Significance
Saponins are plant molecules that are produced as a chemical defense against herbivores and eukaryotic pathogens. They constitute structurally diverse, bioactive compounds composed of a 30-carbon triterpene backbone adorned with multiple functional groups and sugars. Saikosaponins are abundant saponins accumulating in the Asian medicinal plant Bupleurum falcatum, but none of the enzymes involved in their biosynthesis had been characterized. We identified a cytochrome P450 involved in the oxidation of saikosaponins, thereby expanding the enzyme compendium that can generate plant saponins with an extra activity. Using this enzyme compendium, we established a synthetic biology program to reconstitute saponin biosynthesis in the yeast Saccharomyces cerevisiae and developed a cyclodextrin-based culturing strategy to sequester triterpenes from engineered yeast cells and enhance their productivity.
Keywords: cyclodextrin, amyrin, metabolic engineering, triterpenoid
Abstract
The saikosaponins comprise oleanane- and ursane-type triterpene saponins that are abundantly present in the roots of the genus Bupleurum widely used in Asian traditional medicine. Here we identified a gene, designated CYP716Y1, encoding a cytochrome P450 monooxygenase from Bupleurum falcatum that catalyzes the C-16α hydroxylation of oleanane- and ursane-type triterpenes. Exploiting this hitherto unavailable enzymatic activity, we launched a combinatorial synthetic biology program in which we combined CYP716Y1 with oxidosqualene cyclase, P450, and glycosyltransferase genes available from other plant species and reconstituted the synthesis of monoglycosylated saponins in yeast. Additionally, we established a culturing strategy in which applying methylated β-cyclodextrin to the culture medium allows the sequestration of heterologous nonvolatile hydrophobic terpenes, such as triterpene sapogenins, from engineered yeast cells into the growth medium, thereby greatly enhancing productivity. Together, our findings provide a sound base for the development of a synthetic biology platform for the production of bioactive triterpene sapo(ge)nins.
Triterpene saponins are secondary metabolites that exhibit a large structural diversity and wide range of biological activities in many plant species (1, 2). Saponins are glycosides of sapogenins, which are composed of 30 carbon atoms arranged in 4- or 5-ring structures that are “decorated” by functional groups. Saponins are synthesized by multiple glycosylations of the sapogenin building blocks that are produced by multiple cytochrome P450-dependent monooxygenase (P450) or oxidoreductase-mediated modifications of basic backbones, such as β-amyrin (oleanane type), α-amyrin (ursane type), lupeol, and dammarenediol. These backbones are generated by oxidosqualene cyclase (OSC)-mediated cyclization of 2,3-oxidosqualene, which is also an intermediate in the synthesis of sterols in eukaryotes (3, 4). Both saponins and sapogenins include biologically active compounds or serve as starter molecules for the generation of novel, potentially bioactive structures by synthetic modification (5–7).
The genus Bupleurum consists of perennial herbs that are used in Asian traditional medicine, either alone or in combination with other ingredients, for the treatment of common colds, fever, and inflammatory disorders (8). Saikosaponins constitute the largest class of secondary metabolites in Bupleurum and can account for up to ∼7% of root dry weight. Their accumulation can be further stimulated by jasmonate treatment (9). More than 120 closely related oleanane- and ursane-type saikosaponins have been identified from this genus and the oxidations at various positions suggest the presence of multiple enzymes, mainly P450s, capable of catalyzing specific modifications on the amyrin backbones (8, 10). To date, no P450 or oxidoreductase involved in triterpene saponin biosynthesis has been identified from Bupleurum species.
P450s that modify the β-amyrin backbone on C-11; C-12,13; C-16; C-22; C-23; C-28 or C-30 have been characterized from Glycyrrhiza uralensis, Avena strigosa, Medicago truncatula, Glycine max, Vitis vinifera, and Catharanthus roseus (11–18). Hydroxylases from Panax ginseng that oxidize the dammarenediol-II backbone on C-6, C-12, or C-28 (19–21), and a C-20 hydroxylase from Lotus japonicus (22) that modifies lupeol, have also been identified. To characterize these P450s, they have been ectopically expressed in yeast strains either producing β-amyrin or externally supplied with candidate substrates. Similarly, several OSCs have been produced and functionally analyzed in yeast. From these studies, it is clear that yeast cells cannot only be used for the characterization of novel enzymes, but possibly also as a heterologous host for the production of triterpene sapogenins (23). To date only two pilot studies have aimed at engineering of β-amyrin production in yeast (24, 25), but no efforts toward engineering of sustainable production of sapogenins or saponins in yeast have been reported.
Here, we identified and characterized CYP716Y1, a P450 from Bupleurum falcatum that corresponds to a C-16α oxidase, designated according to Nelson’s nomenclature (http://drnelson.uthsc.edu/cytochromeP450.html). By designing triterpene-hyperproducing starter strains, optimizing culturing conditions for triterpene synthesis, and using the CYP716Y1 gene in a combinatorial synthetic biology program, we established a platform that allows us to produce and sequester triterpene sapogenins in culture medium and to reconstitute a full saponin synthetic pathway in yeast cells.
Results
Transcript Profiling of Jasmonate-Treated B. falcatum Roots Reveals CYP716Y1.
To identify saponin biosynthetic genes, expression analysis on roots of methyl jasmonate (MeJA)-treated B. falcatum plants was performed with cDNA-amplified fragment length polymorphism–based transcript profiling (26). The expression of 18,695 transcript tags was monitored over time and 1,771 MeJA-responsive tags (hereafter referred to as “BF tags”) were isolated. Direct sequencing of the reamplified BF tags gave good-quality sequences for 1,217 fragments. BF tags corresponding to genes encoding enzymes known to catalyze early steps in triterpene saponin biosynthesis, such as β-amyrin synthase (bAS), displayed similar expression patterns, suggesting tight coregulation, and reached maximum expression levels 4–24 h post-MeJA elicitation (Fig. 1A). The gene corresponding to tag BF567 was coregulated with these genes and matched a P450 with 48% and 47% sequence similarity with the P. ginseng CYP716A53v2 and M. truncatula CYP716A12, respectively. The full-length open reading frame corresponding to the BF567 tag (hereafter called “CYP716Y1”) was cloned from a B. falcatum cDNA library (27). Phylogenetic analysis confirmed that CYP716Y1 belongs to the CYP716 family of P450s involved in triterpene saponin biosynthesis (Fig. 1B and Fig. S1).
Fig. 1.
Identification of CYP716Y1 by transcript profiling of MeJA-treated B. falcatum roots. (A) Coregulation of CYP716Y1 (in green) with other triterpene saponin biosynthetic enzymes. Cluster of the B. falcatum transcriptome comprising tags corresponding to genes reported to be involved in triterpene biosynthesis. Treatments and time points (in hours) are indicated at the top. Transcriptional activation and repression relative to the average expression level are represented by blue and yellow boxes, respectively. (B) Phylogenetic analysis of CYP716Y1 (in green) and other P450s involved in triterpene saponin biosynthesis. The percentage of replicate trees that clustered together in the bootstrap test is indicated to the left of the branches. The scale bar gives the number of amino acid substitutions per site. The enzymatic activity of the P450s is indicated on the right. The amino acid sequences of the P450s were retrieved from GenBank (www.ncbi.nlm.nih.gov/genbank). As, A. strigosa; Bf, B. falcatum; Cr, C. roseus; Gm, G. max; Gu, G. uralensis; Mt, M. truncatula; Pg, P. ginseng; Vv, V. vinifera.
Generation of Triterpene-Producing Yeast Strains.
To determine the in vivo enzymatic activity of CYP716Y1, we engineered Saccharomyces cerevisiae strains for hyperproduction of triterpenes, analogous to a previously described strain (24). All yeast strains generated in this study are listed in Table S1. First, we replaced the native promoter of lanosterol synthase with a methionine-repressible promoter in BY4742 to generate strain TM1, which in the presence of 1 mM methionine accumulated 60% less ergosterol than the parent strain. Next, the gene encoding a truncated feedback-insensitive copy of isoform 1 of the S. cerevisiae 3-hydroxy-3-methylglutaryl-CoA reductase (tHMG1) was cloned into the high-copy-number plasmid pESC-URA to generate pESC-URA[GAL10/tHMG1], which was transformed into TM1 to create strain TM5. We generated strains TM2 and TM3 from TM1 by expressing bAS of M. truncatula (GenBank accession no. AJ430607) (28) or Glycyrrhiza glabra (GenBank accession no. AB037203) (29), respectively using plasmid pESC-URA[GAL10/tHMG1;GAL1/bAS] (Fig. 2). Gas chromatography–mass spectrometry (GC-MS) analysis of cell extracts derived from 72-h-old TM2 and TM3 cultures revealed a single peak at 27.2 min with an electron ionization (EI) pattern corresponding to a β-amyrin standard, which was not observed in the GC chromatograms of the control strain TM5 (Fig. S2 A and B). β-Amyrin accumulation was higher in TM3 (36.2 ± 3.9 mg/L) than in TM2 (19 ± 1.0 mg/L).
Fig. 2.
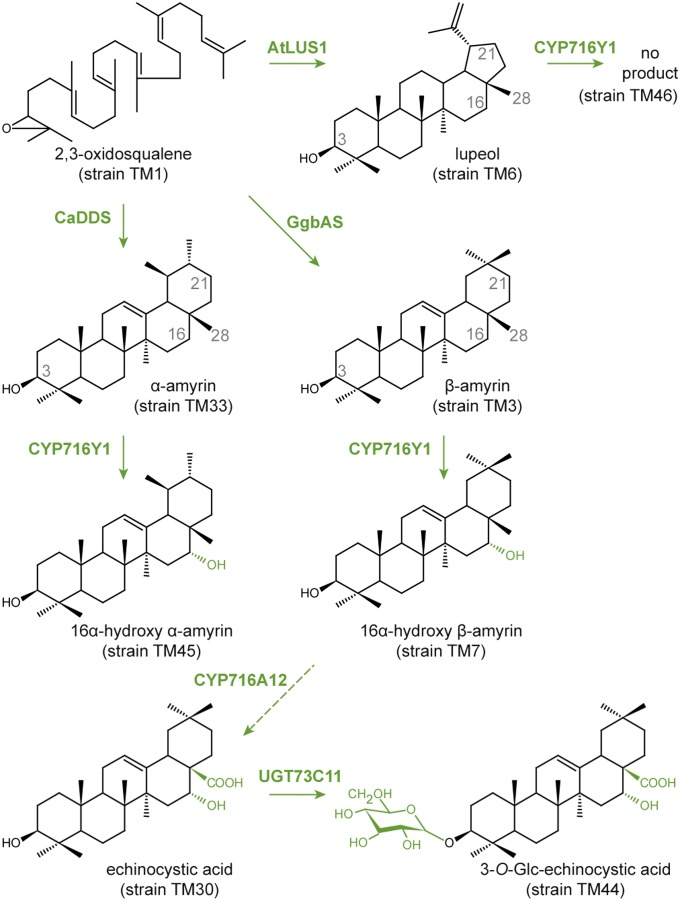
Triterpene saponin biosynthesis pathway engineered in yeast. The triterpene saponin precursor 2,3-oxidosqualene is cyclized to different saponin backbones, such as lupeol, β-amyrin, or α-amyrin. These backbones can be modified by multiple P450s before ultimately being glycosylated by UGTs. The structures of the triterpene saponin intermediates accumulating in the engineered yeasts are shown, and the yeast strains producing these compounds are indicated between parentheses and listed in Table S1. The enzymes catalyzing these reactions and the modifications resulting from their activity are highlighted in green. Dashed arrows mark multiple enzymatic steps. AtLUS1, A. thaliana lupeol synthase 1; CaDDS, C. asiatica dammarenediol synthase; CYP716A12, M. truncatula P450 that catalyzes a multistep oxidation at C-28; CYP716Y1, B. falcatum P450 that hydroxylates C-16; GgbAS, G. glabra β-amyrin synthase; Glc, glucose; UGT73C11, B. vulgaris UGT that catalyzes glucosylation at C-3.
In parallel, we generated a lupeol-producing yeast strain TM6 from TM1 by expressing the Arabidopsis thaliana isoform 1 of lupeol synthase (AtLUS1) (GenBank accession no. U49919) (30) by means of plasmid pESC-URA[GAL10/tHMG1;GAL1/AtLUS1] (Fig. 2 and Fig. S2C). We quantified 46.3 ± 2.4 mg/L lupeol from cells of TM6 that additionally cyclized 2,3-oxidosqualene to 3β,20-dihydroxylupane (31). Strain TM33 was obtained by expressing the Centella asiatica dammarenediol synthase gene (CaDDS) (GenBank accession no. AY520818) (32) from the pESC-URA[GAL10/tHMG1;GAL1/CaDDS] plasmid in strain TM1. In our hands, this strain produced α-amyrin, β-amyrin, and dammarenediol-II in a ratio of 88:11:1 (Fig. 2 and Fig. S2D). Hence, we used TM33 as an α-amyrin–producing strain.
In Vivo Enzymatic Activity of CYP716Y1.
Strains TM7 and TM26 expressing the Arabidopsis cytochrome P450 reductase (CPR) AtATR1 (At4g24520) with or without CYP716Y1, respectively, were generated from the β-amyrin–producing strain TM3. In the GC chromatograms, a unique peak eluting at 31.8 min was observed in cell extracts of TM7 but not of TM26 (Fig. S3A). The EI pattern of this peak corresponded to a D- or E-ring–hydroxylated derivative of β-amyrin (Fig. S3B). As CYP716Y1 was tentatively annotated as a homolog of M. truncatula CYP716A12 (GenBank accession no. FN995113) that encodes a P450 catalyzing the three-step oxidation of β-amyrin at C-28 (16), we compared the EI pattern of this peak with an erythrodiol (28-hydroxy-β-amyrin) standard (Fig. S3D). Additionally, strain TM10 expressing CYP716A12 and AtATR1 was generated and the GC-MS profile of its cell extract was compared with that of TM7. A peak corresponding to the elution time and EI pattern of erythrodiol occurred at 32.5 min in TM10 extracts (Fig. S3C) but not in extracts of TM7 and TM26, suggesting that CYP716Y1 and CYP716A12 hydroxylate β-amyrin at different positions. From the literature we identified the oleanane-type triterpene sapogenins reported in the Bupleurum species (8) and postulated that CYP716Y1 hydroxylates the D- or E-ring of β-amyrin at C-16 or C-21, respectively (Fig. S3B).
Finally, we generated strain TM45 expressing CYP716Y1 and a control strain TM47 from the α-amyrin–producing strain TM33. GC chromatograms showed the accumulation of hydroxylated α-amyrin in TM45, but not in TM47 (Fig. S3 E and F). In contrast, no hydroxylated derivative of lupeol or 3β,20-dihydroxylupane could be detected in strain TM46, expressing CYP716Y1 in the lupeol-producing strain TM6, indicating that lupeol is not a substrate for CYP716Y1.
Engineering Hydroxylated Amyrin Synthesis.
Effect of CPR:P450 ratios on the CYP716Y1 activity.
Hydroxylated amyrins accumulated at low levels in strains TM7 and TM45 and the available amyrin pools seemed hardly consumed, suggesting inefficient in vivo conversion. The endoplasmic reticulum (ER)-localized CPRs are flavoproteins that serve as electron donor proteins for several ER oxygenases, including P450s (33). Optimal interaction between CPR and P450 is essential to allow the reducing equivalents from NADPH to pass from the CPR to the P450 and guarantee P450 functionality (34). To assess whether different CPR:P450 ratios would affect CYP716Y1 performance, we generated strains expressing CYP716Y1 from a high-copy number plasmid together with AtATR1 from either an integrated (1 copy per cell), low-copy (3–5 copies per cell), or high-copy (10–40 copies per cell) number plasmid. Strain TM9 with AtATR1 on the low-copy plasmid accumulated the highest hydroxylated β-amyrin levels, whereas the lowest accumulation occurred in strain TM8 expressing the integrated copy of AtATR1 (Fig. S4). Although the actual CPR:P450 ratios were not biochemically measured, the product accumulation observed after attempting to alter the ratios through the vector copy numbers is in line with the P450 being in excess of the CPR, as reported for mammalian and yeast systems (34). Alternatively, the reaction rate of P450s can be (further) enhanced by interaction with cytochrome b5, as recently demonstrated in a yeast strain engineered for artemisinin production (35), but this was not assessed here.
Cyclodextrin facilitates sequestration and stimulates production of triterpene sapogenins.
Intracellular accumulation of sapogenins might be detrimental to cell viability or alternatively, exert feedback inhibition on the P450 activity as reported for human and Streptomyces P450s (36, 37), possibly accounting for the observed low hydroxy-amyrin accumulation in CYP716Y1-expressing yeasts. Cyclodextrins (CDs) are cyclic oligosaccharides composed of α-d-glucopyranoside units that can be represented by a toroidal topology (Fig. S5). CDs are extensively used for solubilizing and stabilizing pharmaceuticals because their toroidal interior is more hydrophobic, enabling them to host apolar molecules, such as cholesterol, whereas the hydrophilic exterior allows their solubilization in aqueous environments (38, 39). We hypothesized that CDs could also sequester sapogenins, facilitating their export to the growth medium and eventually stimulating their production.
First, we determined whether methyl-β-cyclodextrin (MβCD), the most commonly used CD, could sequester triterpenes. GC-MS analysis of extracts from cells and spent medium of strain TM3 treated or not with 5 or 25 mM MβCD revealed that both ergosterol and β-amyrin could be detected in all of the cell extracts, but only in the spent medium of MβCD-treated samples (Fig. 3A), indicating that MβCD was capable of sequestering and promoting export of both sterol- and sapogenin-type triterpenes. Although MβCD concentration and sequestering capacity correlated positively (Fig. 3A), the lowest concentration was used in further experiments for practical reasons. With 5 mM MβCD, we quantified 20.4 ± 10.5 mg/L and 37.3 ± 3.6 mg/L β-amyrin from cells and spent medium of TM3, respectively (Fig. 3B). The total concentration of β-amyrin in MβCD-treated cultures was 1.6-fold higher than in untreated cultures.
Fig. 3.
Enhanced accumulation of CYP716Y1 products by application of CDs. (A) Sequestration of triterpenes from yeast cells by MβCD. Overlay of GC chromatograms showing ergosterol and β-amyrin in the spent medium of strain TM3 treated with 5 mM (blue) or 25 mM (green) MβCD and untreated control (black). (B) Enhancement of triterpene productivity by MβCD. Quantification of β-amyrin sequestered from cells of strain TM3 upon 5 mM MβCD treatment showed a 1.6-fold higher β-amyrin amount in the MβCD-treated culture with 100% corresponding to 36.2 ± 3.9 mg/L (mean ± SD, n = 3) of β-amyrin in the untreated control. (C) MβCD dose-dependent sequestration of triterpenes. Quantification of β-amyrin from spent medium of strain TM3 treated with MβCD on day 1 (I1); days 1 and 2 (I2); days 1, 2, and 3 (I3); day 2 (R1); days 2 and 3 (R2); day 3 (AR); and an untreated control (C). (D) Effects of MβCD on hydroxy-β-amyrin sequestration and productivity. Relative amounts of hydroxy-β-amyrin quantified from the cells and spent medium of strain TM9 expressing CYP716Y1 treated or not with MβCD. (E) Specific sequestering of triterpenes by MβCD. Relative comparison of the hydroxy-β-amyrin quantified from spent medium of strain TM9 treated with different CD variants (Fig. S5). Quantitation and error bars correspond to mean and SD values, respectively (n = 3).
Next, we investigated whether timing and frequency of MβCD application during the yeast cultivation procedure affected β-amyrin production (Fig. 3C). We applied MβCD at a concentration of 5 mM at different times during culturing, thereby obtaining seven treatment conditions, including the mock-treated control. On day 1, to samples I1, I2, and I3, MβCD was added concomitantly with culture inoculation in galactose medium to induce heterologous gene expression. On day 2, to samples I2 and I3, MβCD was added again simultaneously with methionine; and on day 3, 24 h after the methionine repression, it was supplemented to sample I3 only. To samples R1, R2, and AR1, MβCD was added on day 2 only, days 2 and 3, and day 3 only, respectively. The spent medium of all samples was extracted on day 4 and β-amyrin quantified (Fig. 3C). The frequency of MβCD application and triterpene amounts correlated directly. For practical reasons, condition R2 was judged the most effective and used in subsequent assays to assess MβCD’s capacity to sequester hydroxylated β-amyrin. When MβCD was applied to strain TM9, hydroxylated β-amyrin occurred only in the spent medium (Fig. 3D), suggesting that the sapogenins were sequestered completely from the yeast cells. In addition, 5.5-fold more sapogenin could be extracted from the MβCD-treated than from untreated TM9 cells.
Finally, we compared the efficiency of sequestration with different types of CDs on strain TM9. The most common CD variants are α, β, and γ with 6, 7, and 8 d-glucopyranoside units, respectively (Fig. S5). We applied αCD, βCD, γCD, or MβCD at a concentration of 5 mM under condition R2. Sequestering was observed only with βCD and its methylated derivative with the highest amount extracted from the MβCD-treated culture (Fig. 3E). The specificity of βCD could be a consequence of the variable internal toroidal diameters of the CD variants, resulting from the number of glucopyranoside units they contain (Fig. S5). Likewise, the high affinity of sapogenins for the methylated βCD compared with the nonmethylated βCD could be related to their higher interior hydrophobicity.
Combinatorial Biochemistry Toward in Vivo Production of Saponins in Yeast.
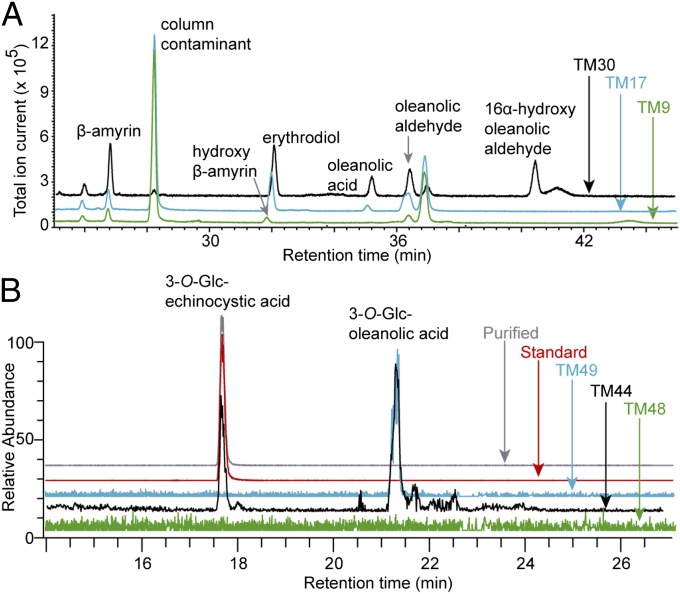
In combinatorial biochemistry, genes from different organisms are combined in a heterologous host to produce novel bioactive compounds (40). As CYP716Y1 and CYP716A12 oxidize β-amyrin at different positions, we reasoned that combining the enzymes in strain TM3 could result in a combinatorial compound corresponding to an intermediate of the saponin biosynthetic pathways from M. truncatula or B. falcatum that normally does not accumulate in these species. We generated strain TM30 in which CYP716Y1 and CYP716A12 were expressed from a high-copy number plasmid to produce a self-processing polyprotein in which the two enzymes are linked via a 2A oligopeptide (41). GC chromatograms of the extracts from the spent medium of TM30, TM9, and TM17 cultured with MβCD showed a single unique peak at 40.5 min in strain TM30 but not in TM9 and TM17 (Fig. 4A). This additional peak corresponds to β-amyrin oxidized at two positions, presumably a hydroxy oleanolic aldehyde based on the EI-MS spectra (Fig. S3G), the differences with an available 3β,16α-dihydroxyolean-12-en-28-oic acid (also called “echinocystic acid”) standard (Fig. S3 H and I), and the predicted enzymatic activities of CYP716A12 (aldehyde on C-28) and CYP716Y1 (hydroxyl on C-16 or C-21).
Fig. 4.
In vivo combinatorial production of 3-O-Glc-echinocystic acid in yeast. (A) Overlay of GC chromatograms showing the accumulation of hydroxyl oleanolic aldehyde in strain TM30 expressing CYP716Y1 and CYP716A12, but not in strains TM9 and TM17 expressing either of the P450s alone. (B) Overlay of LC-MS–extracted ion chromatograms of formate adducts showing the accumulation of 3-O-Glc-echinocystic acid and 3-O-Glc-oleanolic acid in strain TM44 expressing both CYP716Y1 and CYP716A12. Strain TM49 expressing only CYP716A12 accumulates 3-O-Glc-oleanolic acid, whereas strain TM48 expressing only CYP716Y1 does not produce any glucosylated product. For 3-O-Glc-echinocystic acid, 3-O-Glc-oleanolic acid, and 3-O-Glc-3β,16α-dihydroxyolean-12-ene the mass ranges 678.0–679.0 Da, 662.9–663.4 Da, and 648.0–649.0 Da, respectively were screened in all chromatograms.
Next, to synthesize saponins, we generated strain TM44 that expressed two self-processing polyproteins, one with the two P450s and one consisting of AtATR1 and the recently reported Barbarea vulgaris UDP-dependent glucosyltransferase (UGT) UGT73C11 (GenBank accession no. JQ291614) (42). In parallel, strains TM48 and TM49 were designed expressing the self-processing polyprotein of AtATR1 with UGT73C11 together with either CYP716Y1 or CYP716A12 alone. Liquid chromatography (LC)–MS chromatograms of cell extracts revealed accumulation of two saponins in strain TM44 (Fig. 4B), demonstrating that yeast can be engineered to produce monoglucosylated saponins. As one of these compounds also occurred in strain TM49 expressing CYP716A12 alone, it probably corresponds to 3-O-Glc-oleanolic acid based on the reported enzymatic activities (Fig. 4B). No MβCD was applied in these experiments, indicating that it was not necessary to allow accumulation of saponins. As no glucosylation of the hydroxy-β-amyrin was observed in strain TM48, β-amyrin might need to undergo a certain number of oxidations before it can be accepted by UGT73C11 as an in vivo substrate.
CYP716Y1 Catalyzes Hydroxylation of Amyrins on C-16 in an α-Configuration.
To unambiguously establish the enzymatic activity of CYP716Y1, we purified the second monoglucosylated saponin from strain TM44 and investigated its structure by 1D and 2D NMR. Complete assignment was not achieved, but the NMR analysis conclusively established that the product of the combined action of bAS, CYP716A12, CYP716Y1, and UGT73C11 was α-hydroxylated at C-16 of the triterpene base structure (Fig. 2). Further validation was obtained by (i) the near identity of the NMR data with that measured for a commercial 3-O-Glc-echinocystic acid standard (Figs. S6 and S7) and (ii) the comigration of the TM44-specific compound and the 3-O-Glc-echinocystic acid standard by LC-MS analysis (Fig. 4B), definitively establishing that CYP716Y1 hydroxylates β-amyrin on C-16 in an α-configuration.
Discussion
The pharmacological activity of the genus Bupleurum, used in Asian traditional medicine, is generally attributed to the presence of saikosaponins, the most abundant secondary metabolites produced by this genus. The saikosapogenins can contain oxidative modifications at the C-11, C-16, C-21, C-23, C-28, C-29, and C-30 positions (8). Through transcript profiling of MeJA-treated B. falcatum roots, we identified CYP716Y1, encoding a P450 that is transcriptionally coregulated with known triterpene saponin biosynthetic genes and that catalyzes the C-16α hydroxylation of amyrin, a catalytic activity previously not linked to any gene product in the plant kingdom. Hence, the P450 compendium that can oxidize the amyrin backbone is expanded, now covering seven positions on the triterpene backbone (11–18) and including two enzymes that catalyze hydroxylation at C-16, in β- (CYP51H10) (12) and α-configuration (CYP716Y1). Like many reported P450s (15, 17), CYP716Y1 can hydroxylate α- and β-amyrin, both pentacyclic triterpenes.
We generated a high-titer β-amyrin–producing S. cerevisiae strain (TM3) by expressing bAS from G. glabra. By means of a previously published engineering strategy (24) but with a more efficient bAS, we improved the β-amyrin production by sixfold and reached levels of 36 mg/L. Even our less efficient strain, expressing a M. truncatula bAS, produced threefold more β-amyrin (19 mg/L) than the previously published strain (24). This hyperproducing strain was used for the ectopic expression of CYP716Y1, causing accumulation of a hydroxylated product of β-amyrin. However, the production levels were low and only a fraction of the available amyrin pool seemed to be converted. Several reasons may account for this observation, such as the suboptimal folding, recycling, or localization of the P450 protein or, alternatively, the inappropriate localization of the catabolic product, thereby inhibiting P450 activity or affecting cell viability. The latter reflects the concerns that triterpene production might not be as amenable to engineering efforts as production of volatile sesquiterpenes and monoterpenes, molecules that can readily diffuse out of cells (24). Here, we have tackled this issue and designed a culturing strategy in which triterpenes are efficiently transferred from yeast cells into the culture medium by the use of MβCD. We show that these molecules are capable of sequestering hydrophobic triterpene sapogenins into the growth medium, thereby relieving the yeast cells from the load of toxic heterologous compounds, which, in turn, dramatically increased production levels. As the application of MβCD allows compound sequestration from living cells without damaging the integrity of the production host (43), this principle could be applied to continuous culture systems to produce large amounts of any valuable hydrophobic compound, including triterpene sapogenins.
Finally, we have advanced triterpene engineering in two additional aspects. First, we have demonstrated combinatorial triterpene sapogenin biosynthesis by combining CYP716Y1 from B. falcatum and CYP716A12 from M. truncatula in our yeast strain expressing G. glabra bAS and A. thaliana CPR, leading to the production of oxidized amyrins, sequestered in the medium after CD application. Second, we have been able to reconstitute a full saponin synthetic pathway in yeast. By supertransforming yeast strains that produce oxidized amyrins with the B. vulgaris UGT73C11, encoding a 3-O-glucosyltransferase (42), the corresponding monoglucosylated saponins accumulated in the yeast cells, independently of a need for CDs.
In conclusion, we have shown the versatile ability of yeast cells to produce natural or nonnatural sapogenins and saponins, through the utilization of MβCD as a sequestering agent for the hydrophobic sapogenins and by reconstitution of a full pathway to synthesize glycosylated saponins by the combined expression of OSC, P450, and UGT genes. Considering that the current strains have undergone little systems engineering, for instance with regard to the fluxes in the endogenous precursor or competing pathways, we trust that there is still much room for improvement of product yields. Hence, we believe that we have set an excellent base for a synthetic biology program toward the establishment of an efficient platform for the commercial production of valuable bioactive sapo(ge)nins.
Materials and Methods
Chemicals.
β-Amyrin, erythrodiol, and 3-O-Glc-echinocystic acid were purchased from Extrasynthese; random MβCD from CAVASOL; and αCD, βCD, and γCD from Cyclolab.
Cultivation and Elicitation of B. falcatum.
B. falcatum seeds (purchased from www.sandmountainherbs.com) were germinated in soil. Two-week-old seedlings were transferred to aerated hydroponic medium containing 1 g/L 10-30-20 salts (Scotts). The pH of the medium was monitored daily and maintained at 6.5 with potassium hydroxide (KOH). Plants were grown in a 16-h:8-h light:dark regime at 21 °C. After 3 wk, the plants were treated with 50 μM MeJA or an equivalent amount of ethanol as control by adding the solution directly to the hydroponic medium. For transcript profiling (SI Materials and Methods), roots were harvested 0, 0.5, 1, 2, 4, 8, and 24 h after treatment, frozen in liquid nitrogen, and stored at −70 °C. For each sample, three individual plants were pooled.
Cloning of CYP716Y1.
Primers P11+P12 were used to screen for the full-length coding sequence of CYP716Y1 in a B. falcatum Uncut Nanoquantity cDNA library (custom made by Invitrogen) as reported (27). The full-length CYP716Y1 was amplified by PCR for Gateway cloning into pDONR221 with primers P21+P22. All primers used are listed in Table S2. Phylogenies were analyzed as described (SI Materials and Methods).
Generation and Culturing of Yeast Strains.
Yeast strain TM1 and all derivatives were generated from the BY4742 strain (SI Materials and Methods). Precultures were grown at 30 °C with shaking at 250 rpm for 18–20 h in synthetic-defined (SD) medium with appropriate dropout (DO) supplements (Clontech). To induce gene expression, the precultures were washed and inoculated into SD Gal/Raf medium with appropriate DO supplements (Clontech) to a starting optical density of 0.25 on day 1. The cultures were further incubated for 24 h before addition of 1 mM methionine on day 2 and incubated for 48 h. For CD treatment, MβCD was added at a final concentration of 5 or 25 mM; and αCD, βCD, or γCD were added at a final concentration of 5 mM.
Metabolite Extraction for GC-MS.
Organic extracts of yeast cells or spent medium were prepared for identification and quantification of sapogenins. Cells were separated from the culture and lyzed with equal volumes of 40% (wt/vol) KOH and 50% (vol/vol) ethanol by boiling for 10 min. The supernatant corresponded to the spent medium. The lyzed cells and spent medium were extracted thrice with hexane. The organic phases were pooled, vaporized to dryness, and trimethylsilylated for GC-MS analysis (SI Materials and Methods).
Metabolite Extraction for LC-MS.
Yeast cells were lyzed with the YeastBuster protein extraction reagent (Novagen) and extracted thrice with ethyl acetate. The organic phases were pooled, dried, and dissolved in 100 μL water for LC-MS analysis (SI Materials and Methods).
NMR Analysis.
The 3-O-Glc-echinocystic acid was purified from strain TM44 and its structure determined by NMR analysis (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Wilson Ardiles-Diaz for sequencing, Miguel González-Guzmán and Lander Ingelbrecht for technical advice and assistance, Jan Van Bocxlaer (Ghent University) and Barbara Halkier (University of Copenhagen) for fruitful discussions, and Søren Bak (University of Copenhagen) for providing the UGT73C11 clone. This work was supported by funding from the Agency for Innovation by Science and Technology (Strategisch Basisonderzoek Project SBO040093) and the European Union Seventh Framework Programme FP7/2007–2013 (Grant 222716; SmartCell). T.M. and L.A. are indebted to the VIB International Fellowship Program for a predoctoral fellowship and the European Cooperation in Science and Technology Action FA1006-Plant Engine for a short-term scientific mission grant, respectively. J.P. is a Postdoctoral Fellow of the Research Foundation-Flanders.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. KC963423 (CYP716Y1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323369111/-/DCSupplemental.
References
- 1.Augustin JM, Kuzina V, Andersen SB, Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 2011;72(6):435–457. doi: 10.1016/j.phytochem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Osbourn A, Goss RJM, Field RA. The saponins–Polar isoprenoids with important and diverse biological activities. Nat Prod Rep. 2011;28(7):1261–1268. doi: 10.1039/c1np00015b. [DOI] [PubMed] [Google Scholar]
- 3.Phillips DR, Rasbery JM, Bartel B, Matsuda SP. Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol. 2006;9(3):305–314. doi: 10.1016/j.pbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Abe I. Enzymatic synthesis of cyclic triterpenes. Nat Prod Rep. 2007;24(6):1311–1331. doi: 10.1039/b616857b. [DOI] [PubMed] [Google Scholar]
- 5.Pollier J, Goossens A. Oleanolic acid. Phytochemistry. 2012;77(5):10–15. doi: 10.1016/j.phytochem.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Salvador JAR, Moreira VM, Gonçalves BMF, Leal AS, Jing Y. Ursane-type pentacyclic triterpenoids as useful platforms to discover anticancer drugs. Nat Prod Rep. 2012;29(12):1463–1479. doi: 10.1039/c2np20060k. [DOI] [PubMed] [Google Scholar]
- 7.Sporn MB, et al. New synthetic triterpenoids: Potent agents for prevention and treatment of tissue injury caused by inflammatory and oxidative stress. J Nat Prod. 2011;74(3):537–545. doi: 10.1021/np100826q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashour ML, Wink M. Genus Bupleurum: A review of its phytochemistry, pharmacology and modes of action. J Pharm Pharmacol. 2011;63(3):305–321. doi: 10.1111/j.2042-7158.2010.01170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YS, et al. Gene regulation patterns in triterpene biosynthetic pathway driven by overexpression of squalene synthase and methyl jasmonate elicitation in Bupleurum falcatum. Planta. 2011;233(2):343–355. doi: 10.1007/s00425-010-1292-9. [DOI] [PubMed] [Google Scholar]
- 10.Sui C, et al. Transcriptome analysis of Bupleurum chinense focusing on genes involved in the biosynthesis of saikosaponins. BMC Genomics. 2011;12:539. doi: 10.1186/1471-2164-12-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seki H, et al. Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA. 2008;105(37):14204–14209. doi: 10.1073/pnas.0803876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisler K, et al. Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc Natl Acad Sci USA. 2013;110(35):E3360–E3367. doi: 10.1073/pnas.1309157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukushima EO, et al. Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol. 2013;54(5):740–749. doi: 10.1093/pcp/pct015. [DOI] [PubMed] [Google Scholar]
- 14.Shibuya M, et al. Identification of β-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J. 2006;273(5):948–959. doi: 10.1111/j.1742-4658.2006.05120.x. [DOI] [PubMed] [Google Scholar]
- 15.Fukushima EO, et al. CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 2011;52(12):2050–2061. doi: 10.1093/pcp/pcr146. [DOI] [PubMed] [Google Scholar]
- 16.Carelli M, et al. Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell. 2011;23(8):3070–3081. doi: 10.1105/tpc.111.087312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, et al. Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus. Planta. 2012;236(5):1571–1581. doi: 10.1007/s00425-012-1712-0. [Note added: Planta 236(5)1583] [DOI] [PubMed] [Google Scholar]
- 18.Seki H, et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell. 2011;23(11):4112–4123. doi: 10.1105/tpc.110.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J-Y, Hwang H-S, Choi S-W, Kim H-J, Choi Y-E. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53(9):1535–1545. doi: 10.1093/pcp/pcs106. [DOI] [PubMed] [Google Scholar]
- 20.Han J-Y, Kim H-J, Kwon Y-S, Choi Y-E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52(12):2062–2073. doi: 10.1093/pcp/pcr150. [DOI] [PubMed] [Google Scholar]
- 21.Han J-Y, Kim M-J, Ban Y-W, Hwang H-S, Choi Y-E. The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2013;54(12):2034–2046. doi: 10.1093/pcp/pct141. [DOI] [PubMed] [Google Scholar]
- 22.Krokida A, et al. A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytol. 2013;200(3):675–690. doi: 10.1111/nph.12414. [DOI] [PubMed] [Google Scholar]
- 23.Moses T, Pollier J, Thevelein JM, Goossens A. Bioengineering of plant (tri)terpenoids: From metabolic engineering of plants to synthetic biology in vivo and in vitro. New Phytol. 2013;200(1):27–43. doi: 10.1111/nph.12325. [DOI] [PubMed] [Google Scholar]
- 24.Kirby J, Romanini DW, Paradise EM, Keasling JD. Engineering triterpene production in Saccharomyces cerevisiae–β-amyrin synthase from Artemisia annua. FEBS J. 2008;275(8):1852–1859. doi: 10.1111/j.1742-4658.2008.06343.x. [DOI] [PubMed] [Google Scholar]
- 25.Madsen KM, et al. Linking genotype and phenotype of Saccharomyces cerevisiae strains reveals metabolic engineering targets and leads to triterpene hyper-producers. PLoS ONE. 2011;6(3):e14763. doi: 10.1371/journal.pone.0014763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breyne P, et al. Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics. 2003;269(2):173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- 27.Pollier J, González-Guzmán M, Ardiles-Diaz W, Geelen D, Goossens A. An integrated PCR colony hybridization approach to screen cDNA libraries for full-length coding sequences. PLoS ONE. 2011;6(9):e24978. doi: 10.1371/journal.pone.0024978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Achnine L, Xu R, Matsuda SPT, Dixon RA. A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 2002;32(6):1033–1048. doi: 10.1046/j.1365-313x.2002.01497.x. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi H, et al. Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol Pharm Bull. 2001;24(8):912–916. doi: 10.1248/bpb.24.912. [DOI] [PubMed] [Google Scholar]
- 30.Herrera JBR, Bartel B, Wilson WK, Matsuda SPT. Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene. Phytochemistry. 1998;49(7):1905–1911. doi: 10.1016/s0031-9422(98)00366-5. [DOI] [PubMed] [Google Scholar]
- 31.Kushiro T, et al. Stereochemical course in water addition during LUP1-catalyzed triterpene cyclization. Org Lett. 2006;8(24):5589–5592. doi: 10.1021/ol062310d. [DOI] [PubMed] [Google Scholar]
- 32.Kim OT, et al. Characterization of a dammarenediol synthase in Centella asiatica (L.) Urban. Plant Physiol Biochem. 2009;47(11-12):998–1002. doi: 10.1016/j.plaphy.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Laursen T, Jensen K, Møller BL. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim Biophys Acta. 2011;1814(1):132–138. doi: 10.1016/j.bbapap.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Reed JR, Backes WL. Formation of P450·P450 complexes and their effect on P450 function. Pharmacol Ther. 2012;133(3):299–310. doi: 10.1016/j.pharmthera.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 36.Gavira C, et al. Challenges and pitfalls of P450-dependent (+)-valencene bioconversion by Saccharomyces cerevisiae. Metab Eng. 2013;18(7):25–35. doi: 10.1016/j.ymben.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Walczak RJ, Dickens ML, Priestley ND, Strohl WR. Purification, properties, and characterization of recombinant Streptomyces sp. strain C5 DoxA, a cytochrome P-450 catalyzing multiple steps in doxorubicin biosynthesis. J Bacteriol. 1999;181(1):298–304. doi: 10.1128/jb.181.1.298-304.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stella VJ, He Q. Cyclodextrins. Toxicol Pathol. 2008;36(1):30–42. doi: 10.1177/0192623307310945. [DOI] [PubMed] [Google Scholar]
- 39.Christian AE, Haynes MP, Phillips MC, Rothblat GH. Use of cyclodextrins for manipulating cellular cholesterol content. J Lipid Res. 1997;38(11):2264–2272. [PubMed] [Google Scholar]
- 40.Pollier J, Moses T, Goossens A. Combinatorial biosynthesis in plants: A (p)review on its potential and future exploitation. Nat Prod Rep. 2011;28(12):1897–1916. doi: 10.1039/c1np00049g. [DOI] [PubMed] [Google Scholar]
- 41.de Felipe P, et al. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24(2):68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Augustin JM, et al. UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 2012;160(4):1881–1895. doi: 10.1104/pp.112.202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabater-Jara AB, Pedreño MA. Use of β-cyclodextrins to enhance phytosterol production in cell suspension cultures of carrot (Daucus carota L.) Plant Cell Tissue Organ Cult. 2013;114(2):249–258. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.