that is stronger on the meridian at 4.7 Å. A reflection also is highlighted at 9.5 Å, and a well-oriented signal is seen at 18 Å.
Despite the intensely fibrillogenic nature of the residue 49-127 fragment of Ser52Pro TTR, addition of the fibrils formed by this polypeptide did not seed amyloid fibril formation by either native intact Ser52Pro TTR or wild-type TTR in physiological buffer (Fig. S5).
3D Crystal Structures.
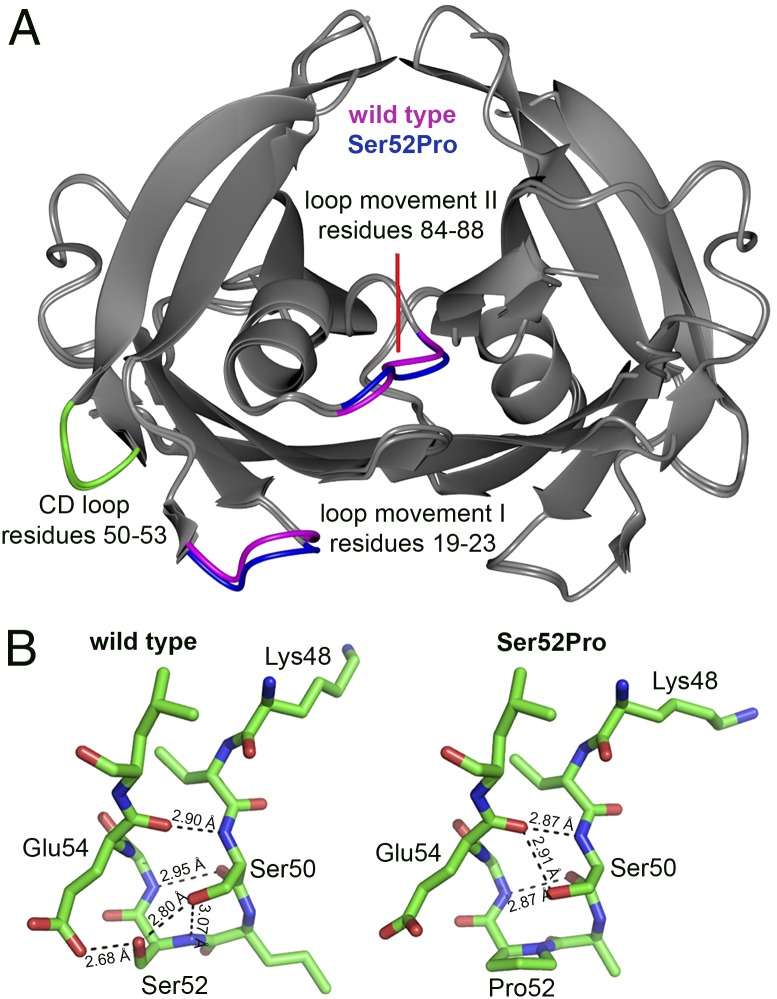
Both wild-type and Ser52Pro TTR crystallized in the commonly reported orthorhombic crystal form (space group P21212) that contains a dimer of TTR subunits in the asymmetric unit. The structures are very similar, and backbone atoms for 221 residues in which the electron density is good superposed with an rmsd of 0.42 Å. The regions showing the largest differences are displayed in Fig. 4A. The region 19-23 of molecule A may be of particular note, because a hydrogen bond between Ala19 CO and Tyr114 NH of a symmetry-related subunit is a contributor to the limited bonding that stabilizes the tetramer. The position of Ala19 CO differs by more than 1.1 Å in the two structures, suggesting some disturbance of the tetramer, but the hydrogen bond with Tyr114 is sustained. The proline substitution was modeled readily into the electron density for the β-turn at position 52 (Fig. 4B). Crucially, however, the hydrogen-bonding network involving Ser52, Glu54, and Ser50 in both TTR subunits in the wild-type structure, and including a water molecule in subunit A, is modified in the variant. In the absence of Ser52, its hydrogen-bonding partner, the Glu54 carboxylate, rotates by 90° and makes new interactions with Lys15 or His56; no interaction can exist between Pro52 and the Ser50 side chain. Only three hydrogen bonds ≤3 Å in length (Ser50 CO and Gly53 NH; Ser50 NH and Glu54 CO; and Ser50 OG and Glu54 CO) remain in the turn of the variant protein, compared with five in wild-type TTR. The loss of two hydrogen bonds would be expected to destabilize the turn and the associated C and D strands substantially. Displacement of this loop region has been proposed as the final common pathway for fibrillogenesis in a selection of amyloidogenic TTR variants by exposing edge strands of the core β-structure of the protein (17). Such movements certainly would increase the accessibility of Lys48 to proteolytic cleavage.
Fig. 4.
3D structure of wild-type and Ser52Pro TTR. (A) Crystal structures of wild-type and Ser52Pro TTR were superposed by secondary structure matching [gray-interpreted and presented by CCP4MG (27)] and show the close similarity of the folds, Rmsd = 0.42Å over 221 residues. Regions of greatest difference are highlighted in blue (Ser52Pro) and magenta (wild type). The CD loops for one subunit of both proteins are shown in green. (B) Stick diagram of the CD loop region (carbon is shown in green, oxygen in red, and nitrogen in blue) for wild-type (Left) and Ser52Pro (Right) TTR showing the diminished hydrogen-bonding network in the vicinity of the proline substitution that may contribute to the enhanced susceptibility of the variant to proteolytic cleavage at Lys48 (28). X-ray data statistics are reported in Table S3.
The mechanism of the observed tetramer destabilization cannot be determined directly from the crystal structure presented, but it is reasonable to suppose that the observed loop I and II changes in the crystal structure of Ser52Pro TTR, situated at or close to subunit interfaces, may be vestiges of a conformer population in solution where more extreme structural fluctuations exist. Transient exposure of isolated subunits, as occurs in subunit exchange (18), could provide a context in which the lack of subunit-stabilizing forces of the tetramer allows full expression of the destabilizing influence of the Pro52 substitution. For the Ser52Pro variant the cleavage of the 48-49 peptide bond should not result in the dissociation of the 1-48 peptide, because many of the bonding interactions remain in place. However, the NH of the 48-49 peptide bond hydrogen bonds to Val28 CO of strand B close to turn I. Although this hydrogen bond might be sustained by the new N terminus generated by cleavage, the separation of this N-terminal end from the new carboxylate of Lys48 must widen from 1.2 to at least 2.8 Å and promote displacement of the CD turn. Departure of the 1-48 peptide caused by shear forces in solution would remove important dimer–dimer interaction regions (19–23) and promote dissociation of the remaining 49-127 fragment.
Proteolysis of Ser52Pro TTR with Bound Ligands.
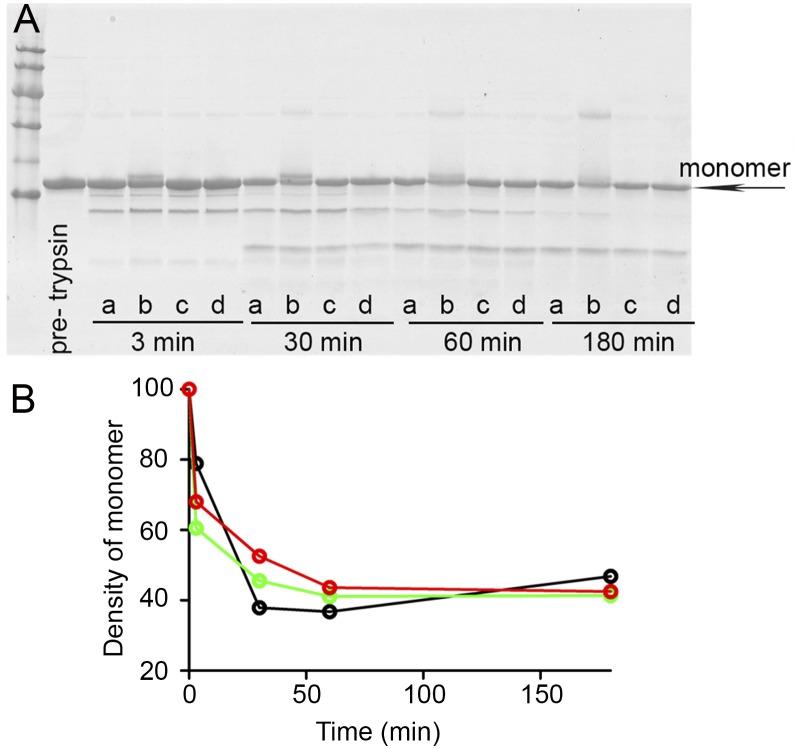
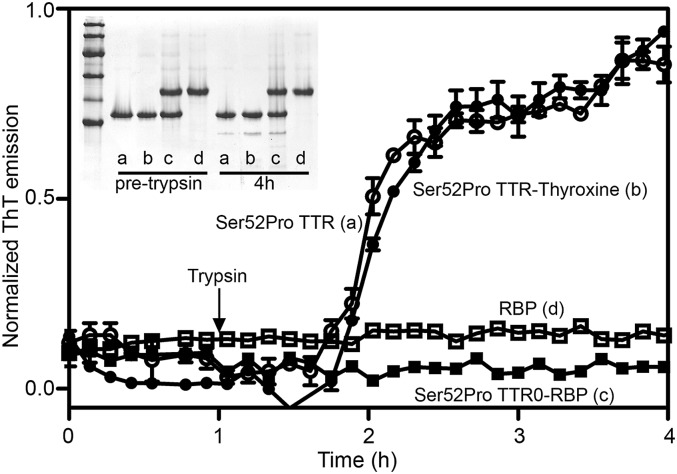
Binding of Tafamidis and mds84 (13, 14), which both enter the TTR inner channel, had no significant effect on the products (Fig. 5A) or kinetics (Fig. 5B) of trypsin digestion of Ser52Pro variant TTR. Epigallocatechin, which binds to the outer molecular surface, similarly did not reduce the susceptibility of Ser52Pro TTR to proteolytic cleavage, although the appearance of TTR oligomers prevented precise quantification of full-length TTR, as previously reported (19). Evidently these various, avidly bound ligands, do not protect the susceptible TTR sequences despite their potent stabilization of the native tetrameric assembly. Finally, we have tested the effect of the two natural ligands of TTR, thyroxine and retinol-binding protein (RBP) (Fig. S6), on the proteolysis of Ser52Pro TTR and its fibrillogenesis (Fig. 6). Neither of the two interactors protected against the selective cleavage (Fig. 6, Inset), and binding of RBP, but not thyroxine, totally inhibited fibrillogenesis. Our preliminary mass spectrometry analysis confirms the capacity of RBP to retain the cleaved monomer within a stabilized native tetramer.
Fig. 5.
Limited proteolysis of Ser52Pro TTR with bound ligands. (A) SDS 15% PAGE under reducing conditions of Ser52Pro TTR alone in presence of DMSO (a) or a fivefold molar excess of epigallocatechin (b), mds84 (c), or Tafamidis (d) after incubation at 37 °C with trypsin at an enzyme:substrate ratio of 1:200 for the times shown. Marker proteins (14.4, 20.1, 30.0, 45.0, 66.0, and 97.0 kDa) and the nontreated Ser52Pro TTR (pretrypsin) are in the first two lanes. (B) Kinetics of proteolytic cleavage of holo Ser52Pro TTR in presence of DMSO (black), mds84 (green), or Tafamidis (red) monitored by density with time of the intact monomeric TTR band (arrow in A).
Fig. 6.
Effect of natural interactors, thyroxine and RBP, on proteolysis/fibrillogenesis by Ser52Pro TTR. Normalized ThT fluorescence emission for trypsin-dependent fibrillogenesis of Ser52Pro TTR alone (a) or in the presence of a twofold excess of thyroxine (b), a twofold excess of RBP (c), or RBP alone (d). (Inset) SDS 15% PAGE under reducing conditions of fibrillogenesis samples before and after 4 h of trypsin digestion. Marker proteins (14.4, 20.1, 30.0, 45.0, 66.0, and 97.0 kDa) are included.
Discussion
The Ser52Pro TTR variant causes devastating, rapidly progressive, hereditary neuropathic systemic amyloidosis with extensive visceral and cardiac involvement which is universally fatal in early- to mid-adult life. Our present characterization of the structure and properties of the Ser52Pro variant provide a compelling biophysical explanation of this aggressive pathology.
The Ser52Pro substitution both destabilizes the native tetramer assembly and makes the protein highly susceptible to a selective, specific proteolytic cleavage at residue 48 to release the C-terminal residue 49-127 fragment, which is potently amyloid fibrillogenic. The 3D structure of the variant TTR shows no gross structural changes affecting the accessibility of Lys48 that would enhance sensitivity to proteases. The structural similarity with the normal wild-type counterpart protein (4) explains the failure of intracellular quality-control mechanisms to censor the secretion of these pathogenic proteins. However, despite the marked overall structural conservation (Fig. 4A), we observed a modest difference in the 3D structure of Ser52Pro TTR (Fig. 4B) that likely is responsible for its crucial sensitivity to proteolytic cleavage.
The variant Pro52 residue is located in the type 1 β-turn that connects β-strands C and D, and the dihedral angles of the wild-type Ser52 residue enable substitution by proline without adjustment of the main-chain path. However, the Ser52 side chain participates in a hydrogen-bonding network across the turn in the wild-type protein. Loss of this network caused by the proline substitution almost certainly destabilizes the turn and perturbs the adjacent strands, increasing their flexibility, abrogating the protection against proteolysis in the wild-type protein, and contributing to the significant observed thermodynamic destabilization.
Exposure of the native Ser52Pro variant TTR tetramer to very low-dose trypsin under physiological conditions rapidly generated the same residue 49-127 fragment that is found in ex vivo TTR amyloid fibrils, specifically those designated as type A, which are present in patients with other amyloidogenic TTR variants and patients with senile systemic amyloidosis caused by deposition of wild-type TTR (20). However, neither wild-type TTR nor the other TTR variants we tested (Val30Met, Leu55Pro, and Val122Ile) were similarly cleaved in vitro, and thus they are notably more stable to proteolysis. Nevertheless, it is plausible that in in vivo conditions, in particular shear flow and exposure to hydrophobic surfaces in the dynamic environment of the interstitial space, may promote local structural destabilization (21, 22) and enable proteolytic cleavage. Indeed focal mechanical destabilization of the polypeptide chain is a well-known mechanism for priming limited physiological proteolytic cleavage in the homeostasis of hemostatic proteins. The classic example of this mechano-enzymatic mechanism involves the multimeric von Willebrand factor in which shear forces induce a local destabilization and permit the proteolytic cleavage by a specific metalloprotease (23).
Our identification, in the amyloid fibrils in the present propositus, of full-length TTR monomers from both the wild-type and variant proteins but the highly fibrillogenic residue 49-127 fragment from only the variant, strongly suggests that the native tetrameric TTR variant, rather than already formed fibrils, is the substrate for proteolytic cleavage in vivo. Furthermore our newly established in vitro method of generating genuine TTR amyloid fibrils after enzymatic cleavage suggests that proteolysis may be a prefibrillar event, at least for this variant, because massive rapid amyloid fibril formation proceeds as soon as the N-terminal 48-residue peptide is released. In contrast, the recombinant residue 49-127 fragment produced by Mizuguchi et al. (24) gave a very low yield of amyloid fibrils in vitro. This discrepancy highlights aspects of the TTR amyloid pathway, suggesting that the type of aggregate is strictly dependent on the structural conformation of the precursor. Our finding that fibril formation is much more efficient when the truncated amyloidogenic precursor is cleaved directly from the folded TTR molecule is consistent with the early observation that the intermediate capable of forming amyloid fibrils retains much of the secondary and tertiary structure of native TTR (25).
In conclusion, the discovery of the peculiar proteolytic processing of the Ser52Pro variant illuminates a potentially general pathway of TTR amyloidogenesis dictated by the formation of the residue 49-127 polypeptide. It will be interesting in the future to investigate the possible presence of cleaved TTR or the residue 49-127 fragment in the circulation of patients and carriers of this mutation. Furthermore, because ligands that potently stabilize the native tetrameric assembly of TTR do not protect Ser52Pro TTR against the amyloidogenic cleavage, additional therapeutic approaches to inhibiting TTR amyloidogenesis may be required. It is worth noting that thyroxine, the natural ligand occupying the inner channel of the TTR tetramer, does not prevent either proteolysis or fibril formation. In contrast, RBP, although unable to protect from cleavage, is a very efficient inhibitor of fibrillogenesis. The different effects of thyroxine and RBP on proteolysis and fibrillogenesis are consistent with a sequential two-phase process. The first phase comprises cleavage of the 48-49 peptide bond but does not itself necessarily release the truncated residue 49-127 polypeptide from the intact native tetramer. In the second phase forces such as those generated by fluid agitation (22) are sufficient to release the fragments with consequent fibril formation. Thyroxine does not stabilize the tetramer sufficiently to prevent fibrillogenesis, whereas the binding of RBP is adequate to block the second phase, impeding release of the fibrillogenic free fragment.
The discovery and characterization of these crucial steps in the pathway of TTR amyloid fibrillogenesis could provide novel drug discovery targets as well as offering insight into the mode of action and potential efficacy of current approaches based on drugs occupying the thyroxine-binding site.
Methods
Limited Proteolysis.
Recombinant wild-type, Ser52Pro variant TTR, Val122Ile, Val30Met, and Leu55Pro TTR, were individually digested at 1 mg/mL in PBS (pH 7.4) at 37 °C containing trypsin at an enzyme:substrate (wt/wt) ratio of 1:200. At different times, aliquots of each incubation mixture were stopped by adding PMSF to 1.5 mM, were boiled immediately in SDS sample buffer, and were stored at −20 °C before SDS 15% PAGE was carried out under reducing conditions. For proteomic characterization of the proteolytic fragments, Coomassie Blue-stained bands were electroeluted, lyophilized, dissolved in 0.2% TFA, mixed with α-cyano-4-hydroxycinammic acid (5 mg/mL in acetonitrile, 0.2% TFA, 7:3, vol/vol), and left to air dry in a metallic sample plate before linear MS analysis (Waters Micromass spectrometer). Horse heart myoglobin was used for mass calibration.
The effect of trypsin also was tested at a 1:200 enzyme:substrate (wt/wt) ratio in the presence of a fivefold molar excess of Tafamidis (14), mds84 (13), and epigallocatechin (19), respectively.
Amyloid Fibrillogenesis.
Samples of recombinant wild-type and Ser52Pro TTR, 100 μL at 1 mg/mL in PBS (pH 7.4) containing 10 µM ThT (26), were incubated at 37 °C in Costar 96-well black-wall plates sealed with clear sealing film and were subjected to 900 rpm double-orbital shaking. Bottom fluorescence was recorded at 5-min intervals (BMG LABTECH FLUOstar Omega). After 1 h, 5 ng/μL of trypsin (Promega Trypsin Gold Mass Spectrometry Grade) or buffer alone was added, and fluorescence was monitored in three or more replicate test and control wells for the next 4 h. The data were normalized to the signal plateau at ∼4 h after the initiation of each reaction. Sample processing for microscopy analyses and fibrillogenesis assays in the presence of seeds, thyroxine, or holo-RBP are described in SI Methods.
Other Methods.
Histology, isolation, and chemical characterization of ex vivo amyloid fibrils, DNA sequencing, recombinant TTR production, thyroxine displacement assay, equilibrium denaturation, electron microscopy, atomic force microscopy and X-ray fiber diffraction, X-ray crystal structure determination, and native mass spectrometry analysis of the TTR–RBP complex are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Amanda Penco for assistance with atomic force microscopy measurements and Beth Jones for processing the manuscript. This study was supported by UK Medical Research Council Grant MR/K000187/1 (to V.B.); the University College London (UCL) Amyloidosis Research Fund and UCL Wolfson Drug Discovery Unit Funds; the Italian Ministry of University and Research Project FIRB RBFR109EOS (to S.G.); and Istituto Nazionale di Biostrutture e Biosistemi and Regione Lombardia (V.B.). Core support for the Wolfson Drug Discovery Unit is provided by the UK National Institute for Health Research Biomedical Research Centre and Unit Funding Scheme.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited the Protein Data Bank, www.pdb.org [PDB ID codes 4MRB (wild-type transthyretin) and 4MRC (Ser52Pro transthyretin)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317488111/-/DCSupplemental.
References
- 1.Pepys MB. Amyloidosis. Annu Rev Med. 2006;57(1):223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 2.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349(6):583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 3.Valleix S, et al. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. N Engl J Med. 2012;366(24):2276–2283. doi: 10.1056/NEJMoa1201356. [DOI] [PubMed] [Google Scholar]
- 4.Palaninathan SK. Nearly 200 X-ray crystal structures of transthyretin: What do they tell us about this protein and the design of drugs for TTR amyloidoses? Curr Med Chem. 2012;19(15):2324–2342. doi: 10.2174/092986712800269335. [DOI] [PubMed] [Google Scholar]
- 5.Hammarström P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299(5607):713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 6.Westermark P, Sletten K, Johnson KH. Ageing and amyloid fibrillogenesis: Lessons from apolipoprotein AI, transthyretin and islet amyloid polypeptide. Ciba Found Symp. 1996;199:205–218, discussion 218–222. doi: 10.1002/9780470514924.ch13. [DOI] [PubMed] [Google Scholar]
- 7.Thylén C, et al. Modifications of transthyretin in amyloid fibrils: Analysis of amyloid from homozygous and heterozygous individuals with the Met30 mutation. EMBO J. 1993;12(2):743–748. doi: 10.1002/j.1460-2075.1993.tb05708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergström J, et al. Amyloid deposits in transthyretin-derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J Pathol. 2005;206(2):224–232. doi: 10.1002/path.1759. [DOI] [PubMed] [Google Scholar]
- 9.Ihse E, et al. Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid. 2013;20(3):142–150. doi: 10.3109/13506129.2013.797890. [DOI] [PubMed] [Google Scholar]
- 10.Ihse E, et al. Amyloid fibril composition is related to the phenotype of hereditary transthyretin V30M amyloidosis. J Pathol. 2008;216(2):253–261. doi: 10.1002/path.2411. [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson S, et al. Amyloid fibril composition as a predictor of development of cardiomyopathy after liver transplantation for hereditary transthyretin amyloidosis. Transplantation. 2012;93(10):1017–1023. doi: 10.1097/TP.0b013e31824b3749. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins PN, Lavender JP, Pepys MB. Evaluation of systemic amyloidosis by scintigraphy with 123I-labeled serum amyloid P component. N Engl J Med. 1990;323(8):508–513. doi: 10.1056/NEJM199008233230803. [DOI] [PubMed] [Google Scholar]
- 13.Kolstoe SE, et al. Trapping of palindromic ligands within native transthyretin prevents amyloid formation. Proc Natl Acad Sci USA. 2010;107(47):20483–20488. doi: 10.1073/pnas.1008255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulawa CE, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109(24):9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, et al. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40(38):11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 16.Blake CCF, Serpell LC. Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous β-sheet helix. Structure. 1996;4(8):989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 17.Serpell LC, Blake CCF. In: Amyloid and Amyloidosis 1993. Kisilevsky R, et al., editors. Pearl River, NY: Parthenon Publishing; 1994. pp. 447–449. [Google Scholar]
- 18.Schneider F, Hammarström P, Kelly JW. Transthyretin slowly exchanges subunits under physiological conditions: A convenient chromatographic method to study subunit exchange in oligomeric proteins. Protein Sci. 2001;10(8):1606–1613. doi: 10.1110/ps.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyata M, et al. The crystal structure of the green tea polyphenol (-)-epigallocatechin gallate-transthyretin complex reveals a novel binding site distinct from the thyroxine binding site. Biochemistry. 2010;49(29):6104–6114. doi: 10.1021/bi1004409. [DOI] [PubMed] [Google Scholar]
- 20.Gustavsson A, et al. Amyloid fibril composition and transthyretin gene structure in senile systemic amyloidosis. Lab Invest. 1995;73(5):703–708. [PubMed] [Google Scholar]
- 21.Bekard IB, Asimakis P, Bertolini J, Dunstan DE. The effects of shear flow on protein structure and function. Biopolymers. 2011;95(11):733–745. doi: 10.1002/bip.21646. [DOI] [PubMed] [Google Scholar]
- 22.Mangione PP, et al. Structure, folding dynamics, and amyloidogenesis of D76N β2-microglobulin: Roles of shear flow, hydrophobic surfaces, and α-crystallin. J Biol Chem. 2013;288(43):30917–30930. doi: 10.1074/jbc.M113.498857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324(5932):1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuguchi M, et al. Unfolding and aggregation of transthyretin by the truncation of 50 N-terminal amino acids. Proteins. 2008;72(1):261–269. doi: 10.1002/prot.21919. [DOI] [PubMed] [Google Scholar]
- 25.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31(36):8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 26.Naiki H, Higuchi K, Hosokawa M, Takeda T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal Biochem. 1989;177(2):244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 27.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLano WL. 2002. in The PyMOL Molecular Graphics System, eds DeLano (Scientific LLC, San Carlos, CA) www.pymol.org. Accessed January 3, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.