Significance
Despite general acceptance of the link between chronic inflammation and cancer, the precise molecular mechanisms underlying the cancer-promoting effects of inflammation remain undefined. Inducible transcription factor NF-κB is the key regulator of inflammation, which is commonly deregulated in cancer cells to become constitutively active. Whether this deregulation contributes to malignant transformation was the main question addressed in this study. We isolated a series of genetic elements encoding artificial intracellular proteins capable of constitutive activation of NF-κB, named NF-κB–activating selectable peptides (NASPs), and demonstrated that all of them had carcinogenic activity in conventional cellular models. Specifically, NASPs made normal rodent cells susceptible to malignant transformation by oncogene Ras, which cannot do it on its own. This result defines chronically active NF-κB as an oncogene.
Keywords: TRAF6, functional genomics
Abstract
Chronic inflammation is associated with increased cancer risk. Furthermore, the transcription factor NF-κB, a central regulator of inflammatory responses, is constitutively active in most tumors. To determine whether active NF-κB inherently contributes to malignant transformation, we isolated a set of NF-κB–activating genetic elements and tested their oncogenic potential in rodent cell transformation models. Genetic elements with desired properties were isolated using biologically active selectable peptide technology, which involves functional screening of lentiviral libraries encoding 20 or 50 amino acid-long polypeptides supplemented with endoplasmic reticulum-targeting and oligomerization domains. Twelve NF-κB–activating selectable peptides (NASPs) representing specific fragments of six proteins, none of which was previously associated with NF-κB activation, were isolated from libraries of 200,000 peptides derived from 500 human extracellular proteins. Using selective knockdown of distinct components of the NF-κB pathway, we showed that the isolated NASPs act either via or upstream of TNF receptor-associated factor 6. Transduction of NASPs into mouse and rat embryo fibroblasts did not, in itself, alter their growth. However, when coexpressed with oncogenic Ras (H-RasV12), NASPs allowed rodent fibroblasts to overcome H-RasV12–mediated p53-dependent senescence and acquire a transformed tumorigenic phenotype. Consistent with their ability to cooperate with oncogenic Ras in cell transformation, NASP expression reduced the transactivation activity of p53. This system provides an in vitro model of NF-κB–driven carcinogenesis and suggests that the known carcinogenic effects of inflammation may be at least partially due to NF-κB–mediated abrogation of oncogene-induced senescence.
Chronic inflammatory conditions are strongly associated with cancer predisposition (1, 2). Inflammatory cells frequently comprise a significant part of the milieu of tumors and promote tumor progression through production of cytokines, chemokines, and growth factors (3–6). The connection between inflammation and cancer is supported by the fact that all major cancer risk factors, including chronic infections, autoimmune diseases, and exposure to alcohol, tobacco, radiation, chemical irritants, and carcinogens, are invariably associated with inflammatory responses (7). Despite strong evidence for, and general acceptance of, the link between inflammation and cancer, the precise molecular mechanism(s) underlying the cancer-promoting effects of inflammation remains undefined. An improved understanding of such mechanisms would enable exploration of numerous potential anticancer therapeutic strategies involving targeting of various aspects of immune responses. Given the paradox of immune surveillance leading to tumor eradication and inflammatory responses leading to tumor promotion, there is strong rationale for development of therapeutic agents capable of activating antitumor immune responses and/or blocking protumorigenic inflammatory pathways.
The NF-κB transcription factor plays a pivotal role in all aspects of immune responses, including inflammation (8, 9). Although NF-κB is activated in normal cells only under conditions requiring an inflammatory response, this strict regulation is lost in the majority of tumors, resulting in constitutive NF-κB activation (10, 11). This phenomenon is usually interpreted as tumor cells taking advantage of NF-κB–driven prosurvival responses, including NF-κB–mediated induction of antiapoptotic and antioxidative proteins and growth factors (8). Although correlative evidence supports prooncogenic properties of NF-κB (12), it remains unclear whether constitutive activation of NF-κB can directly contribute to cellular transformation. Based on the central role of NF-κB in regulating inflammation, definition of the molecular mechanisms connecting NF-κB to tumorigenesis will likely resolve the question of how chronic inflammation promotes cancer.
The significance of NF-κB activity in cancer is further supported by a number of previous studies indicating a functional link between NF-κB and the key mammalian tumor suppressor, p53 (13, 14) (reviewed in refs. 8 and 15). The p53 and NF-κB pathways are the two major pathways that determine cellular and organismal responses to a variety of stresses. Although NF-κB predominantly senses extrinsic stresses such as presence of bacteria and viruses, p53 acts as a guardian against intrinsic stresses such as DNA damage and deregulation of protooncogenes (8, 15–17). Because cellular responses to extrinsic and intrinsic stresses are frequently oppositely directed (e.g., temporary inhibition of apoptosis and stimulation of proliferation of immunocytes in the presence of infection versus induction of apoptosis and inhibition of proliferation in response to DNA damage), it is not surprising that the p53 and NF-κB pathways negatively regulate each other and are deregulated in opposite directions in tumors (8, 14, 15). Mutual negative regulation of p53 and NF-κB is well documented at the phenotypic level, but the mechanism(s) responsible for this phenomenon remains undefined.
The current study was aimed at uncovering the mechanisms involved in associations between inflammation, NF-κB activity, p53 activity, and tumorigenesis. This task required development of an experimental system mimicking the constitutive NF-κB activation that naturally occurs in tumors. Numerous NF-κB–activating agents already in existence could not be used for such studies because physiological activation of NF-κB is typically short-lasting due to induction of potent negative-feedback loops (18, 19). Therefore, we used a functional genomics approach, biologically active selectable peptide (BASP) technology (20), to identify genetic elements encoding peptides capable of causing constitutive activation of NF-κB [NF-κB–activating selectable peptides (NASPs)]. The role of constitutive NF-κB activation in cell transformation was then tested by expressing NASPs alone or together with the activated Ras oncogene in rodent embryo fibroblasts, a conventional in vitro model for testing oncogene cooperation (21). The results of this work demonstrate that constitutive activation of NF-κB makes rodent fibroblasts susceptible to transformation by activated Ras, apparently by attenuating p53-induced senescence, thus defining chronically active NF-κB as an oncogene.
Results
BASP Technology: Establishment of a System for Functional Selection of NF-κB–Activating Genetic Elements.
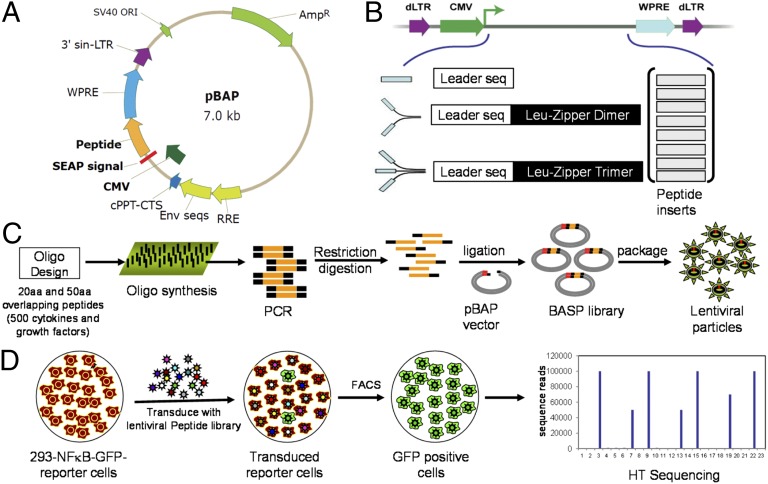
To experimentally induce constitutive NF-κB activation, the common property of tumor cells that links chronic inflammation and tumorigenesis, we used BASP technology (20) to perform a functional screen for genetic elements encoding peptides capable of inducing such NF-κB deregulation. First, we created libraries of genetic elements encoding peptides of 20 or 50 amino acids (aa) built in a lentiviral expression vector (Fig. 1A). Because extrinsic signals, such as cytokines, are common natural activators of NF-κB, we hypothesized that a pool of peptides derived from the extracellular proteome would provide a good source of possible NF-κB activators. Therefore, we created four libraries, each containing 50,000 individual clones, spanning the sequences of 500 well-characterized human and mouse cytokines, chemokines, growth factors, and their receptors. For each of these proteins of interest, we designed oligonucleotides encoding a set of redundant overlapping peptides with lengths of 50 aa (subdomain-like) or 20 aa (epitope-like). Given the secreted nature of most naturally occurring NF-κB activators, a secreted alkaline phosphatase (SEAP) signal sequence was included in the vector upstream of the inserts to facilitate endoplasmic reticulum (ER) targeting and secretion of the library peptides. In addition, oligomerization is necessary for most natural NF-κB activators to trigger receptor activation, and many successful cytokine mimetic peptides bind their cognate receptors as dimers or trimers. Therefore, GCN4 leucine zipper-encoding sequences directing peptide dimerization or trimerization were included (22) in our library vectors between the SEAP signal sequence and the peptide-encoding inserts. Thus, each of the four libraries used for selection of NASPs contained either 20-aa or 50-aa peptide-encoding inserts in one of two variants of the lentiviral expression vector (with dimerization or trimerization leucine zipper sequences) (Fig. 1B). The oligonucleotides were designed through computational analysis of sequences, synthesized on a microarray surface, and then detached and PCR-amplified for cloning into the lentiviral expression vector (Fig. 1C).
Fig. 1.
Schematic illustration of screening strategy for isolation of NASPs. (A) Map of the pBAP lentiviral vector used for library construction. Sequences encoding peptides of 20 aa or 50 aa were cloned downstream of the SEAP-derived leader sequence. (B) Three different versions of the pBAP vector allowed production of peptides as monomers, dimers, or trimers (with the latter two cases including a GCN4 leucine zipper dimerization or trimerization sequence upstream of the peptide-encoding inserts). (C) Procedure followed for library construction. Oligonucleotides encoding overlapping 20-aa- or 50-aa-long peptides derived from 500 different cytokines and growth factors were designed by computational analysis of sequences and synthesized on a microarray surface. Oligonucleotides were then detached and amplified by PCR for cloning into the pBAP-based vectors. Libraries were packaged into pseudoviral particles in HEK293T cells. (D) Procedure for library screening. Lentiviral peptide libraries were transduced into 293-NF-κB-GFP reporter cells, and GFP-positive cells were isolated by FACS (two rounds). Peptide-encoding sequences were PCR amplified from GFP-positive cells from the two pools of genomic DNA using flanking Gex primers and subjected to high throughput (HT) sequencing to identify the most enriched NASPs.
As a readout system for the screen, we generated an NF-κB–responsive reporter cell line, 293-NF-κB-GFP, in which NF-κB–driven expression of the green fluorescent protein (GFP) reporter gene (Fig. S1) can be easily and quantitatively assessed by flow cytometry (Materials and Methods). Upon exposure of the reporter cells to TNF or IL-1 (known NF-κB activators), GFP expression was uniformly induced up to 300-fold over background levels (Fig. S2 A and B), thus indicating a robust readout system capable of detecting even relatively weak NF-κB activators.
The screening procedure was tested using a positive control NASP construct expressing the known NF-κB activator, full-length cytokine TNF, mixed with an insert-free lentiviral vector at a ratio of 1:10,000 to mimic the likely representation of a given peptide within the libraries. After two rounds of FACS sorting of GFP-positive transduced reporter cells, we obtained a population in which 97.5% of the cells expressed both GFP and TNF as assessed by flow cytometry and PCR, respectively (Fig. S3 C and D). Use of TNF-producing clones also allowed us to address the potential concern that secretion of NASPs due to the engineered ER-targeting signal might lead to activation of NF-κB in a paracrine fashion, making it impossible to distinguish between NASP-producing and NASP-responding cells. However, cocultivation of TNF-expressing and TNF-nonexpressing NF-κB reporter cell clones demonstrated that only those clones expressing TNF were GFP-positive (Fig. S3E).
Identification and Functional Validation of NASPs.
A full-scale screening of all four generated BASP libraries was carried out according to the scheme outlined in Fig. 1. We packaged and transduced each library into 106 293-NF-κB-GFP cells at a multiplicity of infection (MOI) of 0.3–0.5, which was expected to yield about 100 transduced cells for each peptide construct. Forty-eight hours after transduction, the cells were sorted by FACS to isolate GFP-positive cells. The isolated population was grown in regular culture medium for 7d and then sorted again to further enrich for GFP-positive cells. For each of the four libraries, we observed that, after the second sorting, up to 50% of the cells expressed GFP (Fig. 2A).
Fig. 2.
Activation of NF-κB–dependent GFP expression upon peptide library or individual NASP transduction. (A) GFP expression in 293-NF-κB-GFP reporter cells transduced with one of three different peptide libraries (monomeric, dimeric, or trimeric 50-aa-long peptides), before and after two rounds of FACS sorting. Photographs of representative fields taken under a fluorescent microscope are shown. (B) GFP expression in 293-NF-κB-GFP reporter cells transduced with NASP validation constructs (rescued NASP inserts cloned into the original lentiviral expression vector used for library construction) or control vector (con. vec). The percentage of GFP-positive cells in each population was determined by flow cytometry 72 h after transduction. Nomenclature of NASP is similar to that indicated in Table 1. (C) Scheme highlighting the overlapping sequences of eight isolated NASPs to the corresponding parent proteins. (D) Photographs taken under a fluorescent microscope of GFP induction by selected NASPs are shown. (E) GFP expression in 293-NF-κB-GFP reporter cells transduced with lentiviral constructs for expression of isolated NASPs (ApoF, PLP, or CCK) as monomers (M), dimers (D), or trimers (T) with or without the SEAP signal sequence (ΔSEAP). The percentage of GFP-positive cells in each population was determined by flow cytometry 48 h after transduction. “Control” cells were nontransduced reporter cells.
To identify the NASPs responsible for activating NF-κB–dependent GFP expression in the reporter cells, genomic DNA was isolated from BASP library-transduced cell populations on the day after library transduction and after the second round of FACS-based enrichment for GFP-positive cells. Peptide-encoding sequences were amplified from the two pools of genomic DNA using flanking Gex primers, and high-throughput (HT) Solexa sequencing was used to determine the relative abundance of individual sequences among amplified PCR products. Statistical analysis revealed sequences that were enriched at least threefold in FACS-sorted GFP-positive cell populations relative to the corresponding cell population just after transduction. The hit rate was similar for all four libraries regardless of whether they expressed 20-aa- versus 50-aa-long peptides or dimeric versus trimeric BASPs. Many peptide sequences that were identified as hits, although not identical, were derived from similar regions of the same protein, thus allowing us to map a minimal functional NF-κB–activating domain of the protein (Fig. 2C and Fig. S4). To confirm the HT sequencing results, we tested the NF-κB–activating ability of 12 putative NASPs by cloning them into the original lentiviral expression vector, transducing the constructs into 293-NFκB-GFP cells, and determining the percentage of GFP-positive cells 48 h after transduction. All twelve individually tested NASPs showed strong GFP reporter induction in a large proportion of cells, thus confirming their NF-κB–inducing activity, and were used for further functional testing (Fig. 2 B and D). The selected NASPs represented (Table 1) four overlapping peptides derived from apoliprotein F (ApoF), two overlapping peptides derived from cartilage oligomeric matrix protein (COMP) and pancreatic lipase-related protein (PLP), and unique fragments of cholecystokinin (CCK), Matrilin-2 (MAT2), subtilisin/kexin type 9 (Subt), and interleukin 27 (IL27).
Table 1.
Origin and structure of NF-κB–activating peptides
| Peptide length, aa | Peptide design | Protein of origin | Abbreviation | Peptide sequence (# of independent clones) | Position |
| 20 | Trimer | Apolipoprotein F | ApoF-T20 | RVGRSLPTEDCENEKEQAVH (1) | 161–180 |
| 20 | Trimer | Cholecystokinin | CCK-T20 | DYMGWMDFGRRSAEEYEYPS (1) | 96–115 |
| 20 | Trimer | Cartilage oligomeric matrix protein | COMP-T20 | HQDSRDNCPTVPNSAQQDSD (1) | 441–460 |
| 50 | Trimer | Apolipoprotein F | ApoF-T50 | LLAREQQSTGRVGRSLPTEDCENEKEQAVHNVVQLLPGVGTFYNLGTALY (1) | 151–200 |
| 50 | Trimer | Pancreatic lipase related protein-1 | PLP-T50 | SLGAHVAGEAGSKTPGLSRITGLDPVEASFESTPEEVRLDPSDADFVDVI (1) | 171–220 |
| 50 | Trimer | Cartilage oligomeric matrix protein | COMP-T50 | DSDQDKDGDGHQDSRDNCPTVPNSAQQDSDSDGQGDACDEDDDNDGVPDS (1) | 431–480 |
| 50 | Trimer | Matrilin 2 | MAT2-T50 | LAEDGKRCVAVDYCASENHGCEHECVNADGSYLCQCHEGFALNPDKKTCT (2) | 315–364 |
| 50 | Trimer | Subtilisin/kexin type 9 | Subt-T50 | LLLGPAGARAQEDEDGDYEEL (stop) (1) | 22–42 |
| 50 | Dimer | Pancreatic lipase related protein-1 | PLP-D50 | GSKTPGLSRITGLDPVEASFESTPEEVRLDPSDADFVDVIHTDAAPLIP (1) | 181–229 |
| 50 | Dimer | IL-27 | IL27-D50 | NLPEEEEEEEEEEEEERKGLLPGALGSALQGPAQVS (frameshift) (2) | 151–198 |
| 50 | Dimer | Apolipoprotein F | ApoF-D50 | QVLIQHLRGLQKGRSTERNVSVEALASALQLLAREQQSTGRVGRSLPTED (1) | 121–170 |
| 50 | Dimer | Apolipoprotein F | ApoF-D50-del | QVLIQHLRGLQKGRSTERNVS——————–RVGRSLPTED (deletion) (1) | 121–170 |
Overlapping sequences of Apo-F peptides are indicated in bold, and those of COMP-peptides are bold and underlined. Overlapping peptides of PLP are in italics and bold. Dotted line indicates deletion corresponding to the parent protein.
Characterization of the Mechanism of Action of NASPs.
For further functional validation, the selected lentiviral constructs of NASPs were expressed individually in a panel of reporter cell lines, including the original 293-NF-κB-GFP cell line used for screening, as well as HeLa-NF-κB-GFP cells (generated with the same reporter construct as 293-NF-κB-GFP) and MEF-NF-κB-LUC cells (culture of mouse embryo fibroblasts isolated from NF-κB reporter mice expressing the firefly luciferase gene under the control of the IκB promoter) (23, 24). All isolated NASPs induced reporter expression in all three reporter-cell types, albeit with variable intensity. The relative NF-κB–activating efficacy of different NASPs correlated among the three reporter cell systems (Fig. 2B and Fig. S5). It is noteworthy that the same NASPs expressed without a leucine zipper domain or without a SEAP-derived signaling peptide completely lacked the ability to activate NF-κB reporters, thus indicating the importance of dimerization or trimerization as well as ER targeting for their functionality (Fig. 2E). Thus, NASPs act similarly to natural NF-κB activators, the majority of which require oligomerization for their activity (25).
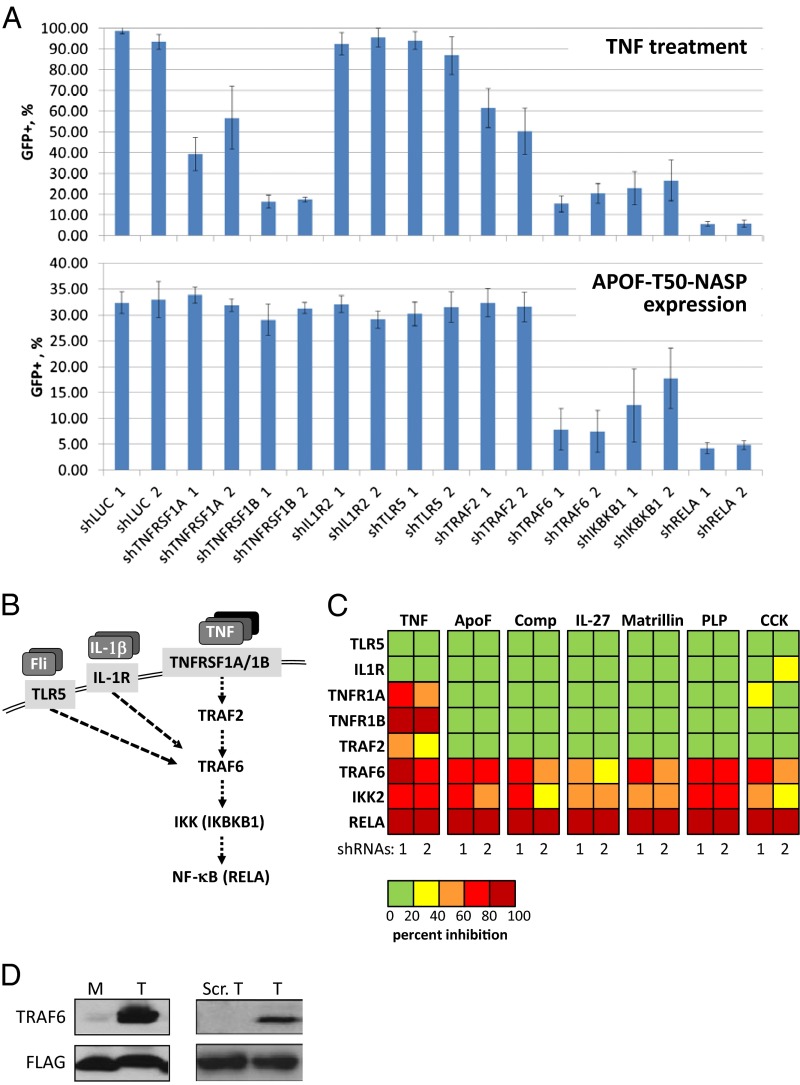
To understand the mechanisms of NF-κB activation by NASPs, we used a set of constructs expressing inhibitory shRNAs against different components of signaling pathways that involve NF-κB activation (two different shRNAs targeting each gene). Specifically, we used shRNAs against two essential subunits of the TNF receptor (TNFRSF1A and TNFRSF1B), the receptor for interleukin-1 (IL1R2), Toll-like receptor 5 (TLR5), two members of the TNF receptor-associated factor protein family (TRAF2 and TRAF6), a protein subunit of IκB kinase essential for the canonical NF-κB activation pathway (IKBKB), and the p65 subunit of NF-κB (RELA) (Fig. 3A). The functionality of shRNAs was tested after transduction into 293-NFκB-GFP cells and selection for puromycin resistance (to obtain a cell population in which all cells express the shRNA), followed by treatment with the NF-κB–activating agent TNF. As expected, shRNAs against known members of the TNF signaling pathway showed strong inhibition of NF-κB reporter induction in this system whereas shRNAs targeting other pathways (i.e., IL1 or TLR5), as well as shRNA against the luciferase gene (negative control), had no effect (Fig. 3 B and C). Each shRNA construct was then transduced into 293-NFκB-GFP cells expressing individual NASPs, and the percentage of GFP-positive cells in each population was determined by flow cytometry 4 d after transduction. Interestingly, the patterns of shRNA activity (i.e., their relative effects on NF-κB–driven GFP expression) were similar for all populations of NASP-expressing cells (i.e., for all of the different tested NASPs) (Fig. 3C). Only three pairs of shRNAs from the panel tested had an inhibitory effect on NASP-induced NF-κB activity: shRNAs against RELA, IKIKB, and TRAF6. Thus, NASPs act via or upstream of TRAF6, but independently or downstream of TRAF2 and the TNF, IL1, and TLR5 receptors (Fig. 3 A–C). These observations identify TRAF6 as a common mechanistic component and suggest that all NASPs act upon a relatively narrow segment of the NF-κB signaling pathway involving TRAF6. To explore this hypothesis, we performed immunoprecipitation experiments with 293-NF-κB-GFP cells expressing FLAG-tagged trimeric ApoF-20-NASP and anti-FLAG antibodies. The same NASP peptide, expressed without a leucine zipper, monomeric ApoF-20-NASP, which is incapable of NF-κB activation, was used as a control. Another control, consisting of a FLAG-tagged scrambled sequence of the ApoF NASP peptide cloned into the trimeric expression vector, was also incapable of activating NF-κB (Fig. S6). Immunoblotting revealed that TRAF6 protein was immunoprecipitated from trimeric-ApoF-20-NASP–expressing cells, but not from monomeric-ApoF-20-NASP–expressing cells (Fig. 3D). Physical interaction with TRAF6 was not detected for any other NASPs. Therefore, despite their shared dependence on TRAF6 for NF-κB activation, other aspects of their mechanisms of action are clearly different from that of trimeric-ApoF-20-NASP.
Fig. 3.
Individual NASPs were tested with the panel of selected shRNAs against critical regulators of the NF-κB pathway. (A) Individual shRNA constructs against TNFRSF1A, TNFRSF1B, IL1R2, TLR5, TRAF2, TRAF6, IKBKB1, and RELA were prepared (two shRNAs per each gene), packaged into lentiviral particles, transduced into 293-NFkB-GFP cells, and selected for 3 d in puromycin. shRNA agaist luciferase (shLuc) was used as the control. This shRNA set was tested for suppression of TNF-induced NF-κB activation (Upper). TNF (1 μg/mL) was added to the media at 4 d after infection with shRNA constructs. The percentage of GFP-positive cells in each population was determined by flow cytometry 4 d after transduction. The same set of individual shRNAs was transduced into the cells expressing APOF-T50-NASP. The percentage of GFP-positive cells in each population was determined by flow cytometry 4 d after transduction with shRNAs (Lower). (B) Scheme of signaling pathways leading to activation of NFκB by flagellin (Fli), IL-1β, and TNF, highlighting the central role of TRAF6. (C) Comparative heatmap of the effect of indicated shRNAs on TNFα-induced or NASP-induced NF-κB activation. (D) Coimmunoprecipitation of TRAF6 with trimeric ApoF-T20-NASP, capable of NF-κB activation, but not with inactive monomeric (M) or scrambled ApoF-T20-NASP (Scr.T). Flag-tagged peptides were immunoprecipitated fom cell lysates using M2-FLAG agarose beads and immuoblotted to detect TRAF6.
NASPs Overcome Ras-Induced Senescence to Promote Cell Transformation.
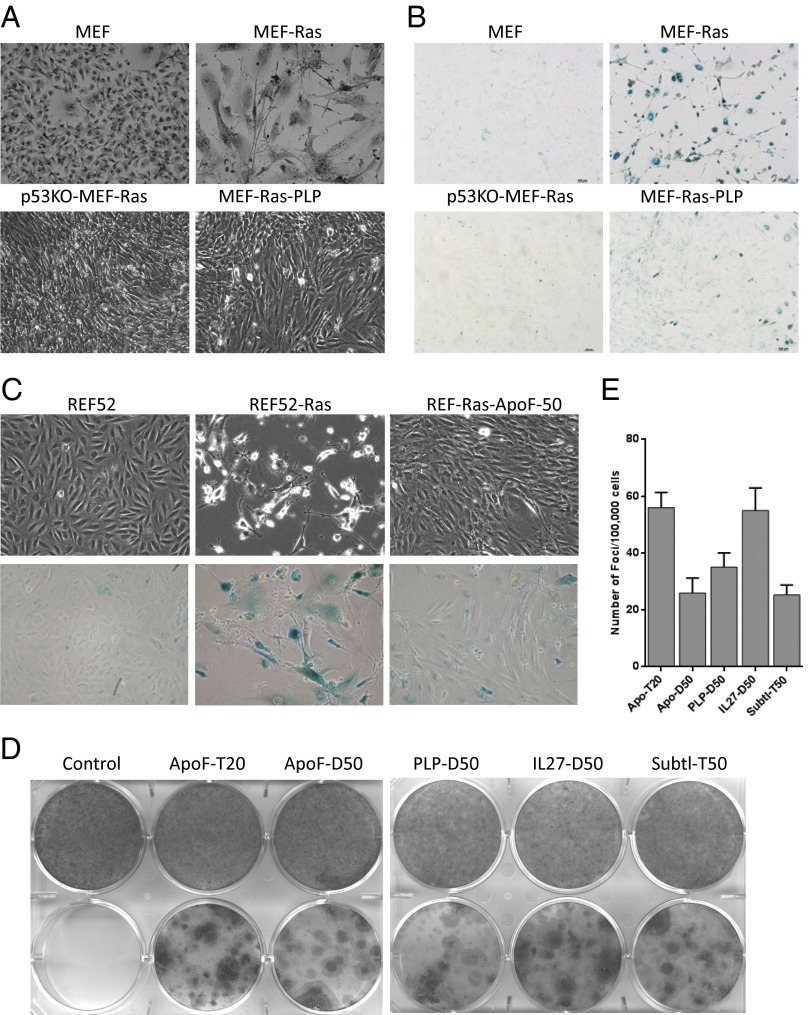
To test our hypothesis that constitutive NF-κB activation by isolated NASPs would mimic chronic inflammatory stimuli and promote malignant transformation, we expressed NASPs in rodent fibroblasts and tested their ability to cooperate with oncogenic Ras (H-RasV12) in inducing transformation (21, 26, 27). The oncogenic effect of H-RasV12 can be observed in mouse embryonic fibroblasts (MEFs) isolated from p53-knockout (p53−/−) mice, which undergo full transformation upon its expression (26). In contrast, expression of H-RasV12 in MEFs from wild-type mice leads to apparent senescence of the cells (based on morphology and senescence-associated β-galactosidase staining), with a “second hit” required to cause transformation (28). In this study, early-passage (passage 3) primary MEFs from wild-type C57BL/6 mice were infected with lentiviral constructs expressing individual NASPs or with the empty lentiviral expression vector as a control. Forty-eight hours later, each transduced cell population was trypsinized and divided into three plates. One plate was then transduced with a lentiviral construct directing expression of H-RasV12 in combination with the bleomycin resistance gene under the control of the H4 promoter (PLV-Ras-bleo). The second plate in each set was transduced with the PLV-bleo vector (containing the bleomycin resistance gene, but not H-RasV12) as a control, and the third plate was left uninfected to monitor the effect of NASP expression alone. All cultures transduced with PLV-Ras-bleo or PLV-bleo were subjected to bleomycin selection to eliminate untransduced cells. Cultures were allowed to grow for 3 wk and then assessed under a microscope for cell morphology and colony formation, followed by staining for senescence-associated β-galactosidase (29). As a control to assess the quality of H-RasV12 lentiviral stocks, MEFs isolated from p53-knockout mice, which are permissive for Ras-induced transformation, were also transduced in the second round (with PLV-Ras-bleo or PLV-bleo), selected in bleomycin, and evaluated as described for wild-type MEFs.
As expected, PLV-Ras-bleo–transduced p53 −/− MEFs formed fully transformed cultures (Fig. 4A). In contrast, wild-type MEFs expressing oncogenic H-RasV12 from the PLV-Ras-bleo construct exhibited morphological features of senescent cells and showed senescence-associated beta-galactosidase staining (Fig. 4 A and B). Coexpression of H-RasV12 with NASPs, however, resulted in transformation rather than senescence of wild-type MEFs. As shown for one NASP (PLP) in Fig. 4, wild-type MEFs expressing both H-RasV12 and the NASP formed colonies of proliferating cells with transformed phenotypes and showed a substantial reduction in senescence-associated β-galactosidase staining.
Fig. 4.
NASP expression allows MEF and REF52 cells to overcome Ras-induced senescence and undergo transformation. (A) Photographs taken under light microscope, and (B) β-galactosidase staining of primary MEFs that were transduced with the empty vector PLV-bleo (MEF), transduced with the lentiviral H-RasV12 expression construct PLV-rasV12-bleo (MEF-Ras), MEFs (p53−/−) transduced with PLV-rasV12-bleo (p53KO-MEF-Ras) or transduced with PLV-rasV12-bleo and a lentiviral NASP (PLP) expression construct (MEF-Ras-PLP). Transduced cells were selected in bleomycin, allowed to grow for 21 d, and then photographed/stained. Note transformation phenotype in cultures of p53−/− MEFs expressing Ras or in those coexpressing Ras with PLP, but not in those expressing Ras alone. (C) Phase contrast images (Upper) and β-galactosidase staining (Lower) of REF52 cells that were (in order from left to right) (i) transduced with the empty vector PLV-bleo, (ii) transduced with the lentiviral H-RasV12 expression construct PLV-rasV12-bleo, or (iii) transduced with PLV-rasV12-bleo and a lentiviral NASP (IL-27-T50) expression construct. Transduced cells were selected in bleomycin, allowed to grow for 18 d, and then photographed/stained. (D) Representative images of methylene blue staining of REF52 cells transduced with NAGE vector construct (control) or different NAGE expression constructs either with PLV-Bleo (Upper) or with PLV-rasV12-bleo (Lower). Tested NAGEs included those derived from apolipoprotein F (ApoF), pancreatic lipase-related protein-1 (PLP), interleukin-27 (IL27), and subtilisin/kexin type 9 (Subtl). Here and in E, the library from which the NAGE was isolated is indicated by D or T (for dimer or trimer) and/or the number of amino acid residues in the peptide inserts (20 or 50). (E) The frequency of foci formation was calculated from triplicate plates (one of which is shown in D).
Because primary MEFs in culture are often prone to spontaneous transformation, we used a second system to test the effect of NASPs on transformation, the rat embryonic fibroblast cell line REF52. This cell line has an undetectable background level of spontaneous transformation and has been shown to undergo senescence-like growth arrest in response to H-RasV12 expression similar to MEFs (26, 30). In addition, like MEFs, REF52 cells lacking p53 activity undergo transformation upon H-RasV12 expression, which we confirmed using shRNA-mediated knockdown of p53 expression (26). Using the protocol outlined above for MEFs, we transduced REF52 cells with combinations of H-RasV12 and NASP lentiviral expression constructs and the corresponding empty vectors and subsequently evaluated cell morphology, formation of transformation foci, and senescence-associated β-galactosidase staining. The effect of H-RasV12 expression alone or in combination with one ApoF-derived NASP is shown in Fig. 4C. NASP coexpression eliminated the senescent phenotype induced by H-RasV12 expression and resulted in transformed cultures. Expression of NASPs alone had no noticeable effect on the phenotype of either REF52 or other cell types. Cooperation of NASPs with oncogenic Ras in inducing REF52 cell transformation was also observed with an independent batch of H-RasV12 lentivirus and all five NASPs (Fig. 4 D and E) tested. The frequency of transformation foci in H-RasV12 + NASP cotransduced cell populations ranged from 2 x10−4 to 6 × 10−4, depending upon the particular NASP (Fig. 4E).
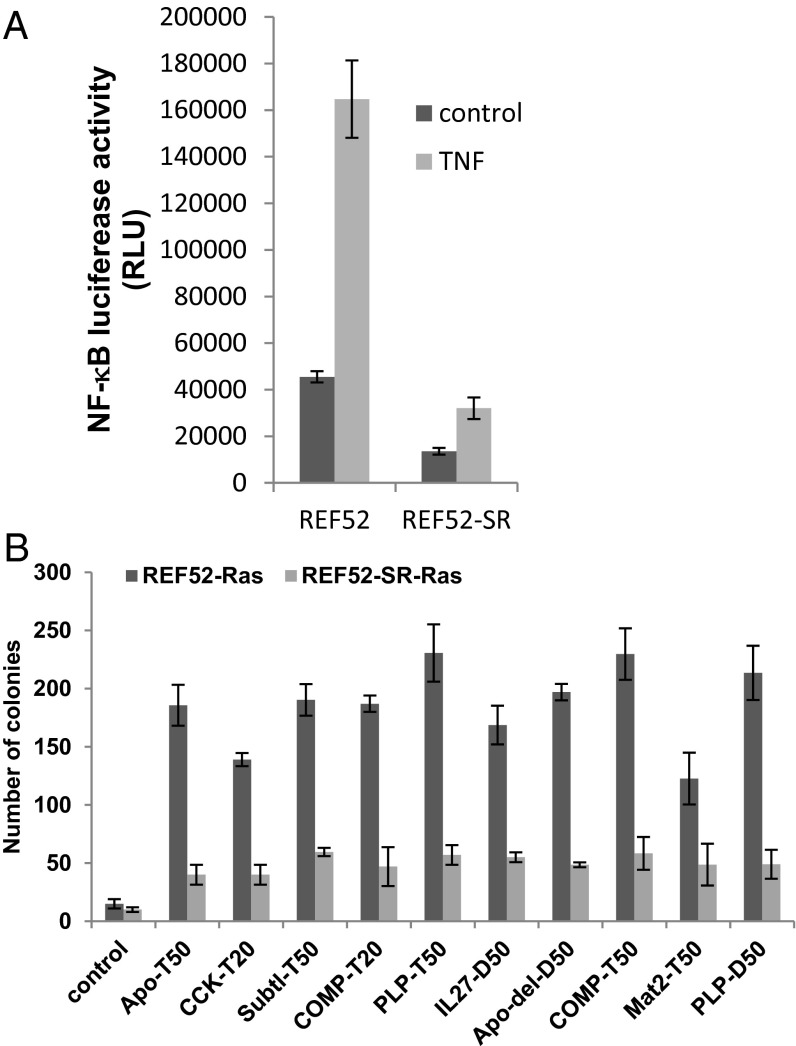
To confirm the requirement of NF-κB for NASP-mediated transformation, we generated REF52 cells stably expressing a superrepressor (SR), a stable IκB mutant lacking both phosphorylation sites, which blocks NF-κB activation (14, 31). We confirmed suppression of basal and TNFα−induced NF-κΒ activation in these cells using reporter assays (Fig. 5A). Parental REF52 cells and REF52-IκBα-SR cells were transduced with individual NASPs and, 2 d later, transduced again either with H-RasV12 or control PLV-bleo lentivirus and selected in bleomycin after 48 h of infection. At 10 d postinfection, REF52-NASP cells exhibited transformed morphology whereas REF52-IκBα-SR cells expressing the same NASP had the morphology of uninfected cells, correlating with their relative ability to form colonies in semisolid agar (Fig. 5B). These results confirm that the ability of NASPs to cooperate with Ras to promote cell transformation requires NF-κB activation.
Fig. 5.
NF-κB is required for NASP cooperation with Ras to induce transformation. (A) Suppression of NF-κB by IκB-super repressor (SR). NF-κB luciferase activity was measured in parental REF52-NF-κB luciferease reporter cells or its derivative stably expressing IkB-super repressor (SR) with or without the addition of TNF after 6 h. (B) REF52 cells or REF52-IkB-SR cells expressing different NASPs were transduced with the empty vector PLV-bleo or with PLV-rasV12-bleo, selected in bleomycin for 5 d postinfection and allowed to grow in soft agar for an additional 2 wk. Data represent mean number of colonies appeared per 50,000 cells plated in triplicate.
To demonstrate the tumorigenic potential of cells transformed by the combined expression of NASPs and oncogenic Ras in vivo, we injected 5 × 106 cells of each transformed REF52 cell population s.c. into both flanks of athymic nude mice (n = 8 mice per group). By 10 d postinjection, the majority of animals that were injected with cells cotransduced with H-RasV12 and NASP expression constructs had developed large tumors requiring euthanasia due to tumor burden according to Institutional Animal Care and Use Committee regulations. The frequency of tumor formation by day 10 postinjection was 100%, 87.5%, and 62.5% in groups of animals coexpressing H-RasV12 with ApoF, PLP, and IL-27–derived NASPs, respectively. In contrast, 0% (0/8) of the control mice that were injected with REF52 cells transduced with NASP expression constructs alone or with the empty lentiviral expression vector developed any visible tumors during the same period. These results confirm that cells showing transformed, nonsenescent phenotypes following coexpression of H-RasV12 and NASPs are truly transformed, with potent tumor-forming capacity.
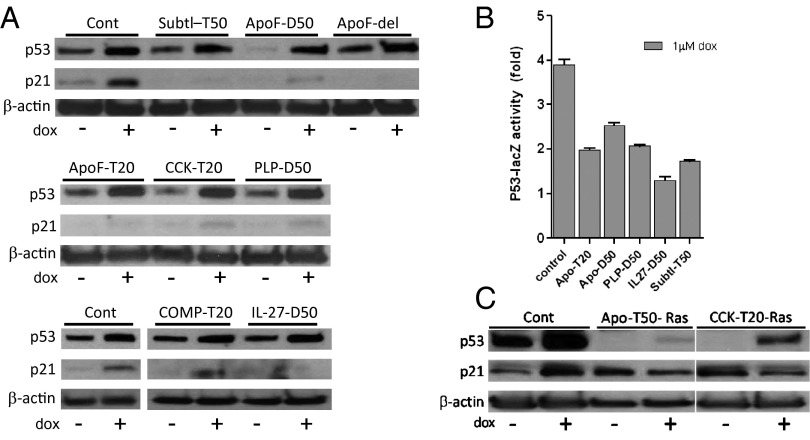
Given the fact that p53 activity is the major mechanism that prevents oncogenic Ras-induced transformation of rodent fibroblasts (inducing senescence instead), we anticipated that suppression of p53 activity caused by NASP expression might underlie the observed ability of NASPs to promote transformation in cooperation with Ras. To test this hypothesis, we assessed the effect of NASP expression on p53 activity induced by doxorubicin, a DNA-damaging chemotherapeutic agent that is a known potent activator of p53. REF52 cells stably expressing individual NASPs were treated with doxorubicin for 18 h, and induction of p21 was assessed. For all eight tested NASPs, doxorubicin-induced expression of p21, a well-known p53-dependent phenomenon, was reduced by NASP expression (Fig. 6A). For additional confirmation of p53 inhibition, we transduced Con A cells, spontaneously immortalized mouse fibroblasts carrying the p53-dependent LacZ reporter (32), with five of the NASP-expressing lentiviral constructs and compared reporter activation in them following doxorubicin treatment with that of control vector-transduced cells. All five NASPs showed suppression of p53 reporter activity (Fig. 6B). It is noteworthy that failure of p53 to induce p21 following DNA damage, even though p53 itself remained responsive to genotoxic stress, was also seen in NASP-expressing cells following transformation with Ras (Fig. 6C). These results suggest that NASP-mediated activation of NF-κB ultimately leads to inhibition of p53, which allows cell transformation in the presence of oncogenic Ras. This conclusion is consistent with accumulated data from our laboratory and others demonstrating reciprocal regulation of the NF-κB and p53 pathways (8, 13) (Discussion).
Fig. 6.
NF-κB–activating peptides inhibit p53 activity. (A) REF52 cells expressing indicated NASPs or control vector (Cont) were treated with 1 μM doxorubicin for 18 h, and expression of p53 and p21 was analyzed by Western blot. β-actin is used as a loading control. (B) Con A-reporter cells carrying a p53-dependent lacZ reporter gene (encoding β-galactosidase) transduced with lentiviral expression constructs for the indicated NASPs or with empty library vector (control). After 48 h, cells were treated with 1 μM doxorubicin for 18 h, and β-galactosidase reporter activity was quantified and expressed as fold increase over reporter activity in the absence of doxorubincin. Tested NASPs included those derived from apolipoprotein F (ApoF), interleukin-27 (IL27), pancreatic lipase related protein-1 (PLP), and subtilisin/kexin type 9 (Subtl). Numbers after the peptide name indicate the peptide length (20 or 50 aa). (C) REF52 cells expressing indicated NASPs or “empty” vector (Cont) were transduced with PLV-rasV12-bleo. Following establishment of transformed (NASP-expressing cells) or senescent (control cells) phenotypes, cultures were treated with 1 μM doxorubicin for 18 h, and expression of p53 and p21 was analyzed by Western blot. β-actin is used as a loading control.
Discussion
The strong link between chronic inflammation and cancer is illustrated by numerous observations. Clinical and epidemiologic studies have revealed associations between infectious agents and chronic inflammatory disorders and cancer (33, 34). Specific examples of inflammation- and infection-associated cancers include gastric cancer associated with Helicobacter pylori infection, hepatocellular cancer associated with chronic hepatitis virus infection, bladder cancer associated with Schistosoma hematobium infection, and lung cancer associated with Mycobacterium tuberculosis infection (8). Epidemiological data indicate that over 20% of the mortality in cancer patients is linked to underlying chronic infections and inflammatory responses. Indeed, given the fact that inflammatory components constitute approximately half of the tumor microenvironment (2), it is likely that inflammatory cells and cytokines present in the tumor milieu might contribute to tumor initiation and progression.
Additional support for a mechanistic link between cancer and inflammation is provided by the fact that the NF-κB pathway is constitutively activated in many types of cancer and is also a central regulator of immune responses that is activated by proinflammatory cytokines and microbial components during infections (10, 11). Constitutively activated NF-κB might promote tumorigenesis through its known ability to induce expression of antiapoptotic factors (35) and positive regulators of proliferation such as cytokines and growth factors (36). In addition, NF-κB may promote tumor angiogenesis and invasiveness (37). However, whether constitutive NF-κB activation is a primary contributor to tumor development or a byproduct of the inflammatory milieu surrounding tumors has not been definitively answered. Therefore, this study aimed to determine whether constitutive activation of NF-κB directly promotes malignant transformation. To reach this goal, we designed a functional screen to identify genetic elements capable of inducing stable constitutive NF-κB activity in cells (NF-κB–activating genetic elements, NASPs). We hypothesized that use of a cell-based phenotypic readout for the screen would allow us to isolate NF-κB activators that bypass the negative feedback mechanisms that lead to rapid suppression of NF-κB activity induced by physiological activators and, thus, would be capable of conferring constitutive NF-κB activity such as that seen under conditions of chronic inflammation or in tumors. Indeed, we succeeded in identifying 12 independent NASPs that, upon expression in reporter cells, caused constitutive expression of an NF-κB–dependent reporter gene.
Sequence analysis of the obtained NASPs indicated that they all encoded peptides derived from proteins not previously implicated in NF-κB activation and that, therefore, they were likely to work through different mechanisms than known NF-κB activators. This fact is consistent with our finding that, although NASPs were selected from libraries comprised of peptides representing secreted proteins, they appeared to act via cell-autonomous intracellular mechanisms (as opposed to many natural NF-κB activators that are secreted and act through interaction with receptors on the surfaces of other target cells). A seeming paradox was presented by our findings that functionality of NASP-encoded peptides required ER-targeting sequences characteristic of secreted proteins, yet was cell-autonomous (no paracrine activity of NASPs on neighboring cells was detectable in coculture experiments). A possible explanation for these observations is that NASPs interact with receptors or other proteins involved in NF-κB activation within the ER. All isolated NASPs also appeared to require oligomerization to activate NF-κB because they were functional only when expressed from lentiviral vectors including leucine zipper dimerization or trimerization sequences. This requirement for oligomerization is shared by many known NF-κB–activating cytokine ligands (38) as well as intracellular mediators of NF-κB signaling, including TRAF family members (39).
TRAF6 was found to be essential for NF-κB activation by all characterized NASPs. This finding suggests existence of a highly “druggable” mechanism of regulation of the canonical NF-κB pathway (40) that involves activation of TRAF6, presumably via its di- or trimerization. In fact, structural studies revealed a highly conserved motif consisting of Pro-X-Glu-X-X (aromatic/acidic residue) (41) that is present within the common region of all four ApoF-derived NASPs, thus pointing at a potential mechanism of NF-κB activation by these peptides. Interestingly, activated TRAF6 has been considered a therapeutic target in multiple myeloma (42). This observation suggests that TRAF6 is a potentially a “weak link” within the NF-κB pathway and that NASPs acting via TRAF6 may mimic naturally occurring mechanisms of constitutive NF-κB activation by endogenous factors.
Having generated NASPs as tools capable of inducing constitutive NF-κB activation, we next used them to evaluate the effect of NF-κB activation on transformation of rodent fibroblasts expressing oncogenic Ras. In both mouse and rat fibroblasts, expression of all tested NASPs, regardless of what proteins they are derived from, reversed cellular senescence induced by oncogenic Ras expression and induced cell transformation. Cells coexpressing NASPs with oncogenic Ras not only lacked senescence-associated β-galactosidase staining and formed proliferative transformation foci but also rapidly formed tumors when injected s.c. into nude mice.
In previous work, inactivation of p53 was shown to be the only requirement that must be met to overcome cellular senescence induced by oncogenic Ras and establish a transformed phenotype in rodent fibroblasts (28). Indeed, we found that all NASPs shown capable of cooperation with Ras also inhibited doxorubicin-induced p53-mediated transactivation. In this regard, NASPs resemble genetic suppressor elements described in our earlier work that encode fragments of p53 protein that act as dominant-negative p53 mutants also capable of cooperation with Ras (27). Even more relevant is our recent work on the association of mycoplasma infection with cancer predisposition where we found that the chronic, typically asymptomatic infections established by this class of bacteria lead to activation of NF-κB, suppression of p53, and promotion of oncogenic Ras-induced malignant transformation (30). Thus, sustained NF-κB activation resulting from infectious agents (such as mycoplasma) might promote cancer development not only through known NF-κB–induced factors (e.g., antiapoptotic factors and growth-promoting cytokines), but also through suppression of p53, which could impact multiple pathways of tumorigenesis, including sensitivity to oncogene-induced transformation.
Activation of NF-κB by oncogenic Ras has been shown to be necessary to suppress Ras-induced apoptosis in rodent fibroblasts (43). Another report indicates that wild-type p53 inhibits cellular transformation by regulating glucose metabolism through NF-κB (44). We propose that the NF-κB–activating peptides identified in this study probably function by augmenting the NF-κB–activating effect of oncogenic Ras such that apoptosis is efficiently blocked while simultaneously suppressing p53 to promote complete transformation.
Suppression of p53 activity by NASPs is consistent with the reciprocal negative regulatory relationship between p53 and NF-κB that has been reported by our group and several others (15, 45–48). Our recognition of this interplay between p53 and NF-κB stemmed from the finding that treatment of a particular cell line with genetic suppressors of p53 or with an NF-κB–activating agent (TNF) resulted in strikingly similar changes in gene-expression profiles. Consistent with these data, p53-null mice exhibit enhanced inflammatory responses, increased susceptibility to septic shock, and high levels of NF-κB activation (13). Moreover, spontaneous inflammation-associated lethality is observed in 25% of p53 knockout mice (49). We have shown that p53 negatively regulates the NF-κB pathway, which provides a further advantage to tumor cells (13). The underlying mechanism has been reported to involve p53-mediated suppression of NF-κB binding to p300 (47). A similar molecular mechanism is likely involved in the suppression of p53 by other aforementioned infectious agents and the NASPs identified in this study.
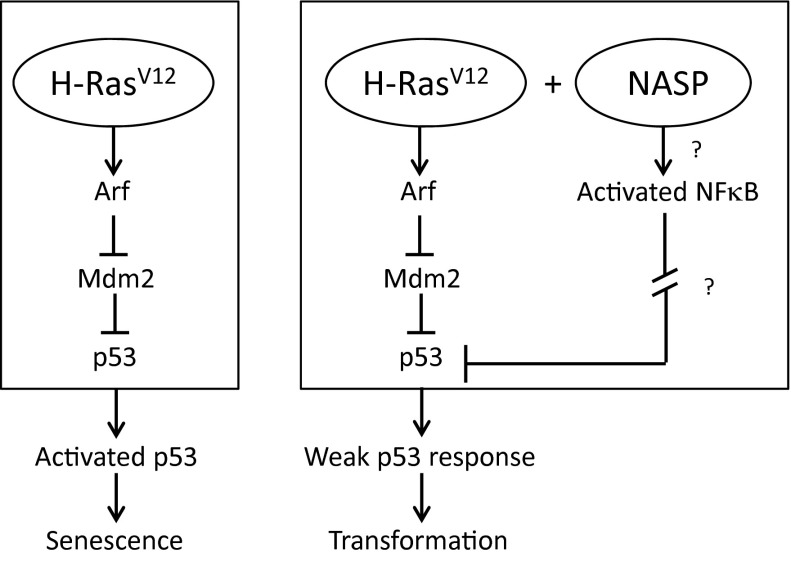
In summary, our work supports a model in which constitutively active NF-κB functions as an oncogene, presumably through attenuation of p53 function, which, in cooperation with oncogenic Ras, drives malignant transformation (Fig. 7). Although activated NF-κB induces expression of antiapoptotic proteins and inhibitors of reactive oxygen species (ROS), suppressed p53 reduces levels of proapoptotic factors, thus providing a collective growth advantage to transformed cells. Chronic infections permit selection and expansion of cell clones with such a dual advantage, thus leading to cancer. Therefore, inhibition of NF-κB activity (through, for example, anti-inflammatory therapies) may be clinically relevant not only for preventing tumor progression, but also for preventing tumor initiation. Besides demonstrating that constitutive NF-κB activation promotes tumorigenesis through p53 suppression, the NASPs identified here present a set of research tools for future studies on NF-κB regulation and activity. In addition, this study illustrates the general feasibility of using pooled lentiviral peptide libraries in cell-based assays to isolate biologically active peptides via functional selection and offers a technological platform for the isolation of bioactive peptides. Our BASP approach to isolate activating genetic elements parallels the principles of genetic suppressor element (GSE) selection to identify functional genes (50). We anticipate that BASP technology should permit unbiased screening of functional peptides to identify novel drug targets without any prior knowledge of cell-signaling mechanisms and could broadly be exploited in other biological systems using appropriate functional assays.
Fig. 7.
Hypothetic model for NASP cooperation with oncogenic Ras.
Materials and Methods
Primary MEFs were prepared as described earlier (51). MEF transformation by vectors expressing oncogenic Ras and p53-inhibitory constructs a well as cell transformation assays were done as previously reported (26). ConA reporter cells carrying p53-dependent lacZ reporter gene were described previously (32). Details of other methods used in this study, including cell culture, lentivirus preparation and transduction, development of 293-NF-κB-GFP reporter cells, construction of peptide libraries, senescence-associated β-gal assays (29), reporter assays, Ras cooperation assays, soft agar assays, and in vivo tumorigenicity assays are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kellee Greene and Michael Yeluashvily for technical help and Patricia Stanhope Baker for help in manuscript preparation. This work was supported by National Institutes of Health Grants R01AI080446 and RC2AI087616 (to A.V.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311945111/-/DCSupplemental.
References
- 1.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 4.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4(1):11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 5.Mantovani A. Cancer: Inflammation by remote control. Nature. 2005;435(7043):752–753. doi: 10.1038/435752a. [DOI] [PubMed] [Google Scholar]
- 6.Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Curr Opin Genet Dev. 2008;18(1):3–10. doi: 10.1016/j.gde.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Mantovani A. Cancer: Inflaming metastasis. Nature. 2009;457(7225):36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 8.Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A tale of two stresses. Genes Cancer. 2011;2(4):503–516. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Hayden MS. Celebrating 25 years of NF-κB research. Immunol Rev. 2012;246(1):5–13. doi: 10.1111/j.1600-065X.2012.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M, Greten FR. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 11.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komarova EA, et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005;19(8):1030–1032. doi: 10.1096/fj.04-3213fje. [DOI] [PubMed] [Google Scholar]
- 14.Gurova KV, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci USA. 2005;102(48):17448–17453. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ak P, Levine AJ. p53 and NF-κB: Different strategies for responding to stress lead to a functional antagonism. FASEB J. 2010;24(10):3643–3652. doi: 10.1096/fj.10-160549. [DOI] [PubMed] [Google Scholar]
- 16.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Lane D, Levine A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 19.Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324(5924):242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chenchik A, Gudkov A, Komarov A, Natarajan V. 2010. US Patent Appl 20100305002 A1.
- 21.Hunter T. Cooperation between oncogenes. Cell. 1991;64(2):249–270. doi: 10.1016/0092-8674(91)90637-e. [DOI] [PubMed] [Google Scholar]
- 22.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N, et al. Regulation of IkBa expression involves both NF-kB and the MAP kinase signaling pathways. J Inflamm (Lond) 2005;2:10. doi: 10.1186/1476-9255-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdelya LG, et al. Central role of liver in anticancer and radioprotective activities of Toll-like receptor 5 agonist. Proc Natl Acad Sci USA. 2013;110(20):E1857–E1866. doi: 10.1073/pnas.1222805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- 26.Boiko AD, et al. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20(2):236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ossovskaya VS, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93(19):10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 29.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logunov DY, et al. Mycoplasma infection suppresses p53, activates NF-kappaB and cooperates with oncogenic Ras in rodent fibroblast transformation. Oncogene. 2008;27(33):4521–4531. doi: 10.1038/onc.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldi L, Brown K, Franzoso G, Siebenlist U. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I kappa B-alpha. J Biol Chem. 1996;271(1):376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 32.Komarova EA, et al. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 1997;16(6):1391–1400. doi: 10.1093/emboj/16.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karin M. The IkappaB kinase: A bridge between inflammation and cancer. Cell Res. 2008;18(3):334–342. doi: 10.1038/cr.2008.30. [DOI] [PubMed] [Google Scholar]
- 34.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell. 2006;124(4):823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 35.Chu ZL, et al. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci USA. 1997;94(19):10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFkappaB activation in cancer. Oncogene. 2004;23(12):2138–2145. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- 37.Iosef C, et al. Inhibiting NF-κB in the developing lung disrupts angiogenesis and alveolarization. Am J Physiol Lung Cell Mol Physiol. 2012;302(10):L1023–L1036. doi: 10.1152/ajplung.00230.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Baud V, et al. Signaling by proinflammatory cytokines: Oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13(10):1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12(8):695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25(11):1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, et al. TRAF6 activation in multiple myeloma: A potential therapeutic target. Clin Lymphoma Myeloma Leuk. 2012;12(3):155–163. doi: 10.1016/j.clml.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayo MW, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278(5344):1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 44.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10(5):611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 45.Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: A role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1(5):493–503. doi: 10.1016/s1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 46.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404(6780):892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 47.Ravi R, et al. p53-mediated repression of nuclear factor-kappaB RelA via the transcriptional integrator p300. Cancer Res. 1998;58(20):4531–4536. [PubMed] [Google Scholar]
- 48.Kawauchi K, Araki K, Tobiume K, Tanaka N. Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification. Proc Natl Acad Sci USA. 2009;106(9):3431–3436. doi: 10.1073/pnas.0813210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gudkov AV, Komarova EA. Pathologies associated with the p53 response. Cold Spring Harb Perspect Biol. 2010;2(7):a001180. doi: 10.1101/cshperspect.a001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roninson IB, et al. Genetic suppressor elements: New tools for molecular oncology—thirteenth Cornelius P. Rhoads Memorial Award Lecture. Cancer Res. 1995;55(18):4023–4028. [PubMed] [Google Scholar]
- 51.Zou X, et al. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 2002;16(22):2923–2934. doi: 10.1101/gad.1033002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.