Significance
Neural stem/progenitor cells (NSPCs) restrict their differentiation potential by developmental stage-dependent temporal specification. Thereby, specific and efficient induction of homogeneous target cell populations remains a challenge in stem cell biology. Here, we provided a potential solution by identifying the molecular machinery responsible for neurogenic-to-gliogenic transition of NSPCs, a process we termed “competence change.” We identified the microRNA-17/106–p38 axis as a critical regulator of the competence change, although epigenetic regulation seemed to be the regulatory program behind it, by our previous Coup-tf study. NSPCs sustained and restored neurogenic potential by competence regulation. Moreover, at least a part of neuron-subtype specification was regulated independent of it. Control of these multilayered regulatory programs seems promising for rigorous manipulation of cytogenesis from NSPCs.
Keywords: neural development, glia, fate determination, differentiation, neurogenesis
Abstract
Neural stem/progenitor cell (NSPC) multipotency is highly regulated so that specific neural networks form during development. NSPCs cannot respond to gliogenic signals without acquiring gliogenic competence and decreasing their neurogenic competence as development proceeds. Coup-tfI and Coup-tfII are triggers of these temporal NSPC competence changes. However, the downstream effectors of Coup-tfs that mediate the neurogenic-to-gliogenic competence transition remain unknown. Here, we identified the microRNA-17/106 (miR-17/106)–p38 axis as a critical regulator of this transition. Overexpression of miR-17 inhibited the acquisition of gliogenic competence and forced stage-progressed NSPCs to regain neurogenic competence without altering the methylation status of a glial gene promoter. We also identified Mapk14 (also known as p38) as a target of miR-17/106 and found that Mapk14 inhibition restored neurogenic competence after the neurogenic phase. These results demonstrate that the miR-17/106–p38 axis is a key regulator of the neurogenic-to-gliogenic NSPC competence transition and that manipulation of this axis permits bidirectional control of NSPC multipotency.
Treatments of central nervous system (CNS) injury and diseases have become more promising with advances in modern medicine. Recent progress in stem cell biology has drawn attention to stem cells as innovative resources for transplantation therapies and individualized drug screenings (1, 2). Multipotent neural stem/progenitor cells (NSPCs) that give rise to all types of neural cells can now be readily obtained from induced pluripotent stem cells. However, specific and efficient induction of homogeneous target cell populations from NSPCs remains difficult because of the complex mechanisms that regulate NSPC development and differentiation. Therefore, further elucidation of how specific cell types can be generated from NSPCs is required to facilitate therapeutic applications.
We recently used a newly developed embryonic stem cell (ESC)-derived neurosphere culture system to investigate the molecular mechanisms that govern NSPC differentiation (3). Although NSPCs are multipotent, and are thus able to differentiate into neurons and glial cells, neurogenesis largely precedes gliogenesis during CNS development in vertebrates. The neurogenesis-to-gliogenesis switch requires temporal identity transitions of NSPCs (4). Importantly, our neurosphere culture system recapitulates neural development in vivo. Using this system, we found that Coup-tfI and Coup-tfII (also known as Nr2f1 and Nr2f2, respectively) are critical molecular switches in the temporal identity transition of NSPCs (3). Remarkably, Coup-tfs do not repress neurogenesis or promote gliogenesis but, instead, change the competence of NSPCs. Although Coup-tfs permit alterations by changing the responsiveness of NSPCs to extrinsic gliogenic signals, the critical regulators and/or drivers of this process remain largely unknown. The aim of this study was to determine the molecular machinery underlying the neurogenic-to-gliogenic competence transition of NSPCs.
Results
Identification of miR-17/106 as Downstream Effectors of Coup-tfs.
We first attempted to identify candidate genes that are downstream of Coup-tfs. We searched for genes that are responsible for inhibition of gliogenesis and/or sustained generation of early-born neurons in Coup-tf-knockdown (KD) cultures. To this end, we investigated the direct DNA binding sites of Coup-tfs by performing chromatin immunoprecipitation sequencing. We then compared global gene and microRNA (miRNA) expression profiles between control and Coup-tf-KD neurospheres from mouse ESCs, using microarray analyses. Candidate genes were defined as genes with expression levels that were down-regulated as development proceeded in control neurospheres but that remained in a steady state in Coup-tf-KD neurospheres. We cloned and constructed lentiviral libraries for overexpression (OE) and KD of the candidate genes: 150 transcription factors and 83 miRNAs, which also included miRNA clusters. We then used the ESC-derived neurosphere culture system to functionally screen the candidate genes, as previously described (SI Appendix, Tables S1 and S2 and SI Appendix, Materials and Methods) (3). After screening the candidate genes, we identified the miR-17-92 miRNA cluster as a functional downstream effector of Coup-tfs that regulated NSPC competence, although not as their direct target.
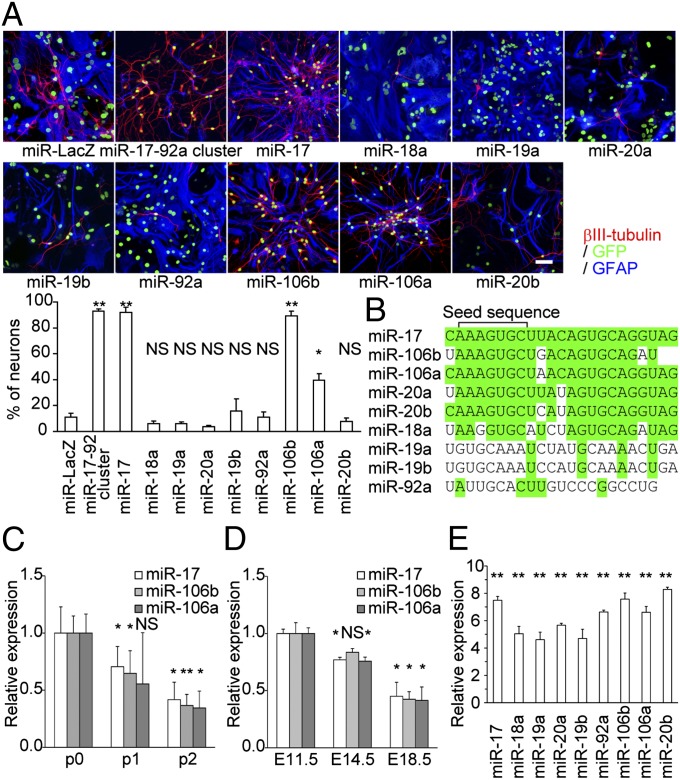
The miR-17-92 cluster was originally identified as a cluster of oncogenic miRNAs and is associated with the development of various organs (5, 6). The microarray results showed that the expression levels of all of the miRNAs that compose this cluster were higher in Coup-tf-KD neurospheres that were passaged twice (p2) than in control p2 neurospheres. Normally, the majority of cells in p2 neurospheres differentiate into astrocytes, and fewer than 20% become neurons. However, OE of the miR-17-92 cluster caused these cells to differentiate almost exclusively into neurons under the differentiation conditions without mitogen (Fig. 1A).
Fig. 1.
OE of miR-17/106 inhibits the neurogenic-to-gliogenic transition of NSPCs in vitro. (A) Representative immunocytochemical images of differentiated p2 neurospheres infected with lentiviruses of the indicated miRNAs and GFP at p0 stage. MiR-LacZ was used as a control. (Scale bar, 50 μm.) The table indicates the percentage of βIII-tubulin + neurons among the total number of virus-infected cells (n = 3). (B) Sequence comparison of miRNAs encoded by the miR-17-92, miR-106b-25, and miR-106a-363 clusters. Nucleotides that are identical to the corresponding nucleotides in miR-17 are shown in green. (C) qPCR of miR-17/106 in neurospheres at the p0, p1, and p2 stages (n = 3). (D) qPCR of miR-17/106 in developing NSPCs sorted from the cortex of Nestin-EGFP transgenic mouse embryos at E11.5, E14.5, and E18.5 (n = 3). (E) qPCR of each miRNA in miRNA-OE cells (compared with miR-LacZ-OE cells; n = 3). Results are shown as mean ± SEM in A and E and mean ± SD in C and D. NS (not significant), P > 0.05; *P < 0.05; **P < 0.01.
The miR-17-92 cluster encodes six distinct miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a), as well as their star (*) strands. To determine which of these miRNAs are responsible for the neurogenic phenotype, we individually overexpressed each miRNA by infecting NSPCs with lentiviruses. The lentiviruses permitted simultaneous OE of the miRNA of interest and green fluorescent protein (GFP). Only miR-17(-5p) OE replicated the phenotype induced by the intact miR-17-92 cluster (Fig. 1A).
MiR-17 has two additional paralogs with high similarity and identical 7mer seed sequences (nucleotides 2–8; Fig. 1B) that are critical for binding to target mRNAs (Fig. 1B), miR-106b and miR-106a, which are encoded by the miR-106b-25 and miR-106a-363 clusters, respectively (7). The target mRNAs of these two miRNAs should have highly overlapping sequences. These paralogs were individually overexpressed in NSPCs to examine whether they also affect the competence transition. OE of either miRNA led to a neurogenic phenotype, although the effects of miR-106b were stronger than those of miR-106a (Fig. 1A). MiR-20a and miR-20b are also extremely similar to miR-17, but we did not observe any phenotype when it was overexpressed (Fig. 1 A and B). We hypothesized that the total amounts of mature miR-17 and miR-106 are critical for the regulation of neurogenic competence in NSPCs at early developmental stages. Therefore, we assessed the expression levels of miR-17/106 in neurospheres by quantitative PCR (qPCR). All three miRNAs were down-regulated during development, and the expression levels at the p2 stage were less than half of the levels observed at p0 (Fig. 1C). Comparable declines in miR-17/106 were observed in NSPCs sorted from the cerebral cortex of Nestin-EGFP transgenic mice (Fig. 1D) (8). However, forced OE of miR-17 resulted in a 7.5-fold increase in the expression level of miR-17 in p2 neurospheres compared with that observed in control miR-LacZ-OE neurospheres (Fig. 1E).
MiR-17 Regulates the Neurogenic-to-Gliogenic Transition, but Not the Neuron-Subtype Specification.
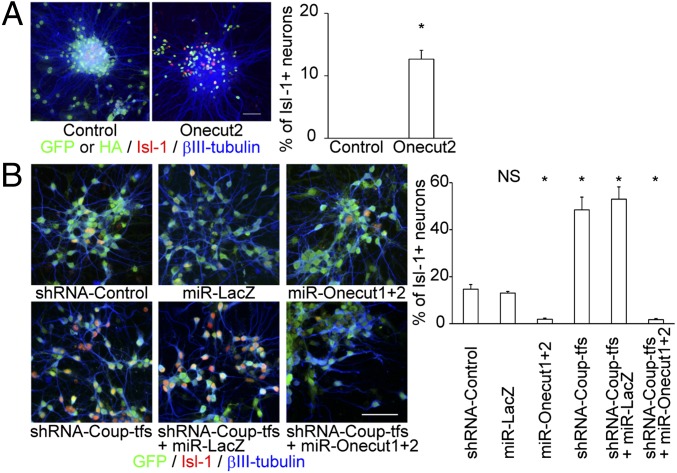
Coup-tf-KD causes sustained generation of Islet 1 (Isl-1)-expressing neurons, a subpopulation of early-born neurons that cannot be observed at the normal p2 neurosphere stage (3). Notably, in contrast to Coup-tf-KD, the miR-17 OE did not result in cytogenesis of Isl-1-positive neurons (SI Appendix, Fig. S1). However, during the candidate gene screening, the transcription factors Onecut1/2 were identified as positive regulators of Isl-1-positive cell production. OE of Onecut2 increased production of Isl-1-positive neurons, and KD of Onecut1/2 caused early termination of the production of Isl-1-positive neurons (Fig. 2 A and B). In support of these findings, a recent study demonstrated that Onecut factors directly regulate Isl-1 expression and maintain Isl-1 production during motor neuron diversification (9). These results suggest that the neurogenic-to-gliogenic transition in NSPCs, and at least a part of neuron-subtype specification, are independently regulated downstream of Coup-tfs.
Fig. 2.
Onecut1/2 regulate production of Isl-1-positive neurons downstream of Coup-tfs. (A) OE of HA-tagged Onecut2 in p2 neurospheres resulted in prolonged production of Isl-1 + neurons (n = 3). The control with nuclear-localized EGFP is also shown. (Scale bar, 50 μm.) (B) Repression of Onecut1 and Onecut2 by specific artificial miRNAs reduced the production of Isl-1 + neurons and eliminated the Coup-tf-KD phenotype in p1 neurospheres (n = 3). GFP fluorescence stemmed from the lentiviral construct containing the indicated miRNA/shRNA. (Scale bar, 50 μm.) Results are shown as mean ± SEM. NS, P > 0.05; *P < 0.05.
MiR-17 Regulates NSPC Competence Without Altering the Methylation Status of the Gfap Promoter.
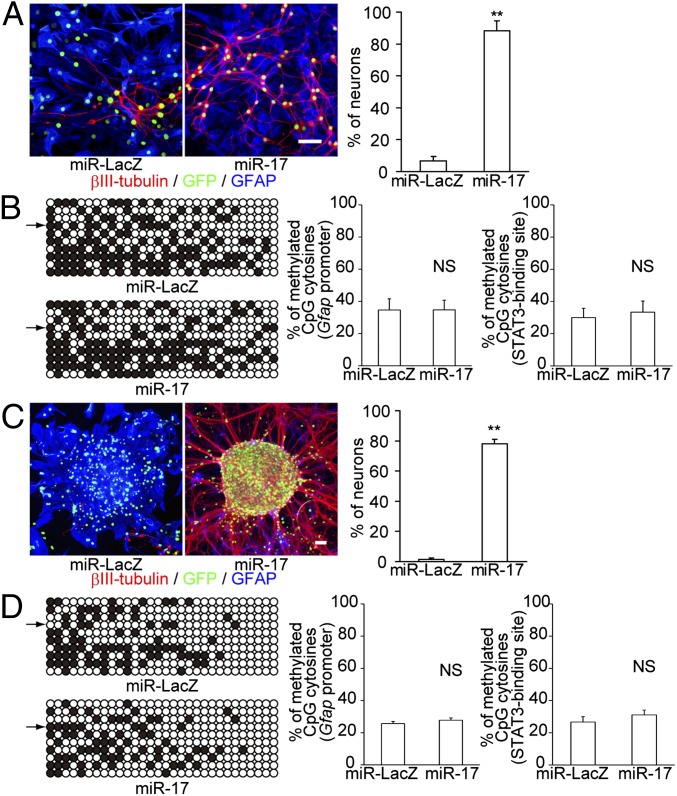
The transition from early developmental neurogenic competence to late developmental gliogenic competence can be identified by changes in NSPC responsiveness to gliogenic cytokines (3). Leukemia inhibitory factor (LIF) and bone morphogenetic protein 2 (BMP2) are well-studied extrinsic gliogenic factors that promote gliogenesis in the later stages of NSPC development by activating the JAK-STAT pathway and BMP signaling, respectively (10–12). LIF does not act as a gliogenic factor in early developmental NSPCs, as the STAT3-binding sites in the promoters of glial-associated genes are epigenetically silenced by DNA methylation (13). Furthermore, BMP signaling promotes neuronal, but not glial, differentiation in the early stages of NSPC development (12, 14). Therefore, to examine whether miR-17 alters the responsiveness of NSPCs to gliogenic cytokines, we exposed miR-17-OE NSPCs to LIF and BMP2. The miR-17-OE neurospheres were strongly resistant to cytokines and exclusively differentiated into neurons at the p2 stage (Fig. 3A).
Fig. 3.
MiR-17 regulates NSPC competence without altering the epigenetic status of the Gfap promoter. (A) MiR-17-OE p2 neurospheres did not undergo gliogenesis in response to LIF (10 ng/mL) and BMP2 (100 ng/mL) (n = 3). (Scale bar, 50 μm.) (B) The CpG methylation status of the Gfap promoter was analyzed by bisulfite sequencing. A total of 10 possible CpG cytosines in the Gfap promoter, including the CpG site in the STAT3-binding sequence (indicated by the arrows), were available for methylation. The percentages of methylated CpG cytosines in the Gfap promoter (Left) and the STAT3-binding region (Right) are shown (n = 5). (C) MiR-17 OE restored neuropotency in p3 neurospheres (n = 3). (Scale bar, 50 μm.) (D) The CpG methylation status of the Gfap promoter was analyzed by bisulfite sequencing, as described for B (n = 5). Results are shown as mean ± SEM. NS, P > 0.05; **P < 0.01.
We next investigated the temporally regulated DNA methylation status of the STAT3-binding site and its surrounding CpG sites in the Gfap promoter. Glial fibrillary acidic protein (GFAP) is commonly used as a marker of astrocytes. Coup-tfs are transiently up-regulated in developing NSPCs during midgestation. NSPCs then lose their plasticity and only produce late-born neurons and glial cells. Coup-tf-KD in ESC-derived neurospheres maintains the early epigenetic status of the Gfap promoter (3). Therefore, we initially expected that the molecular effectors/drivers responsible for changes in NSPC competence would regulate the epigenetic status of neuronal or glial specification-associated genes. However, surprisingly, no significant changes in CpG methylation were observed in miR-17-OE p2 neurospheres (Fig. 3B). These results suggest that the changes in methylation status of glial genes are not enough for the abolition of neurogenic potential of NSPCs and that neurogenic competence can be maintained independent of the acquisition of gliogenic competence by the activation of miR-17/106 pathway.
MiR-17 OE Forces Restoration of Neurogenic Competence in Gliogenic NSPCs.
We next sought to determine whether miR-17/106 OE could restore neurogenic competence in stage-progressed NSPCs. To address this question, we used lentiviruses to overexpress miR-17 in neurospheres at the onset of the p3 stage. Normally, neurospheres exclusively differentiate into glial cells at this time. We previously found that Coup-tf-KD results in restoration of neuropotency in limited populations of p3 NSPCs, which have not yet lost their plasticity (3). Surprisingly, miR-17 OE led to a marked restoration of neuropotency in p3 neurospheres within 1 wk. In contrast, the vast majority of cells in the control p3 neurospheres differentiated into astrocytes (Fig. 3C). A neurogenic phenotype similar to that induced by miR-17 OE was observed in primary cultured neurospheres derived from the subventricular zone (SVZ) of the mouse forebrain at postnatal day 30 (P30) (SI Appendix, Fig. S2). Furthermore, similar to our observations in neurospheres that overexpressed miR-17 from stage p0 to p2 (Fig. 3B), the DNA methylation status of the Gfap promoter did not significantly change (Fig. 3D). These results further support the idea that competence changes are essential prerequisites for gliogenesis.
MiR-17/106 Determine the Neurogenic Competence of NSPCs.
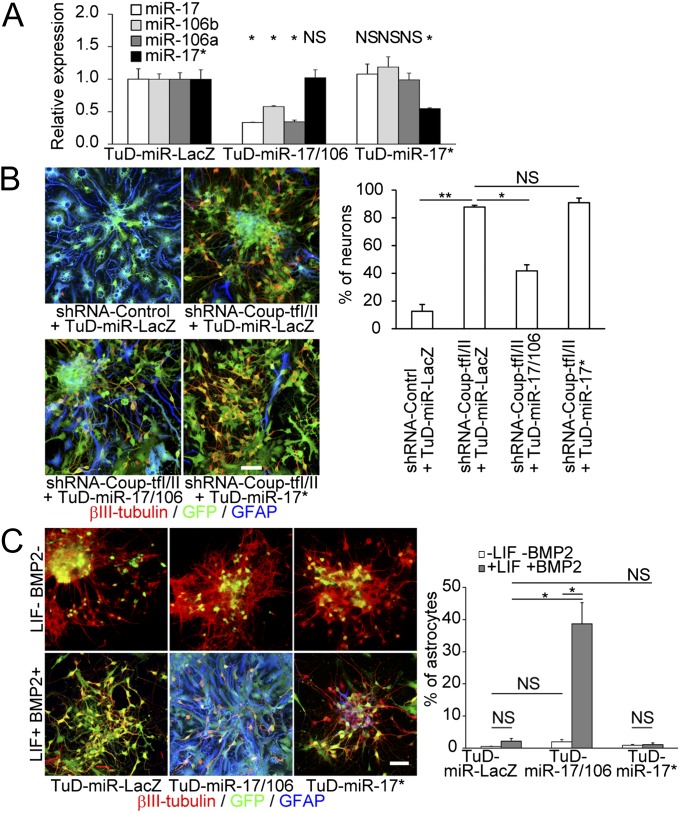
We next used loss-of-function experiments to confirm whether miR-17/106 are downstream effectors of Coup-tfs. We used RNA decoys, referred to as “tough decoy” RNAs (TuDs), to efficiently and stably suppress the activity of specific miRNA family members (15). Lentivirus vectors were constructed that expressed a miR-17 family-specific TuD (TuD-miR-17/106), a miR-17*-specific TuD (TuD-miR-17*), or a control TuD (TuD-miR-LacZ), and we confirmed that these TuDs induced degradation of each target miRNA (Fig. 4A). In these experiments, Coup-tfs were silenced in neurospheres with a Coup-tf-specific short hairpin RNA (shRNA) from the p0 stage onward. Similar to miR-17-OE neurospheres, the neurogenesis-to-gliogenesis transition was inhibited in these Coup-tf-silenced neurospheres (3). TuD-miR-17/106 expression largely abolished the neurogenic phenotype induced by Coup-tf-KD at the p2 stage, whereas the TuD-miR-17* and TuD-miR-LacZ controls had no significant effect (Fig. 4B). Normally, p0 neurospheres barely differentiate into glia even in the presence of gliogenic cytokines. However, OE of TuD-miR-17/106 (but not TuD-miR-17*) inhibited endogenous miR-17/106 (Fig. 4A) and resulted in the induction of precocious gliogenesis, but only in the presence of LIF and BMP2 (Fig. 4C). These results demonstrate that miR-17/106 are responsible for the early developmental neurogenic competence of NSPCs downstream of Coup-tfs. Furthermore, these data show that down-regulation of miR-17/106 is an essential prerequisite for initiation of gliogenesis.
Fig. 4.
Down-regulation of miR-17 eliminates the Coup-tf-KD phenotype in p2 neurospheres and causes precocious gliogenesis from p0 neurospheres. (A) qPCR of endogenous miR-17, 106b, 106a, and 17* in TuD-expressing p0 neurospheres (n = 3). (B) TuD-miR-17/106 OE eliminated the neuronal phenotype in Coup-tf-KD p2 neurospheres and restored the astrocytic phenotype, as evidenced by the reduced number of neurons and increased GFAP immunostaining (n = 3). (Scale bar, 50 μm.) (C) TuD-miR-17/106 OE caused abnormal early gliogenesis from p0 neurospheres only in the presence of LIF and BMP2 (n = 3). (Scale bar, 50 μm.) Results are shown as mean ± SD in A and mean ± SEM in B and C. NS, P > 0.05; *P < 0.05; **P < 0.01.
Functional Roles of miR-17/106 in Developing NSPCs.
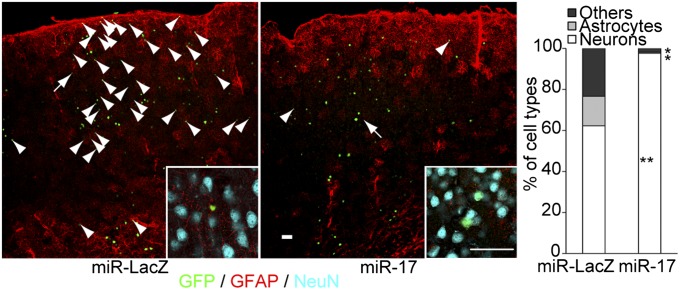
To better understand the functional roles of miR-17/106 in vivo, we first investigated their expression patterns in the developing CNS. The in vivo expression patterns of miR-17 and miR-106b were similar at embryonic day 11.5 (E11.5) and P0. The levels of both miRNAs in the brain [including the ventricular zone (VZ)] declined as development progressed (SI Appendix, Fig. S3). Next, we analyzed the in vivo roles of miR-17/106 by overexpressing miR-17 in the developing mouse brain via in utero lentiviral microinjections. Consistent with the in vitro results, most cells (97.6 ± 0.6%, mean ± SEM) infected with the miR-17-OE lentivirus at E10.5 were fated to become neurons in the cerebral cortex by P30. In contrast, only 62.2 ± 2.8% of the cells infected with the control miR-LacZ lentivirus were fated to become neurons (Fig. 5). Thus, miR-17 OE inhibited gliogenesis in vivo in a cell-autonomous fashion. In contrast, we observed no precocious gliogenesis by TuD-miR-17/106 OE in vivo. TuD-miR-17/106 promoted precocious gliogenesis only when we supplied gliogenic cytokines in vitro. Therefore, the effects of TuD-miR-17/106 may have been suppressed by an insufficient amount of gliogenic factors (16) and the presence of antigliogenic factors (17–19) during the neurogenic period when miR-17/106 are highly expressed.
Fig. 5.
In vivo roles of miR-17/106 in developing mouse forebrains. Lentiviruses that permitted OE of miR-LacZ or miR-17 were microinjected in utero into the cerebral ventricle of mouse embryos at E10.5, and the fates of the infected cells were examined in the cerebral cortex at P30 by immunohistochemistry (n = 3). Neuronal nuclei were stained with an antibody against NeuN. Arrowheads indicate nonneuronal cells, which include GFAP+ astrocytes. Most GFAP-negative nonneuronal cells (defined as “Others” in the graph) appeared to be immature astrocytes. Higher magnification images of the cells indicated with arrows are shown as insets. (Scale bars, 50 μm.) *P < 0.05; **P < 0.01.
Identification of p38 as a Target of miR-17/106.
Normally, miRNAs regulate the translation and/or degradation of multiple target mRNAs (20). To further identify the molecular mechanisms that underlie competence changes of NSPCs, we next attempted to identify the target mRNAs of miR-17/106. We first carried out proteomics analyses using the Isobaric Tags for Relative and Absolute Quantitation method (21) to identify proteins that were down-regulated by miR-17 OE in p2 neurospheres. Candidate genes were then scanned using our original computer program. This program searched for mRNA sequences that were complementary to miRNA seed sequences of particular miRNAs. An initial list of 40 genes was obtained (SI Appendix, Table S3). We then shortened the list by bioinformatic evaluation with Ingenuity Pathway Analysis and focused on 10 genes that were associated with the TGF-β signaling pathway. Finally, we functionally screened 10 candidate genes with the neurosphere culture system. Through these procedures, we identified the mRNA encoding p38 (also known as mitogen-activated protein kinase 14 or Mapk14) as a direct target of miR-17/106 during the regulation of the NSPC competence transitions (SI Appendix, Materials and Methods).
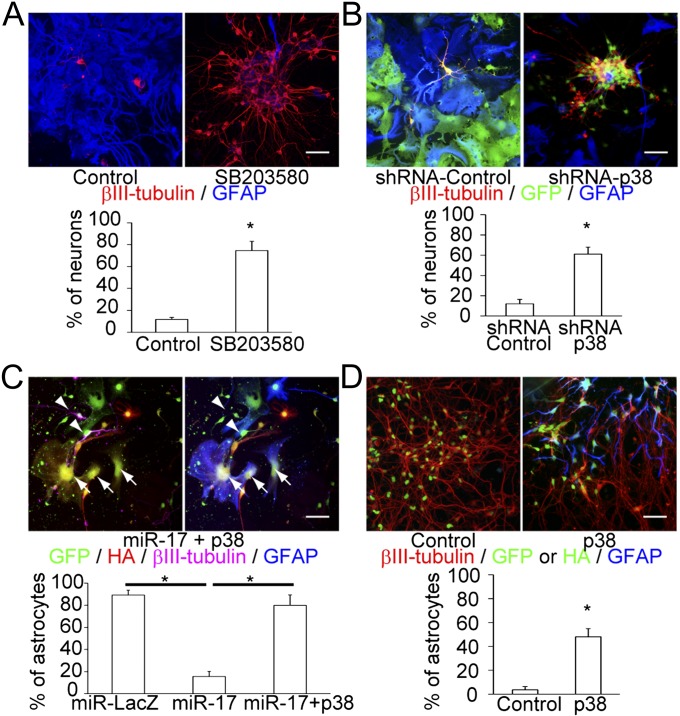
p38 protein expression increased during neurosphere passaging and was 2.78-fold and 3.16-fold higher in p2 neurospheres than in p0 neurospheres by Western blot and proteomics analyses, respectively (SI Appendix, Fig. S4A and Table S3). We also confirmed the increase in p38 expression in developing NSPCs by analyzing Nestin+ cell populations of the SVZ of wild-type mouse forebrains at E11.5, E14.5, and E18.5, using fluorescence-activated cell sorting and immunohistochemical analysis (SI Appendix, Fig. S4 B and C). Furthermore, p2 neurospheres exhibited higher neuropotency than controls on exposure to SB203580, a specific p38 inhibitor, during their growth (Fig. 6A). p2 neurospheres subjected to KD of p38 using a p38-specific shRNA also exhibited higher neuropotency than control neurospheres (Fig. 6B). In addition, a reporter assay conducted using the miR-17/106-binding site in the 3′ untranslated region of p38 mRNA revealed a direct interaction between p38 mRNA and miR-17 (SI Appendix, Fig. S5). Finally, OE of miR-17/106-resistant p38 mRNA that lacked the entire 3′ untranslated region reversed the neurogenic phenotype induced by miR-17 OE to a gliogenic phenotype (Fig. 6C) and induced precocious gliogenesis in p0 neurospheres (Fig. 6D).
Fig. 6.
p38 is a direct target of miR-17/106 and is responsible for the NSPC competence transition. (A) Treatment of p2 neurospheres in the growth phase with SB203580 increased the percentage of neurons that differentiated from the neurospheres (n = 3). (Scale bar, 50 μm.) (B) p38-specific shRNA introduced by lentiviral infection increased the neuropotency of p2 neurospheres (n = 3). (Scale bar, 50 μm.) (C) Simultaneous OE of miR-17 and miR-17/106-resistant p38 mRNA (tagged with 3× HA) eliminated the neuronal phenotype of miR-17 OE in p2 neurospheres (n = 3). Arrows indicate GFP, HA-p38, and GFAP triple-positive astrocytes. Arrowheads indicate GFP and βIII-tubulin double-positive neurons. (Scale bar, 50 μm.) (D) OE of p38 caused abnormal early gliogenesis from p0 neurospheres in the presence of LIF and BMP2 (n = 3). (Scale bar, 50 μm.) Results are shown as mean ± SEM. NS, P > 0.05; *P < 0.05; **P < 0.01.
In Vivo Functional Roles of p38 in Developing NSPCs.
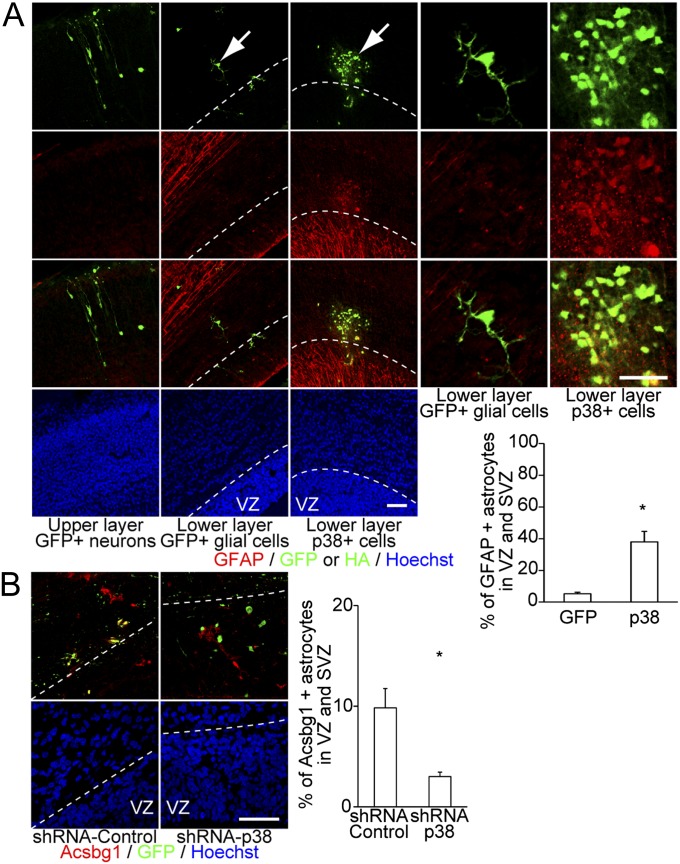
To determine the in vivo roles of p38, we performed gain- and loss-of-function studies via in utero lentiviral microinjection. Consistent with the in vitro results, precocious GFAP expression was observed in the cerebral cortex at E17.5 when p38 was overexpressed. In utero injections of the p38-OE lentivirus resulted in cell clusters with low GFAP expression in the SVZ. In contrast, control cells that overexpressed enhanced GFP did not form cell clusters or express GFAP (Fig. 7A). p38 KD experiments using a lentivirus that expressed p38-specific shRNA revealed a decrease in the number of VZ and SVZ cells that expressed acyl-CoA synthetase bubblegum family member 1 (Acsbg1) at E17.5 (Fig. 7B). Acsbg1 is a gray matter astrocyte marker that is useful for the detection of GFAP-positive mature astrocytes and GFAP-negative immature astrocytes in the cerebral cortex (22). These observations suggest that p38 is critically involved in the initiation of gliogenesis. Taken together, our results strongly suggest that the miR-17/106–p38 axis is the major regulator of the neurogenic-to-gliogenic competence transition in developing NSPCs and that manipulation of this axis permits bidirectional control of NSPC multipotency (SI Appendix, Fig. S6).
Fig. 7.
p38 regulates the acquisition of gliogenic competence in the developing mouse forebrain. (A) OE of p38 resulted in cell clusters with abnormal early GFAP protein expression in the VZ and SVZ (n = 5) (three left panels). Higher magnification images of the cells indicated by the arrows are shown in the two right panels. (Scale bars, 50 μm.) (B) shRNA-mediated p38-KD significantly decreased the percentage of Acsbg1-expressing astrocytes in the VZ and SVZ (n = 5). (Scale bars, 50 μm.) Results are shown as mean ± SEM *P < 0.05.
Discussion
The miR-17-92 cluster is associated with tumorigenesis and the development of various organs (5–7, 23). A recent study of the functions of the miR-17-92 cluster has shown that miR-19 and miR-92a repress PTEN and Tbr2 (also known as Eomes), respectively, and suppress the transition from radial glial cells to intermediate progenitors (24). These two miRNAs have strong oncogenic effects by regulating PTEN and Bim (also known as Bcl2l11) (6, 23). Therefore, they have often been the focus of miR-17-92 cluster studies, whereas the functions of miR-17/106 in developing NSPCs have remained obscure. We identified the miR-17/106-p38 axis as a key effector of the neurogenic-to-gliogenic competence transitions in NSPCs, and our previous study identified Coup-tfs as the triggers of this competence transition. We initially hypothesized that epigenetic regulation is the most fundamental program behind the competence transition of NSPCs for three reasons: the expression of Coup-tfs in NSPCs peaks at the midgestation stage and then declines, this expression peak is required for acquisition of gliogenic competence and changes in the epigenetic status of the Gfap promoter, and epigenetic regulation of the polycomb-group complex restricts neurogenic competence by terminating the expression of the proneural Neurogenin gene, which in turn regulates responsiveness to Wnt signals (25). However, our present results suggest that these miRNAs form a distinct regulatory layer that is essential for neurogenic competence of NSPCs independent of the acquisition of gliogenic competence (Fig. 3). Forced miR-17 OE not only maintained but also restored neurogenic competence in stage-progressed NSPCs without changes in the methylation status of the Gfap promoter. Moreover, miR-17 loss of function (via TuD-miR-17/106 OE) resulted in precocious gliogenesis in p0 neurospheres, albeit with the addition of gliogenic cytokines (Fig. 4B). These observations suggest that multilayered systems independently govern the neurogenic/gliogenic competence of NSPCs. Identification of other pro- and antigliogenic/neurogenic signals in developing NSPCs and subsequent in vivo correlations with the timing of neurogenic competence termination and gliogenic competence acquisition are necessary to further advance the understanding of these systems.
Our data also provide insight into the molecular mechanisms underlying the multilayered regulation programs of cytogenesis from NSPCs. The Onecut2 gene was identified as a regulator of NSPC differentiation into early-born Isl-1-positive neurons (Fig. 2). This regulation occurred independent of miR-17/106-mediated NSPC competence changes. These data suggest that the temporally regulated neurogenic-to-gliogenic competence transition and neuron-subtype specification of NSPCs are not totally regulated via a common molecular pathway. However, timing of these events is synchronized within and across competence phases. Therefore, integration of outputs from multiple dynamic regulatory programs seems to be important for the sequential generation of various cell types from NSPCs. Indeed, the competence regulation may also be involved in the neuron-subtype specification indirectly. For example, Foxg1 is constitutively required for the suppression of the earliest-born Cajal-Retzius neuron fate, and Foxg1-KD can reset the timing of the neuron subtype specification of cortical progenitors only within the neurogenic phase (4, 26).
p38 was responsible for the neurogenic-to-gliogenic competence transition in NSPCs (Figs. 6 and 7). However, it is unclear how p38 is linked with other gliogenic factors. Recent studies concerning the Oasis-Gcm1 axis (27) and the RAF/MEK/ERK pathway (28) found that these pathways also control gliogenesis. Deficiency of the transcription factor Oasis results in astrocytogenesis-specific disturbances and increases the number of Nestin-positive NSPCs. Therefore, Oasis seems to be more important for the differentiation or maturation steps of astrocytogenesis than the competence regulation or cell fate determination steps of NSPCs. In addition, OE of constitutively active Mek1 dramatically increases gliogenesis from radial progenitors, and Mek1/2 deletion results in problems in the maintenance of the glial-like properties of radial progenitors, severe loss of gliogenesis, and a prolonged neurogenesis at late embryonic stages. It will be interesting to investigate whether the p38 signaling pathway interacts with the RAF/MEK/ERK pathway to regulate the neurogenic-to-gliogenic competence transition. Further investigations of the upstream and downstream effectors of p38 and of the crosstalk between p38 and other signaling pathways will help identify novel molecular mechanisms that enable rigorous manipulation of cytogenesis from NSPCs.
Materials and Methods
Cell Culture and Neurosphere Differentiation Assay.
Mouse ESC culture, embryoid body formation, neurosphere formation, and neurosphere differentiation were performed as previously described (3). Identification of miR-17/106 and p38 were performed as described in SI Appendix, Materials and Methods.
Lentivirus Preparation.
Lentiviral particles were produced by transient transfection of human embryonic kidney 293T (HEK293T) cells with lentivirus constructs (provided by H. Miyoshi, RIKEN BioResource Center) that contained the gene of interest. The sequences of the shRNAs, artificial miRNAs, and TuDs used in this study are shown in SI Appendix, Table 6. The KD efficiencies are shown in Fig. 4A and SI Appendix, Fig. S7 and in our previous report (3).
Mice and in Utero Virus Injection.
Experiments were performed with the Institute of Cancer Research (ICR) strain of mice. Animal care and experiments were performed according to the guidelines of the Experimental Animal Care Committee of Keio University School of Medicine and the Yokohama Safety Center of RIKEN. In utero microinjections of lentivirus particles into E10.5 ICR mouse brains were guided by an in vivo ultrasound real-time scanner (Vevo660; VisualSonics).
Immunostaining.
Immunocytochemistry and immunohistochemistry were performed as previously described (3), using antibodies against βIII-tubulin (1:1,000, Covance MMS-435P; 1:2,000, Covance PRB-435P), NeuN (1:100, Chemicon 377), GFAP (1:400, DAKO Z0334), Acsbg1 (1:200, Abcam ab118154), and hemagglutinin (HA, 1:1,000, Roche 11867423001).
qPCR Analyses.
qPCR analyses were performed using miScript II RT kits, miScript Primer Assays (Hs_SNORD61, Mm_miR-17, Mm_miR-106a, and Mm_miR-106b), miScript SYBR Green PCR Kits (Qiagen), and the StepOne Plus Real-Time PCR System (Applied Biosystems).
Statistical Analyses.
A minimum of three independent experiments were included in each statistical analysis. Statistical significance was determined by two-tailed t tests. Throughout the study, P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. H. Miyoshi (Riken BioResource Center) for the lentiviral constructs, Dr. H. Iba (University of Tokyo) for the TuD constructs, and Ms. R. Iijima for technical assistance. This study was supported by the Young Chief Investigator Program at RIKEN (H.N.-K.), by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (to H.N.-K., T.S., and H.O.), by a Grant-in-Aid for Scientific Research on Innovative Areas entitled “Neural Diversity and Neocortical Organization” from MEXT (to T.S.), and by the Funding Program for World-Leading Innovative R&D on Science and Technology from the Japan Society for the Promotion of Science (H.O.).
Footnotes
Conflict of interest statement: H.O. is a scientific consultant for San Bio, Inc.; Eisai Co., Ltd.; and Daiichi Sankyo Co., Ltd.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315567111/-/DCSupplemental.
References
- 1.Okano H. Neural stem cells and strategies for the regeneration of the central nervous system. Proc Jpn Acad, Ser B, Phys Biol Sci. 2010;86(4):438–450. doi: 10.2183/pjab.86.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nori S, et al. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci USA. 2011;108(40):16825–16830. doi: 10.1073/pnas.1108077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11(9):1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 4.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9(6):743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 5.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18(3):262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi A, et al. Nestin-EGFP transgenic mice: Visualization of the self-renewal and multipotency of CNS stem cells. Mol Cell Neurosci. 2001;17(2):259–273. doi: 10.1006/mcne.2000.0925. [DOI] [PubMed] [Google Scholar]
- 9.Roy A, et al. Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development. 2012;139(17):3109–3119. doi: 10.1242/dev.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koblar SA, et al. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc Natl Acad Sci USA. 1998;95(6):3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakashima K, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284(5413):479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 12.Nakashima K, et al. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci USA. 2001;98(10):5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takizawa T, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Cogswell CA, LoTurco JJ. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J Neurosci. 1998;18(21):8853–8862. doi: 10.1523/JNEUROSCI.18-21-08853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37(6):e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnabé-Heider F, et al. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48(2):253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 17.Schmid RS, et al. Neuregulin 1-erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex. Proc Natl Acad Sci USA. 2003;100(7):4251–4256. doi: 10.1073/pnas.0630496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127(1):185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier AS, et al. Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron. 2007;54(2):245–262. doi: 10.1016/j.neuron.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: Ups and downs in cancer and aging. Biogerontology. 2010;11(4):501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009;9(5):1374–1384. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian S, et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3(5):1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63(5):600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303(5654):56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 27.Saito A, et al. Unfolded protein response, activated by OASIS family transcription factors, promotes astrocyte differentiation. Nat Commun. 2012;3:967. doi: 10.1038/ncomms1971. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. MEK Is a Key Regulator of Gliogenesis in the Developing Brain. Neuron. 2012;75(6):1035–1050. doi: 10.1016/j.neuron.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.